Abstract
Synthesis of the plasminogen activator streptokinase (SK) by group A streptococci (GAS) has recently been shown to be subject to control by two two-component regulators, covRS (or csrRS) and fasBCA. In independent studies, response regulator CovR proved to act as the repressor, whereas FasA was found to act indirectly as the activator by controlling the expression of a stimulatory RNA, fasX. In an attempt at understanding the regulation of SK production in the human group C streptococcal (GCS) strain H46A, the strongest SK producer known yet, we provide here physical and functional evidence for the presence of the cov and fas systems in GCS as well and, using a mutational approach, compare the balance between their opposing actions in H46A and GAS strain NZ131. Sequence analysis combined with Southern hybridization revealed that the covRS and fasCAX operons are preserved at high levels of primary structure identity between the corresponding GAS and GCS genes, with the exception of fasB, encoding a second sensor kinase that is not a member of the GCS fas operon. This analysis also showed that wild-type H46A is actually a derepressed mutant for SK and streptolysin S (SLS) synthesis, carrying a K102 amber mutation in covR. Using cov and fas mutations in various combinations together with strain constructs allowing complementation in trans, we found that, in H46A, cov and fas contribute to approximately equal negative and positive extents, respectively, to constitutive SK and SLS activity. The amounts of SK paralleled the level of skcH46A transcription. The most profound difference between H46A and NZ131 regarding the relative activities of the cov and fas systems consisted in significantly higher activity of a functional CovR repressor in NZ131 than in H46A. In NZ131, CovR decreased SK activity in a Fas+ background about sevenfold, compared to a 1.9-fold reduction of SK activity in H46A. Combined with the very short-lived nature of covR mRNA (decay rate, 1.39/min), such differences may contribute to strain-specific peculiarities of the expression of two prominent streptococcal virulence factors in response to environmental changes.
The presence of the gene for the plasminogen activator streptokinase (SK) is a stable trait in group A streptococci (GAS) and human isolates of group C and G streptococci (GCS and GGS, respectively). In addition to its omnipresence in these organisms (11), the linkage relationships of this gene are preserved in a chromosomal region in which it is interspersed among five unrelated genes transcribed in the opposite direction (6, 7, 19). Its product, translated from a 440-codon monocistronic mRNA (16, 19), has no known homologues outside the genus Streptococcus. This protein, originally termed streptococcal fibrinolysin (3, 30), binds plasminogen with high affinity and, after activation of the zymogen (33), causes not only fibrino(geno)lysis but also the hydrolysis of various other substrates of the active enzyme, plasmin. The absence of fibrin in spreading streptococcal lesions has long been associated with the action of streptokinase as a virulence factor that contributes to the invasiveness of the pathogen, a notion supported by the failure of knockout mutants for the streptokinase gene to acquire cell-associated plasmin activity in the presence of plasminogen (15).
SK activities found in cell-free culture fluids can vary considerably among strains, ranging from exceedingly low fibrinolytic activities seen in, e.g., GAS strain NZ131 (≈3 U/ml in stationary-phase cultures) to very high activities (≈80 U/ml) as measured in the culture medium of the GCS strain H46A. In fact, in his early studies of SK-plasminogen interactions, Christensen (3) chose the latter strain as the source of the protein because H46A produced the most active fibrinolytic filtrates among more than 100 strains tested.
Recently, three lines of investigations have shed some light on the regulatory systems involved in the expression control of the SK gene. First, Northern hybridization analysis of the SK gene (skc) region of H46A revealed that of the six genes examined, skc is transcribed most abundantly (19) from a core promoter region resembling Escherichia coli σ70 consensus promoter sequences. With a TG motif one base upstream of the −10 region, this promoter qualifies as a promoter with an extended −10 region that directs transcription initiation predominantly at a G located 32 bases upstream of the skc translational start site (8). The −35 regions of skc and the oppositely oriented lrp gene adjacent to skc are separated by 202 bp of intergenic sequence that is intrinsically bent (9). Circular permutation analysis combined with computer modeling placed the bending center at position −98 relative to the major transcription initiation site of skc. Despite sequence differences in the intergenic regions between GCS and GAS, this bending locus is also present in NZ131, the curvature maps revealing a high degree of congruence at homologous positions (9).
The functional significance of the sequence features in the skc-lrp intergenic regions was analyzed with nested deletions that were inserted as single-copy promoter-′lacZ transcriptional fusions into the chromosome. Reporter gene activities revealed that full skc promoter activity depends strongly on the region containing the bending locus. Interestingly, deletion of the bending locus did not alter skc promoter strength in the heterologous E. coli background, where skc is generally highly expressed from its own promoter (8, 17, 21). In addition, reporter gene constructs containing the wild-type promoter upstream regions from H46A and NZ131 were exchanged in allele swap experiments between the GCS and GAS strains, with results showing strikingly that reporter gene activity expressed from the GAS construct is upregulated in the GCS strain background and, vice versa, expression of the GCS construct is downregulated in the GAS background. Taken together, these results show that the host genetic background dictates SK gene expression levels and suggest the existence of a trans-acting factor(s) with strain-specific activity that contacts the bending locus, thereby modulating SK gene expression (17).
Evidence for the existence in GAS of regulators that control ska expression has recently come from two additional lines of investigation. Several groups of investigators from different laboratories have identified the two-component signal transduction system CovRS, also designated CsrRS, as a pleiotropic repressor that negatively regulates the transcription of at least five virulence genes, among them ska (5, 10, 13, 20). Specifically, mutational analysis has also included strain NZ131 among the GAS strains in which ska expression is repressed by CovRS (27). Very recently, another group of investigators (12) has discovered, in GAS, the fasBCA two-component system that encodes, in addition to the response regulator (FasA), two histidine protein kinases (FasB and FasC). This system, which acts growth phase dependently and requires the RNA product of the fasX gene for activity, regulates ska expression positively (12). In NZ131, the four genes of the fas operon exhibit differential transcription modes, depending on nutritional conditions. Under amino acid starvation conditions, polycistronic fasBCAX mRNA accumulates, whereas under normal conditions, fasX is preferentially transcribed separately from one of its two operon-internal promoters, P2 (28).
The skc gene found in the GCS strain H46A is the first SK gene that was cloned and sequenced (14, 16). As this strain has also been exploited in industrial SK production for fibrinolytic therapy, it is of obvious interest to understand the mechanism(s) which controls the synthesis of this protein in H46A. Here we show that the cov and fas systems also function in this strain. Whereas their actions in GAS have been studied independently of one another, we analyzed the balance between their opposing actions and show that wild-type H46A is actually a derepressed mutant for Skc production. As the cov and fas systems also target the streptolysin S (SLS) operon, sag (5, 10, 12, 22), we included the control of SLS production in our analyses.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The GCS strain H46A and the GAS strains NZ131 and SF370 were grown in ambient air at 37°C without agitation in brain heart infusion (BHI) broth (Difco) or chemically defined medium (CDM) buffered with 26 mM morpholinepropanesulfonic acid (18). The complete genomic sequence of SF370 (6) allowed derivation of oligonucleotide primers for the synthesis of specific PCR-generated genes or gene fragments from the streptococcal chromosomal DNAs as templates. E. coli JM109 was used as the host for plasmid constructions and was grown at 37°C in rotary flasks in standard Luria broth (LB). Concentrations of antibiotics used were as follows: chloramphenicol, 3 μg/ml for streptococci and 100 μg/ml for E. coli; erythromycin, 2.5 μg/ml for streptococci and 200 μg/ml for E. coli; kanamycin, 100 μg/ml for streptococci and 50 μg/ml for E. coli; and spectinomycin, 100 μg/ml for both streptococci and E. coli.
Nucleic acid techniques.
Plasmid isolation, DNA fragment isolation and ligation, electrotransformation of E. coli, random-primed DNA labeling, Southern hybridization, and DNA sequencing by the dideoxynucleotide chain termination method were performed by standard procedures (25) or according to the manufacturer's recommendations. Chromosomal DNA from streptococci was isolated by the method of Caparon and Scott (2). Electrotransformation of streptococci was carried out as described previously (8). PCR, performed with Pwo DNA polymerase (Peqlab Biotechnologie, Erlangen, Germany), was used to amplify DNA fragments for gene isolation, sequence analysis, insertion mutagenesis, DNA probe production, and confirmation of the constructed plasmids and streptococcal insertion mutants. Total RNA isolation from streptococci, Northern hybridization of streptococcal RNA, and determination of mRNA chemical half-lives have been described in detail elsewhere (17). Hybridization reactions were quantified by PhosphorImager (Fujix BAS1000) analysis with software TINA 2.08c from Raytest (Straubenhardt, Germany).
Construction of cov plasmids.
Plasmid pMcovRSF, containing the complete SF370-derived covR gene, including its promoter, was constructed by generation of an amplicon with primers covRF1 (ccggaattcCAAGGGTTGTTTGATGAATA) and covRR1 (ccggaattcATGACTTATTTCTCAC) (lowercase letters indicate sequences added to facilitate cloning.) The EcoRI cleavage product of this DNA was cloned into pMG36e (31) with JM109 as the host. The resultant plasmid, pMcovRSF, was isolated from JM109 and introduced into H46A by electrotransformation, and transformants were selected with erythromycin.
To construct pVAcovRSNZ, containing the complete covRS genes from NZ131, including the promoter of the operon, NZ131 template DNA was amplified with primers covRF1 (see above) and covSR1 (cgcctgcagCTTAAGCTACTCTAACTCTC). The EcoRI plus PstI cleavage product of this amplicon was inserted into pVA8912 (26), and plasmid transformants of JM109 were selected with erythromycin. The resultant pVAcovRSNZ plasmid was linearized with EcoRI and used for ligation with an EcoRI fragment containing the 3′ end of the dexB gene from H46A to form pVAcovRSNZdI and pVAcovRSNZdII containing ′dexB in either orientation. These suicide plasmids, unable to replicate in streptococci, were introduced into H46A, and erythromycin-resistant transformants were confirmed by appropriately primed PCR to result from insertion of the plasmids into the resident dexB gene of the H46A chromosome (19).
Construction of plasmid pFC1 containing an internal covRNZ131 fragment in the vector pFW5 (selective marker, spectinomycin resistance; GenBank accession number U41082) has already been described (27). Likewise, NZ131 covR::pFC1 isolates bearing covR truncated at codon 174 were characterized previously (27). For complementation, these strains were transformed with pVA covRSNZ, selecting for spectinomycin plus erythromycin resistance.
Construction of fas plasmids.
For insertional inactivation of fasAH46A, two different fragments internal to the fasAH46A coding region were amplified by PCR with the following primers: fasAF1B (cgcggatccCCAGCAGACACGCATAGAATC), fasAR2H (cgcaagcttCGGTGCTACTGCTTGAATCTCAG), fasAF2B (cgcggatccCTTGAATTAGCTGCAGCTATTCG), and fasAR1H (cgcaagcttCACAAGGTAGGATCTATGGC). The BamHI plus HindIII cleavage products of the amplicons were cloned in pFW5 (see above), pFW15 (erythromycin resistance; GenBank accession number U50977), or pFW14 (chloramphenicol resistance; GenBank accession number U50981; identical to pFW8) to form pFW5fasA1, pFW15fasA1, and pFW14fasA2, respectively. Electrotransformation of H46A with the latter two mutagenic plasmids and verification of the transformants resulted in strains H46AfasA::pFW15fasA1 and H46AfasA::pFW14 fasA2, carrying fasA alleles interrupted at codon positions 194 (fasA1) and 205 (fasA2), respectively. Plasmid pFW15 fasA1 was also used to construct the corresponding fas insertions in NZ131covR::pFC1. Plasmid pFW5fasA1 served to inactivate fasA in H46AdexB::pVAcovRSNZdII.
For complementation of the insertionally inactivated fasAH46A gene, plasmid pFasAXSF was constructed, which contains the complete coding sequences of fasAXSF370, the expression of which is driven by promoter P32 of the vector pMG36 (kanamycin resistance) (31, 32). The fasAXSF370 genes were amplified by PCR with primers fasAF3 (tcccccgggGACAATTGTTAGAAAGGAGATAAAG) and fasXR3 (cgcaagcttGACGTCAGCTACTTATCCCTG). Amplicon DNA restricted with SmaI plus HindIII was inserted into pMG36, and pFasAXSF was primarily established in JM109. Plasmid pFasAXSF DNA was then used to transform H46AfasA::pFW15fasA1, with selection for erythromycin plus kanamycin resistance.
To facilitate comprehension, the relevant characteristics of the strains and plasmids used in this study are compiled in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Streptococcus dysgalactiae subsp. equisimilis | Human serogroup C | |
| H46A | Cov− (K102 amber) Fas+ | 3; this study |
| H46A(pMcovRSF) | Cov+ Fas+; Emr | This study |
| H46AdexB::pVAcovRSNZdI | Cov+ Fas+; Emr | This study |
| H46AdexB::pVAcovRSNZdII | Cov+ Fas+; Emr | This study |
| H46AfasA::pFW15fasA1 | Cov− Fas−; Emr | This study |
| H46AfasA::pFW14fasA2 | Cov− Fas−; Cmr | This study |
| H46AfasA::pFW15fasA1(pFasAXSF) | Cov− Fas+; Emr Kmr | This study |
| H46AdexB::pVAcovRSNZdII, fasA::pFW5fasA1 | Cov+ Fas−; Emr Spcr | This study |
| Streptococcus pyogenes | Human serogroup A | |
| NZ131 | Cov+ Fas+ | J. J. Ferretti |
| NZ131covR::pFC1 | Cov− Fas+; Spcr | 27 |
| NZ131covR::pFC1, covR::pVAcovRSNZ | Cov+ Fas+; Spcr Emr | This study |
| NZ131covR::pFC1, fasA::pFW15fasA1 | Cov− Fas−; Spcr Emr | This study |
| Plasmids | ||
| pMG36, pMG36e | Vectors; Kmr and Emr respectively | 31 |
| pMcovRSF | pMG36e containing covRSF370 under its natural promoter; Emr | This study |
| pVA8912 | Suicide vector; Emr | 26 |
| pVAcovRSNZ | pVA8912 containing covRSNZ131 under its natural promoter; Emr | This study |
| pVAcovRSNZdI | pVA8912 containing covRSNZ131 under its natural promoter and the 3′ end of dexBH46A (orientation I) for genomic integration into H46A; Emr | This study |
| pVAcovRSNZdII | As pVA covRSNZdI but 3′ end of dexBH46A in orientation II; Emr | This study |
| pFW5, pFW14, pFW15 | Vectors; Spcr, Cmr, Emr, respectively | 24 |
| pFW5fasA1 | pFW5 containing an internal fragment of fasAH46A; Spcr | This study |
| pFW15fasA1 | As pFW5 fasA1; Emr | This study |
| pFW14fasA2 | pFW14 containing an internal fragment of fasAH46A; Cmr | This study |
| pFasAXSF | pMG36 containing fasAXSF370 under vector promoter P32; Kmr | This study |
Probes used for Southern and Northern hybridizations.
Probes were generated with the following primer sets: covRF1 and covRR1 (see above) for covR; CCAGCAGACACGCATAGAATC and CACAAGGTAGGATCTATGGC for fasA; CAGTATGGCTTTATTCTATGCTACC and GGAATTCATGATACTGCGAATCAC for fasC; GTCGCCATTGCTAATTGATCCTC and GTTGTGCTTCAAGAGCTGCCTC for fasB; GAGAGCAATAACATTTTAGGAC and GACGTCAGCTACTTATCCCTG for fasX; ggcgccaagcttTTTTAGCTCCATAGCCATTCC and ccgtggTTATTTGTCGTTAGGGTTATCAGG for ska and skc; GCTACTAGTGTAGCTGAAACAAC and CCTGAGACGTTAGCATCAAGAAG for sagA; and CAAGTCAACAGTGGAGAGAAC and CGGTAGTGTTATGAAGGATGAC for mga.
Protein activity assays.
The plasminogen activation assay on microtiter plates as described by Tewodros et al. (29) was used to measure SK activity in BHI culture supernatant fluids of the various wild-type and mutant strains. The release of para-nitroaniline from the chromogenic substrate H-d-valyl-leucyl-lysin p-nitroaniline (Sigma) was measured at an optical density at 405 nm (OD405) over time, and activity rates were calculated from the linear parts of absorbance versus time plots. SK activities were also visualized as caseinolytic zones on agarose well plates containing plasminogen and casein as described previously (14). Standard streptokinase came from Sigma and its definition was used to determine specific activities.
SLS activity was measured with cultures grown in BHI containing 10% horse serum. The SLS activity assay of culture supernatant fluids was performed with rabbit erythrocytes according to Heath et al. (10). The release of free hemoglobin was measured at an OD540 in the presence of cholesterol (10 μg/ml) to inhibit streptolysin O and in some samples in the presence of trypan blue, an inhibitor of SLS, to control the specificity of the assay.
Nucleotide sequence accession numbers.
The nucleotide sequences of the cov and fas genes from Streptococcus dysgalactiae subsp. equisimilis strain H46A have been submitted to the DDBJ/EMBL/GenBank databases under accession numbers AY075106 and AY075107, respectively.
RESULTS
Identification of the covRS locus in S. dysgalactiae subsp. equisimilis.
In search of Streptococcus pyogenes covRS homologues in strain H46A, the heterologous primer pair covRF1 and covSR1, designed on the basis of the genomic sequence of SF370 (6), was used to produce by PCR from the chromosomal DNA of H46A a weak DNA band corresponding in size to the entire covRS locus of SF370 (≈2.5 kb). Sequence determination of this DNA with walking primers covered 1,141 nucleotides counting from the covRF1 forward primer. This sequence revealed a high degree of homology to the three S. pyogenes covRS operons sequenced so far from different strains, SF370 (6), Manfredo (http://www.sanger.ac.uk), and DLS003 (13). Our sequence represented the entire covRH46A gene, including its promoter region, the very short intergenic region between covRH46A and covSH46A (5 nucleotides), and the first 59 codons of covSH46A.
The coding sequences of covRH46A and covRSF370 and the derived amino acid sequences showed 84 and 98% identity, respectively. Similarly, the partial sequence of covSH46A was 88% and the deduced amino acid sequence was 95% identical to the corresponding sequences from SF370. Most importantly, the covRH46A sequence contained a translational stop codon at codon position 102, which altered the corresponding AAA triplet present in all known GAS covR genes to read TAG in H46A. Sequence analysis of this position done repeatedly with independently isolated H46A chromosomal DNAs confirmed that this mutation, which requires 2 base substitution events, is a genuine H46A trait rather than being introduced during DNA manipulations. Given that the full-size CovR protein contains 228 amino acids, its truncated form in H46A, presumably missing the DNA-binding domain, was highly likely to be functionally inactive. In the context of this work, however, we refer to H46A with the identified amber mutation in covR as the wild type.
Similar to the high degree of coding sequence conservation, the covRSH46A promoter, resembling E. coli σ70 promoters, was found to be identical in both the −10 region (TATACT) and the −35 region (TTGAAA) to the corresponding GAS promoters. Likewise, the putative transcriptional start site at a G eight bases downstream of the −10 hexanucleotide was identical to that determined for GAS strain MGAS166 (20). However, the relatively long sequence between the −10 region and the translational start codon, which is almost identical among the GAS strains, differed from them in 79 of 212 base positions.
Properties of covRS mRNA.
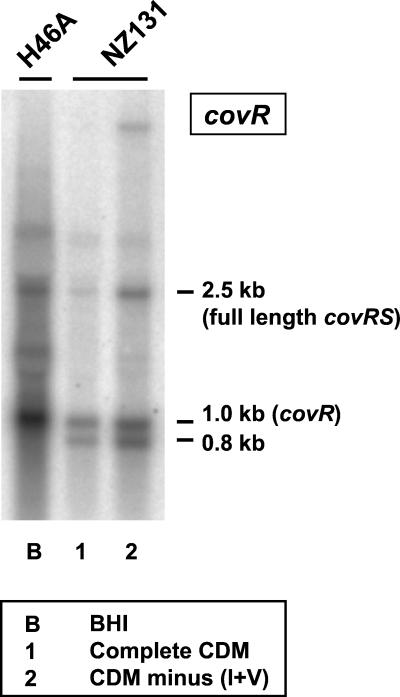
Similar to what has been found for transcription of covRSDLS003 (13), Northern hybridization analysis of covRSH46A mRNA probed with covR revealed cotranscription of the two genes in the GCS strain as well (Fig. 1). However, in addition to the 2.5-kb dicistronic transcript, a prominent 1.0-kb transcript corresponding in size to monocistronic covR mRNA was detected in both H46A and NZ131. Since no terminator sequence was visible downstream of covR, this transcript might result from degradation of the dicistronic message, as might a distinct 0.8-kb RNA seen in NZ131 but not in H46A (Fig. 1). Loss of about 200 nucleotides from the 5′ end would still allow functional activity of covR mRNA, as judged by the retained ribosome-binding site.
FIG. 1.
Detection of covRS mRNA in H46A and NZ131 incubated in the indicated media. Shown is a Northern blot of total RNA probed with covR (boxed).
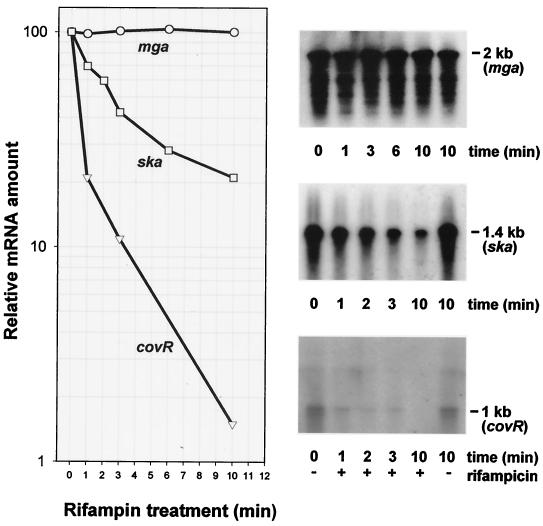
The stability of covR mRNA was determined in NZ131 rather than H46A because the nonsense mutation in covRH46A could lead directly to decreased message stability. Decay kinetics were measured in comparison to those of ska and mga mRNAs during a time period within which new synthesis of mRNA was inhibited by rifampin (Fig. 2). The chemical half-lives of the three messages amounted to 0.5, 2.5, and >10 min, respectively, corresponding to decay rates of 1.39/min, 0.28/min, and <0.07/min, respectively. This indicates the extremely short-lived character of covR mRNA in contrast to mga mRNA, which proved to be very stable. It is interesting that the mRNA half-lives of two important pleiotropic regulatory genes can differ more than 20-fold.
FIG. 2.
Determination of mRNA half-lives of the indicated NZ131 genes under conditions in which new mRNA synthesis is inhibited by rifampin. Estimation of decay kinetics of the mRNAs (left) is based on Northern blots, representative results of which are shown on the right.
Identification of fasCAX genes in S. dysgalactiae subsp. equisimilis.
Using the SF370-based heterologous primer sets for fasA, fasX, and fasAX (see the Materials and Methods section), we were able to produce from H46A chromosomal DNA three amplicons, the DNA sizes of which agreed with those predicted from the fasAX sequence of SF370 (580, 322, and 1,095 bp). Using standard PCR conditions, we failed, however, to produce any amplicons that corresponded to the proximal fasBC genes from H46A template DNA when SF370-specific primers for these genes were employed. With temperature gradient PCR and fasBSF370-specific primers (see Materials and Methods), a 600- to 700-bp fragment was obtained that deviated from the predicted length of 931 bp. Sequence analyses of this DNA ruled out its relationship to fasBSF370 but revealed homology to the Spy1148 gene product of SF370, a predicted ATP-binding protein of an ABC transporter (6).
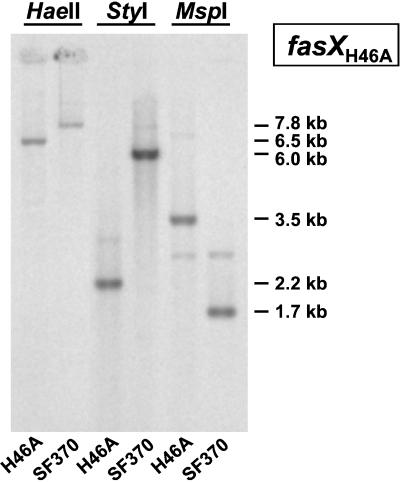
Evidence for the existence of a fasCSF370 homologue and for linkage of fasCAX in H46A was obtained by Southern hybridization analysis. The gene-specific probes used included both heterologous and homologous DNAs, namely fasCSF370, fasASF370, fasAH46A, fasXSF370, and fasXH46A. All probes detected hybridizing H46A DNA fragments and, as controls, SF370 DNA fragments, with fasCSF370 giving the weakest signal with H46A DNA. Hybridizing fasCAXH46A DNA was associated with a ≈3.5-kb MspI fragment, a ≈2.2-kb StyI fragment, and a ≈6.5-kb HaeII fragment (Fig. 3). For comparison, fasCAXSF370 DNA has a length of 2.3 kb (6).
FIG. 3.
Detection of the fasCAX operon by Southern hybridization of H46A and SF370 chromosomal DNAs restricted with the indicated enzymes and probed with fasXH46A (boxed).
Determination of the complete coding sequences of the fasAXH46A genes by primer walking revealed a high degree of homology to the corresponding genes from SF370. The fasAH46A gene and deduced protein showed, respectively, 81 and 90% sequence identity to fasASF370, and the sequence of fasXH46A was 95% identical to that of fasXSF370. Taken together, these results provided physical evidence for the presence of homologues of the fasCAXSF370 genes in H46A but failed to detect a proximal fasB gene encoding a second sensor kinase.
Functional analysis of covRS in H46A.
To answer the question of whether or not heterologous covR or covRS genes are capable of complementing in trans the putatively inactive covRH46A gene, two complementation systems were established by transformation of H46A. In the first system, functional covRSF370 was delivered in multicopy plasmid form by the construction of H46A(pMcovRSF). In the second system, covRSNZ131 was introduced into H46A in single-copy form by integration into the resident dexB gene in either orientation to form H46AdexB::pVAcovRSNZdI and H46AdexB::pVAcovRSNZdII. The three transformed strains were used to determine the amounts of SK and SLS in culture supernatant fluids relative to the amounts of these proteins in comparable cultures of the original H46A strain.
Depending on the particular transformant strain examined, repression of SK production ranged from 68 to 48%, and that of SLS ranged from 75 to 24% (Fig. 4). In general, cov activity delivered in plasmid form tended to have a greater repressive effect than that specified by cov integrated into the chromosome. Furthermore, in the integrated form, covRS was more active in orientation dII than dI, suggesting some dexB promoter contribution to the expression level of covRS in the dII orientation. We conclude from these results that, regardless of their copy number, the covRS genes from GAS were functionally active as repressors of H46A SK and SLS synthesis. Moreover, the heterologous CovSH46A and CovRSF370 protein pair is suggested to interact normally in strain H46A(pMcovRSF), as might be expected from the high degree of sequence identity between CovRH46A and CovRSF370.
FIG. 4.
SK and SLS activities in cell-free supernatant fluids of BHI cultures of the specified H46A wild-type and mutant strains containing comparable numbers of CFU. For SK determination, cultures were sampled after 16 h of growth, whereas log-phase cultures grown to an OD600 of 0.30 were used for SLS determination. Relative activities of 100% correspond to 80 U of SK per ml or 0.75 OD540 hemolytic units produced by 200 μl of a 1:40 diluted supernatant fluid obtained from an H46A culture. (For comparison, an OD540 value of 1.15 corresponds to complete hemolysis produced by the same volume of water.)
In NZ131covR::pFC1 isolates, inactivation of the resident covR gene increases SK production about 10-fold (27). At the level of skaNZ131 transcription, derepression of ska increased the ska mRNA level about 100-fold (Fig. 5). Complementation of NZ131covR::pFC1 with single-copy pVA covRSNZ restored the repression of SK production close to the very low NZ131 wild-type level, indicating high CovR repressor activity in this strain (Fig. 5). In comparison, the corresponding complementation system caused strain H46A to retain 40 to 50% of its wild-type SK production level (Fig. 4).
FIG. 5.
CovR-regulated SK production by wild-type NZ131, the covR mutant, and complemented covR mutant strains. SK activities of culture supernatant fluids were compared by measuring plasminogen activation with a plasmin-specific chromogenic substrate as a function of time. The inset shows a slot blot hybridization of total RNA extracted from the indicated strains and probed with ska.
Functional activity of the fas system in H46A.
We used insertional mutagenesis of fasAH46A to explore whether this response regulator influences the levels of SK and SLS production in H46A. The results showed that, relative to protein activities measured in culture supernatant fluids of H46A, SK activities produced by H46AfasA::pFW15fasA1 and H46AfasA::pFW14fasA2 were reduced by about 50% and SLS activities dropped to an average of about 60% (Fig. 4). Complementation in trans of the interrupted fasA gene by introduction of plasmid pFasAXSF restored the wild-type levels of both proteins (Fig. 4). We conclude from these data that the fas system positively regulates the skc and sag genes in H46A. Combination of the functional covR repressor with an inactivated fasA gene in strain H46A(dexB::pVAcovRSNZdII, fasA::pFW5fasA1) decreased the SK and SLS production capacities of H46A to 3 and 4%, respectively, showing that in the absence of positive regulation, the negative covR regulator almost completely abolished the capability of H46A to produce the two proteins (Fig. 4).
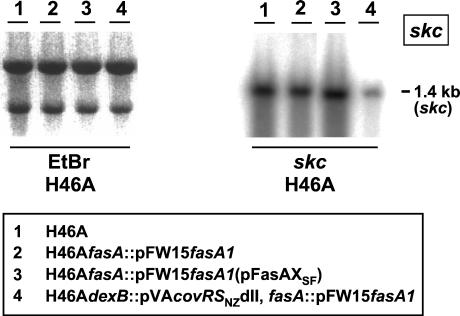
The results obtained at the protein level for the combined effects of the cov and fas regulatory systems correlated with quantitative skc transcript analysis. The Northern blot in Fig. 6 shows that inactivation of fasAH46A reduced skc mRNA abundance to 79%, and restoration of covR activity decreased the amount of skc mRNA further to 16% of that seen in the wild type.
FIG. 6.
Transcriptional regulation of skc expression by the cov and fas regulatory systems demonstrated by Northern hybridization of total RNA extracted from the indicated strains and probed with skc (boxed). The ethidium bromide (EtBr)-stained gel on the left demonstrates equal loading of the slots and integrity of the 23S and 16S rRNAs.
Transcriptional analysis of fas system in H46A.
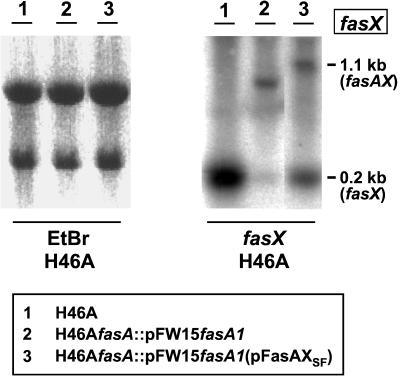
According to previous results, regulation of fas operon transcription in GAS occurs in a complex manner. The transcriptional profile has been found to be growth phase dependent (12), influenced by nutritional conditions (28), and also determined by the presence of at least two operon-internal promoters for fasX (12, 28), the relative activities of which are responsive to amino acid availability (28). In agreement with the situation found previously in strain NZ131 (28), H46A RNA isolated from log-phase or stationary-phase cells grown in BHI or CDM and probed with fasX yielded only one strongly hybridizing band at 0.2 kb, corresponding to fasX RNA produced from the downstream P2 operon-internal fasX promoter (Fig. 7). Probing the same RNAs with a homologous fasA or heterologous fasCNZ131-specific DNA failed to detect any transcripts corresponding to these genes. However, total RNA isolated from strain H46AfasA::pFW15fasA1(pFasAXSF), in which transcription of fasAXSF was driven from the strong P32 promoter of the vector pMG36 (32), yielded, in addition to the 0.2-kb monocistronic fasX transcript, a prominent 1.1-kb dicistronic transcript corresponding to fasAX.
FIG. 7.
Cotranscription of fasAX and dependence of separate transcription of fasX from its own promoter on the presence of FasA, demonstrated by Northern hybridization of total RNA extracted from the indicated strains and probed with fasX (boxed). The ethidium bromide (EtBr)-stained gel on the left demonstrates equal loading of the slots and integrity of the 23S and 16S rRNAs.
These results corroborate our previous observation that fasBCAXNZ131 can be cotranscribed but that, in addition to transcriptional linkage, there is strong independent transcription of fasX from its P2 promoter. Expectedly, RNA isolated from H46AfasA::pFW15fasA1 and probed with fasX did not show the 1.1-kb fasAX transcript. Importantly, this RNA also lacked the 0.2-kb fasX transcript (Fig. 7). Its absence cannot be attributed to the polar nature of the fasA insertion; rather, it confirms the previous finding of Kreikemeyer et al. (12) showing that fasX transcription from its own promoter(s) is dependent on the fasA gene product.
SK production levels of H46A and NZ131 strains with comparable genetic backgrounds.
The H46A and NZ131 mutant strains constructed for the cov and fas systems enabled us to compare their relative efficiencies in the two strains and to detect strain-specific differences (Table 2). In a Fas+ background, repression of SK synthesis by CovRS was approximately sevenfold stronger in NZ131 than in H46A where it only halved the SK production. Accordingly, when both systems were active, SK production by H46A exceeded that of NZ131 by a factor of about 13. In contrast, in a Cov− background, the positive action of the Fas system did not differ substantially between the two strains, promoting SK production by factors of 2 and 1.5, respectively. In the absence of both repression and activation, i.e., in a Cov− Fas− background, the remaining SK activity would appear to reflect mainly the strength of cis-active elements. This constitutive level of SK expression appeared to be greater by merely a factor of 1.3 in H46A relative to that seen in N131 (Table 2).
TABLE 2.
Comparison of SK production by strains H46A and NZ131 as a function of the genetic background
| Phenotype | SK activitya (U/ml)
|
Activity ratio
|
||
|---|---|---|---|---|
| H46A | NZ131 | H46A | NZ131 | |
| Cov+ Fas+ | 42 | 3.2 | ||
| Cov− Fas+ | 80 | 44 | ||
| Cov+ Fas− | 2.4 | NDa | ||
| Cov− Fas− | 39 | 30 | ||
| Cov− Fas+/Cov+ Fas+ | 1.9 | 13.8 | ||
| Cov− Fas+/Cov− Fas− | 2.0 | 1.5 | ||
ND, not determined.
DISCUSSION
In this study, we provided physical and functional evidence for the existence of the cov and fas two-component regulatory systems in the genome of GCS. In GAS, recent electrophoretic mobility shift and footprinting experiments have revealed the binding sites of CovR in the promoter regions of the regulated genes (1, 20). The footprints in the ska promoter region are large, measuring >130 bp and containing AT-rich DNA. The search of common sequence tracts in the footprints of five CovR-regulated genes led to the identification of a conserved 16-bp sequence with runs of A's and T's (20). For the skaNZ131 and skcH46A promoter regions, the sequences that matched the 16-bp motif most closely are given in Table 3.
TABLE 3.
Similarity of streptococcal promoter-upstream sequences to the 16-bp consensus sequence proposed (20) to be contained in CovR target sequences
| Promoter | Sequence | Location | Adherence to consensus (no. of bases/total) |
|---|---|---|---|
| PskaNZ131 | TCATTTAAAAACAATT | −64 to −49 | 10/16 |
| PskcH46A | TCATTTTAAAAAAATC | −64 to −49 | 13/16 |
| PcovRH46A | TTTTATTTGAAAAAAG | −42 to −27 | 12/16 |
| Consensus | TTATTTTTAAAAAAAC | Variable | |
| ATA |
Since covR is autoregulated (5), we also inspected the promoter region of covRH46A and found a sequence tract that showed some adherence to the 16-bp motif (Table 3). However, CovRMGAS166 was found not to bind to its own promoter region (20), which makes it difficult to understand the mechanism of autoregulation. Moreover, as CovR recognition of its target promoters requires AT-rich DNA segments much longer than 16 bp, direct evidence for the involvement of this motif in CovR binding is missing (20). In a similar vein, comparison of the 16-bp sequence between the promoter regions of skcH46A and skaNZ131 does not suggest any explanation of why CovRNZ131 represses transcription of skaNZ131 more strongly than that of skcH46A (Table 2). The fact that we restored repressor activity in H46A by complementation with the heterologous covRNZ131 gene does not seem to explain the strain-specific CovR activities, given the high degree of sequence identity between the CovR proteins of different strains (≈98%). At the target DNA level, the biggest sequence difference between skcH46A and skaNZ131 consists in the presence or absence of a 9-bp direct repeat (CATTATCAT) located between the bending center and the −35 region. The homologue of this sequence is present only once in H46A, and this offers a testable hypothesis about its possible involvement in differential CovR binding.
Of considerable interest is our finding that the covRS system in H46A is inactive because of a naturally acquired K102 amber mutation in covR. A systematic search of spontaneous covRS mutations in an in vivo mouse model of GAS infection led Engleberg et al. (4) to the discovery of frameshift, missense, nonsense, and IS1548 insertion mutations in the gene tandem. These authors also detected mutated covRS loci in a high proportion of a panel of clinical GAS isolates (4). Expectedly, spontaneous mutations such as those created by site-directed mutagenesis have proven to enhance the virulence of the mutant strains in animal models of GAS infection (4, 10). The reported hypermutability of the covRS system and the idea that the course of a streptococcal infection could be determined by the clonal fluctuation of more virulent subpopulations of the original wild-type strain (4) is worth pursuing and would also need to include human GCS. Given the disparity of the mutations found in the covRS locus, the mechanism(s) that renders it hypermutable remains a mystery. An alternative explanation involves random mutation associated with a strong in vivo selection for the loss of covRS expression, which could confer a selective advantage on covRS mutants.
The number of times that an mRNA is translated, and hence the amount of protein it produces, will be determined by its half-life. This means of gene regulation has not been explored before in the pathogenic streptococci, and our results concerning the stability of covRS, ska, and mga mRNAs bear on this problem. Whereas the half-life of ska mRNA falls in the range typical of many bacterial mRNAs, covRS mRNA is extremely short-lived, and in contrast, mga mRNA is very long-lived. The mga and covRS pleiotropic regulators have distinct arrays of target genes, mga controlling genes related predominantly to the production of cell surface proteins and covRS-regulated gene products being released into the extracellular environment. Grossly different stabilities of the messages of these regulators will render the surface proteins more rigidly expressed than the released proteins, the levels of which may quickly respond to changing environmental conditions. As a case in point, the level of covRS mRNA but not that of mga mRNA increases by an as yet undetermined mechanism during amino acid starvation (27) (see Fig. 1). Thus, unfavorable nutritional conditions will stabilize repression of the covRS regulon. As the same conditions will also restrict the expansion of variant subpopulations, potentially short-term environmental changes for the better may induce rapid responses of the CovR regulatory circuit at both the population and gene expression level.
In this study, the fas operon of GCS was found to differ from the GAS fas system (12) by lacking the proximal fasB gene encoding a second histidine protein kinase. Because, in the absence of repression, the fasCAXH46A operon stimulates skcH46A expression to approximately the same extent as the fasBCAXNZ131 operon stimulates skaNZ131 expression (Table 2), one sensor kinase, FasC, would appear to be sufficient for full activity of the operon. Thus, FasC and FasA appear to be the genuinely mated pair, and FasB would assume an orphan state rather than forming a heterodimer with FasC or sensing different environmental signals to communicate with FasA, as discussed by Kreikemeyer et al. (12). Thus, since the overwhelming majority of the two-component systems are organized in dicistronic operons, fasB accretion may be a secondary feature of the fasGAS system, the possible functional significance of which remains to be explored. Our previous observation that there is some degree of transcriptional independence of fasBNZ131 from the downstream genes of the operon (28) might also be taken to lend some support to our notion that the occurrence of fasB in the fasGAS operon is a derived feature. In any case, at the sequence level, FasB and FasC are very dissimilar (sequence identity, ≈28%), and it would not be surprising if FasA were able to tell them apart.
The present results confirm for GCS our previous observation with GAS that fasX RNA, in addition to its monocistronic form, can occur at lower abundance as part of the fas operon transcript. They also extend to GCS the original finding by Kreikemeyer et al. (12) that the high transcription rate of fasXGAS from its own promoter(s) depends on the products of the promoter-proximal fas genes, in particular fasA. These authors also showed that deletion of fasXGAS alone evokes the same regulatory behavior of the target genes as inactivation of fasA, lending strong support to the notion that fasX is the effector of the fas regulon. As such, fasXH46A RNA appears to act primarily at the transcriptional level, as judged by reduced skc mRNA levels in H46A fas mutants (Fig. 7). However, a search of complementarity between the 280-bp region upstream of the skc start codon and the fasXH46A sequence did not reveal potential target DNA tracts of more than 10 contiguous nucleotides. A similar observation was made as a result of screening the SF370 genome sequence for regions matching the fasXGAS sequence (12). However, long matching regions between the target and the effector might be dispensable, given the possibility that extended secondary-structure formation of the effector RNA (23), target DNA bending (9), or additional protein(s) (23) could assist in binding of fasX RNA to its DNA targets.
Seeing that the manner in which the much longer known staphylococcal analogue of fasX, RNAIII, regulates target genes still awaits understanding (23), elucidation of the mechanism of fasX action may present a formidable problem. For the time being, the present results provide appropriate explanations of differential SK activities in distinct streptococcal isolates and of the previously observed epistatic effects of different host backgrounds in SK allele swapping (17).
Acknowledgments
We thank M. Völkel and U. Wrazidlo for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (Ma 1330/2-1) and the Fonds der Chemischen Industrie (10046).
Editor: E. I. Tuomanen
REFERENCES
- 1.Bernish, B., and I. van de Rijn. 1999. Characterization of a two-component system in Streptococcus pyogenes which is involved in regulation of hyaluronic acid production. J. Biol. Chem. 274:4786-4793. [DOI] [PubMed] [Google Scholar]
- 2.Caparon, M. G., and J. R. Scott. 1991. Genetic manipulation of pathogenic streptococci. Methods Enzymol. 204:556-586. [DOI] [PubMed] [Google Scholar]
- 3.Christensen, L. R. 1945. Streptococcal fibrinolysis: a proteolytic reaction due to a serum enzyme activated by streptococcal fibrinolysin. J. Gen. Physiol. 28:363-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engleberg, N. C., A. Heath, A. Miller, C. Rivera, and V. J. DiRita. 2001. Spontaneous mutations in the CsrRS two-component regulatory system of Streptococcus pyogenes result in enhanced virulence in a murine model of skin and soft tissue infection. J. Infect. Dis. 183:1043-1054. [DOI] [PubMed] [Google Scholar]
- 5.Federle, M. J., K. S. McIver, and J. R. Scott. 1999. A response regulator that represses transcription of several virulence operons in the group A streptococcus. J. Bacteriol. 181:3649-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferretti, J. J., W. M. McShan, D. Ajdic, D. Savic, K. Lyon, S. Sezate, A. N. Suvorov, S. Clifton, S. Kenton, H. S. Lai, S. P. Lin, F. Z. Najar, L. Song, J. White, X. Yuan, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frank, C., K. Steiner, and H. Malke. 1995. Conservation of the organization of the streptokinase gene region among pathogenic streptococci. Med. Microbiol. Immunol. 184:749-758. [DOI] [PubMed] [Google Scholar]
- 8.Gase, K., T. Ellinger, and H. Malke. 1995. Complex transcriptional control of the streptokinase gene of Streptococcus equisimilis H46A. Mol. Gen. Genet. 247:749-758. [DOI] [PubMed] [Google Scholar]
- 9.Gross, S., K. Gase, and H. Malke. 1996. Localization of the sequence-determined DNA bending center upstream of the streptokinase gene skc. Arch. Microbiol. 166:116-121. [DOI] [PubMed] [Google Scholar]
- 10.Heath, A., V. J. DiRita, N. L. Barg, and N. C. Engleberg. 1999. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect. Immun. 67:5298-5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, T. T., H. Malke, and J. J. Ferretti. 1989. Heterogeneity of the streptokinase gene in group A streptococci. Infect. Immun. 57:502-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreikemeyer, B., M. D. P. Boyle, B. A. Leonard Buttaro, M. Heinemann, and A. Podbielski. 2001. Group A streptococcal growth phase-associated virulence factor regulation by a novel operon (Fas) with homologies to two-component-type regulators requires a small RNA molecule. Mol. Microbiol. 39:392-406. [DOI] [PubMed] [Google Scholar]
- 13.Levin, J. C., and M. R. Wessels. 1998. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol. Microbiol. 30:209-219. [DOI] [PubMed] [Google Scholar]
- 14.Malke, H., and J. J. Ferretti. 1984. Streptokinase: cloning, expression, and excretion by Escherichia coli. Proc. Natl. Acad. Sci. USA 81:3557-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malke, H., U. Mechold, K. Gase, and G. Gerlach. 1994. Inactivation of the streptokinase gene prevents Streptococcus equisimilis H46A from acquiring cell-associated plasmin activity in the presence of plasminogen. FEMS Microbiol. Lett. 116:107-112. [DOI] [PubMed] [Google Scholar]
- 16.Malke, H., B. Roe, and J. J. Ferretti. 1985. Nucleotide sequence of the streptokinase gene from Streptococcus equisimilis H46A. Gene 34:357-362. [DOI] [PubMed] [Google Scholar]
- 17.Malke, H., K. Steiner, K. Gase, and C. Frank. 2000. Expression and regulation of the streptokinase gene. Methods 21:111-124. [DOI] [PubMed] [Google Scholar]
- 18.Mechold, U., and H. Malke. 1997. Characterization of the stringent and relaxed responses of Streptococcus equisimilis. J. Bacteriol. 179:2658-2667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mechold, U., K. Steiner, S. Vettermann, and H. Malke. 1993. Genetic organization of the streptokinase region of the Streptococcus equisimilis H46A chromosome. Mol. Gen. Genet. 241:129-140. [DOI] [PubMed] [Google Scholar]
- 20.Miller, A. A., N. C. Engleberg, and V. J. DiRita. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976-990. [DOI] [PubMed] [Google Scholar]
- 21.Müller, J., H. Reinert, and H. Malke. 1989. Streptokinase mutations relieving Escherichia coli K-12 (prlA4) of detriments caused by the wild-type skc gene. J. Bacteriol. 171:2202-2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. S. de Azavedo. 2000. Genetic locus for streptolysin S production by group A streptococcus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Novick, R. P. 2000. Pathogenicity factors and their regulation, p. 392-407. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 24.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lütticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Steiner, K., and H. Malke. 1995. Transcription termination of the streptokinase gene of Streptococcus equisimilis H46A: bidirectionality and efficiency in homologous and heterologous hosts. Mol. Gen. Genet. 246:374-380. [DOI] [PubMed] [Google Scholar]
- 27.Steiner, K., and H. Malke. 2000. Life in protein-rich environments: the relA-independent response of Streptococcus pyogenes to amino acid starvation. Mol. Microbiol. 38:1004-1016. [DOI] [PubMed] [Google Scholar]
- 28.Steiner, K., and H. Malke. 2001. relA-independent amino acid starvation response network of Streptococcus pyogenes. J. Bacteriol. 183:7354-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tewodros, W., M. Norgren, and G. Kronvall. 1995. Streptokinase activity among group A streptococci in relation to streptokinase genotype, plasminogen binding, and disease manifestations. Microb. Pathog. 18:53-65. [DOI] [PubMed] [Google Scholar]
- 30.Tillet, W. S., and R. L. Garner. 1933. The fibrinolytic activity of hemolytic streptococci. J. Exp. Med. 58:485-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van de Guchte, M., J. M. B. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Vossen, J. M. B. M., D. van der Lelie, and G. Venema. 1987. Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53:2452-2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang, X., X. Lin, J. A. Loy, J. Tang, and X. C. Zhang. 1998. Crystal structure of the catalytic domain of human plasmin complexed with streptokinase. Science 281:1662-1665. [DOI] [PubMed] [Google Scholar]