Abstract
The gram-negative enteric pathogen Vibrio cholerae requires iron for growth. V. cholerae has multiple iron acquisition systems, including utilization of heme and hemoglobin, synthesis and transport of the catechol siderophore vibriobactin, and transport of several siderophores that it does not itself make. One siderophore that V. cholerae transports, but does not make, is enterobactin. Enterobactin transport requires TonB and is independent of the vibriobactin receptor ViuA. In this study, two candidate enterobactin receptor genes, irgA (VC0475) and vctA (VCA0232), were identified by analysis of the V. cholerae genomic sequence. A single mutation in either of these genes did not significantly impair enterobactin utilization, but a strain defective in both genes did not use enterobactin. When either irgA or vctA was supplied on a plasmid, the ability of the irgA vctA double mutant to use enterobactin was restored. This indicates that both VctA and IrgA transport enterobactin. We also identify the genes vctPDGC, which are linked to vctA and encode a periplasmic binding protein-dependent ABC transport system that functions in the utilization of both enterobactin and vibriobactin (VCA0227-0230). An irgA::TnphoA mutant strain, MBG40, was shown in a previous study to be highly attenuated and to have a strong colonization defect in an infant mouse model of V. cholerae infection (M. B. Goldberg, V. J. DiRita, and S. B. Calderwood, Infect. Immun. 58:55-60, 1990). In this work, a new irgA mutation was constructed, and this mutant strain was not significantly impaired in its ability to compete with the parental strain in infant mice and was not attenuated for virulence in an assay of 50% lethal dose. These data indicate that the virulence defect in MBG40 is not due to the loss of irgA function and that irgA is unlikely to be an important virulence factor.
Vibrio cholerae causes the severe diarrheal disease cholera (18, 32). Many of the genes required for this gram-negative pathogen to cause disease in humans or in animal models have been described. A key virulence factor is the cholera toxin, which is responsible for the severe voluminous diarrhea characteristic of cholera. The toxin-coregulated pilus is essential for colonization of the intestinal epithelium (65). The synthesis of this bundle-forming, type IV pilus is coordinately regulated with the synthesis of cholera toxin, and proper transcriptional control of this regulon is required for virulence. Another V. cholerae gene that is reported to be required for virulence is irgA. The irgA::TnphoA mutant strain MBG40 has a decreased competitive index and nearly a 100-fold-increased 50% lethal dose (LD50) in an infant mouse model relative to its parental strain O395 (24). Other genes required for full virulence have been identified more recently, and these include genes for nutrient acquisition, stress response, and proper colonization of the lower small intestine (see references 18 and 32 for reviews).
Genes for the acquisition of the nutrient iron play a critical role in the ability of a pathogen to establish and maintain an infection in its host (48). A variety of strategies for iron acquisition have evolved in pathogenic bacteria. These include the synthesis and secretion of small iron-chelating molecules termed siderophores (8, 12, 17). After binding iron in the extracellular environment, the iron-siderophore complex is transported back into the cell, where the iron is removed and used for various cellular functions. Another strategy for iron acquisition is the direct use of host iron compounds, including heme, hemoglobin, transferrin, and lactoferrin. Uptake of siderophores and iron from host compounds involves specific, high-affinity outer membrane receptors. The energy for transport of these ligands across the outer membrane is provided by the TonB-ExbBD complex, which transduces energy from the inner membrane (7). Transport through the periplasm and across the inner membrane is facilitated by a periplasmic binding protein-dependent ABC transport system. In these systems the periplasmic binding protein binds the ligand and delivers it to the inner membrane permease. This permease usually consists of two integral inner membrane proteins, each of which is bound to an ATPase subunit. The hydrolysis of ATP by the ATPase subunit provides the energy for transport across the inner membrane (8, 17).
Multiple iron acquisition systems have been identified in V. cholerae. Heme and hemoglobin are efficiently used as iron sources (28, 29, 40, 44, 61). V. cholerae makes and transports the catechol siderophore vibriobactin (10, 25, 33, 34, 38, 68, 69) and can use several siderophores that it does not make, including ferrichrome (25), enterobactin (70), and schizokinen (57). The vibriobactin biosynthesis genes are located in two separate gene clusters, one of which also contains the vibriobactin outer membrane receptor gene, viuA (10, 11, 60). The other vibriobactin biosynthesis gene cluster contains the genes for a periplasmic binding protein-dependent ABC transport system. These proteins, ViuPDGC, function in the utilization of both vibriobactin and enterobactin. Although V. cholerae strains carrying mutations in either viuP or viuG had a reduced ability to use vibriobactin and enterobactin, the utilization of these siderophores could still be detected. This suggested that an additional system for the transport of catechol siderophores across the inner membrane is present in V. cholerae (70).
The outer membrane receptor has been identified for ferrichrome (52) but not for schizokinen or enterobactin. A candidate outer membrane receptor for transport of one of these iron complexes is IrgA. The predicted amino acid sequence of IrgA has homology to those of TonB-dependent outer membrane receptors, and it is an abundant iron-regulated protein in the outer membrane (21). Its expression is negatively regulated by the general iron regulatory protein Fur and is positively regulated by the divergently transcribed upstream gene irgB (22, 23) (Fig. 1A). These data suggested that IrgA may function in the transport of iron into the cell. However, no defect in the utilization of various iron compounds was detected in the irgA mutant, and thus no ligand for this potential receptor was identified (21).
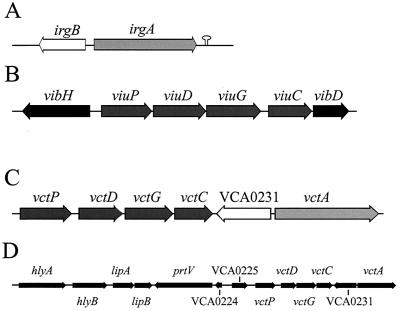
FIG. 1.
Genetic maps of siderophore transport genes in V. cholerae. The arrows indicate the direction of transcription for each of the ORFs. For unnamed reading frames, the locus designation from the TIGR database is used (27). The scale is the same for parts A to C, while part D is drawn on a smaller scale. (A) The irgA irgB region, showing a potential rho-independent terminator downstream of irgA (21, 22); (B) the viuPDGC region (70); (C) the vct region (27); (D) the vct region, showing linkage of these siderophore transport genes with an iron-regulated hemolysin gene and other genes possibly involved in tissue damage (27, 45).
In this work we identified irgA and vctA as the two enterobactin receptor genes present in V. cholerae. We also determined that genes linked to vctA constitute the second periplasmic binding protein-dependent ABC transport system for vibriobactin and enterobactin. Contrary to previous reports, we found that a newly constructed irgA mutant competed efficiently with its wild-type parent for colonization in an infant mouse model of V. cholerae virulence. In infant mice, the LD50 of this irgA mutant was the same as the LD50 of its wild-type parent.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and siderophore utilization assays.
The strains and plasmids used in this study are listed in Table 1. The iron chelator ethylenediamine di(ortho-hydroxyphenylacetic acid) (EDDA) was deferrated by the method of Rogers (51). The antibiotic concentrations used were 250 μg of carbenicillin per ml, 50 μg of kanamycin per ml, and 50 (for E. coli) or 5 (for V. cholerae) μg of chloramphenicol per ml. The bioassay for siderophore utilization was performed as previously described (69).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| CA401 | Classical biotype | 20 |
| CA40130 | Vibriobactin synthesis mutant of CA401 | 25 |
| CA40130N | Nalr derivative of CA40130 | 40 |
| ARM316 | CA40130N irgA::cam | This study |
| AGO310 | CA40130N vctA::kan | This study |
| ARM616 | CA40130N irgA::cam, vctA::kan | This study |
| O395 | Classical biotype | 39 |
| ARM516 | O395 irgA::cam | This study |
| MBG40 | O395 irgA::TnphoA | 24 |
| Lou15 | El Tor biotype | 58 |
| EWV103 | Lou15 viuP::cam | 70 |
| EWV108 | Lou15 vctPD::kan | This study |
| EWV109 | Lou15 viuP::cam, vctPD::kan | This study |
| N16961 | El Tor biotype | J. Kaper |
| E. coli | ||
| DH5α | Cloning strain, Ent+ | 26 |
| SY327(λpir) | Host for pGP704 derivatives | 41 |
| SM10(λpir) | Host for pGP704 derivatives | 41 |
| Plasmids | ||
| pBluescript SK− | Cloning vector | Stratagene |
| pWKS30 | Cloning vector | 66 |
| pACYC184 | Cloning vector | 15 |
| pMTLcam | cam resistance gene | 67 |
| pUC4K | kan resistance gene | Pharmacia |
| pACYCsac | pACYC184 containing sacB | 44 |
| pHM5 | pGP704 carrying sacB | 53 |
| pVIB147 | viuPDGC from Lou15 in pWKS30 | 70 |
| pCAT119 | vctA from CA401 in pWKS30 | This study |
| pCAT120 | vctPDGC from Lou15 in pWKS30 | This study |
| pCAT121 | irgBA from CA401 in pWKS30 | This study |
DNA sequencing.
DNA was sequenced using an Applied Biosystems Prism 377 DNA sequencer (Perkin-Elmer Corp.). Amino acid sequence alignments were performed using the ClustalW feature of the MacVector package (46). The BLAST program (2) was used to search the National Center for Biotechnology Information protein database.
Plasmid construction.
To construct pCAT119, the vctA gene of CA401 was amplified with Pfu DNA polymerase by using primers CatAfor2 (5′-TGGTGGTCATCACATCGCAATC) and CatArev (5′-TGTTTCATCCCAAGTCGCAGG). The resulting PCR product was cloned into the EcoRV site of pWKS30. pCAT120 was constructed by PCR amplification of the vctPDGC genes from strain Lou15 with Pfu DNA polymerase by using primers Liz180 (5′-GCCAAACCATTGCGGAAATAGAAG) and Liz181 (5′-CGTTATCTCAGCACCAAGAGGGAC). The product was cloned into the SmaI site of pWKS30. To construct pAMR18, a fragment containing irgA and irgB was PCR amplified from the strain CA401 by using Pfu DNA polymerase and primers irgBA1 (5′-AGTGAATTCAGCTAAAGAACTGGTGG) and irgBA2 (5′-GGGAATTCTAACCGATACTCTAGGC). The PCR product was digested with EcoRI and cloned into the EcoRI site of pACYC184. The irgBA insert was moved as an EcoRI fragment to pWKS30 to yield pCAT121.
Mutant strain construction.
To construct EWV108 and EWV109, the region carrying vctPDGC was amplified with Taq polymerase by using the primers Liz 174 (5′-TCGGTCACAAAGAGGGGATAGG) and Liz 175 (5′-ATTGCGAAGTAACAGCGAGAGG) with Lou15 DNA as a template. The product was cloned into pGEM-Teasy (Promega) to yield pCAT114. The kan cassette from pUC4K was inserted to replace the internal EcoRV-MscI fragment to yield the plasmid pCAT116. The NotI fragment containing the vct genes was subcloned into pACYCsac to give pCAT117. Allelic exchange mutations were obtained as previously described (69) in Lou15 to give EWV108 or in EWV103 to give EWV109. For the construction of AGO310, the vctA region was amplified using primers CatAfor (5′-TCCATTGCTCAGATTGTCCCTC) and CatArev (listed above) with Taq polymerase. The product was cloned into pGEM-Teasy to give pCAT103. The kan cassette from pUC4K was inserted into the SmaI site to give pAGO-cat1. The SalI-SphI fragment was then cloned into SalI-SphI-digested pHM5, and allelic exchange was performed as previously described (40). To make ARM316, ARM516, and ARM616, the cam cassette from pMTLcam was inserted as a SmaI fragment into the SmaI site of the irgA gene carried on pAMR18. The irgA::cam insert was moved as a Klenow-blunted ClaI fragment into pHM5 digested with EcoRV to yield pAMS5. Allelic exchange was performed as previously described (40) in CA40130N to yield ARM316, in O395 to yield ARM516, and in AGO310 to yield ARM616.
Virulence assays.
In vivo competition assays were performed by a protocol modified from that of Taylor et al. (65). Five-day-old prestarved BALB/c mice were inoculated intragastrically with 50 μl of saline containing 0.02% (wt/vol) Evan's blue dye and 105 CFU of each strain grown to mid-log phase at 37°C. After 24 h, the mice were sacrificed, and the intestines were isolated and homogenized in sterile phosphate-buffered saline. Serial dilutions were plated on selective or differential media to determine the viable counts for each strain. To calculate the competitive index, the ratio of mutant to wild-type bacteria recovered from the intestine was determined and then normalized by dividing by the ratio of mutant to wild-type bacteria in the initial inoculum. The LD50 of each strain was assessed by orally inoculating groups of four 5- to 7-day-old CD-1 mice (Charles River) with a series of 10-fold dilutions of bacteria that had been grown overnight at 30°C in Luria-Bertani broth with a starting pH of 6.5. The inoculum ranged from approximately 10 × 104 through 10 × 108 bacteria. The LD50 is based on the extrapolated dose that would have resulted in a mean survival rate of 50% of the mice after 48 h (49).
Nucleotide sequence accession number.
The GenBank accession number for the sequence of the V. cholerae strain CA401 vctA gene is AY061945.
RESULTS
Identification of enterobactin receptors in V. cholerae.
We previously showed that V. cholerae uses the catechol siderophore enterobactin and that enterobactin transport is independent of the vibriobactin receptor ViuA (70). The recent sequencing of the V. cholerae genome (27) allowed us to identify candidate enterobactin receptors. The genome contains several open reading frames (ORFs) that potentially encode TonB-dependent outer membrane receptors, and the predicted amino acid sequence of each of these ORFs was used to perform a BLAST search (2) of the National Center for Biotechnology Information nonredundant database. Enterobactin receptors were among the highest-scoring proteins in the BLAST search for two of the predicted V. cholerae TonB-dependent receptors, IrgA and a previously uncharacterized protein which we have designated VctA (for Vibrio catechol transport) (see below).
irgA was originally identified as the site of the TnphoA insertion in a strain with reduced virulence in an infant mouse model (24). This putative virulence factor has sequence homology with TonB-dependent outer membrane receptors but no identified ligand. In a BLAST search using IrgA, the top-scoring protein with an identified ligand is the Escherichia coli enterobactin receptor FepA (21) (Table 2). This suggested that enterobactin might be the ligand for IrgA. To test this, an irgA insertion mutation was constructed in CA40130N, a vibriobactin synthesis mutant derived from the classical strain CA401 (40). The vibriobactin mutation was included in the genetic background to reduce the background growth in enterobactin utilization assays. The irgA mutant, ARM316, had no significant defect in enterobactin utilization (Table 3). In addition, ARM316 used vibriobactin, ferrichrome, and heme as efficiently as the parental strain CA40130N (Table 3). This result is in agreement with data for the previously characterized irgA mutant, MBG40, which also used enterobactin, vibriobactin, ferrichrome, and heme as efficiently as its parent (reference 21 and unpublished data).
TABLE 2.
Homologies of V. cholerae enterobactin transport proteins to selected proteins
| Protein (ORF)a and homolog | Ligand | Identity (%) | Similarity (%) | Reference(s) |
|---|---|---|---|---|
| IrgA (VC0475) | ||||
| E. coli IreA | Unknown | 36 | 56 | 54 |
| E. coli CirA | Unknown | 36 | 54 | 43 |
| E. coli O157:H7 Iha | Unknown | 33 | 48 | 63 |
| E. coli FepA | Enterobactin | 26 | 43 | 37 |
| VctA (VCA0232) | ||||
| N. gonorrhoeae FetA | Enterobactin | 22 | 39 | 6, 14 |
| P. shigelloides HugA | Heme | 22 | 41 | 31 |
| S. dysenteriae ShuA | Heme | 22 | 38 | 42 |
| Y. enterolitica HemR | Heme | 21 | 38 | 62 |
| VctP (VCA0227) | ||||
| B. subtilis YclQ | Unknown | 30 | 50 | 36 |
| N. gonorrhoeae FetB | Enterobactin | 28 | 48 | 14 |
| V. anguillarum FatB | Anguibactin | 26 | 48 | 1 |
| C. coli CeuE | Enterobactin | 25 | 47 | 50 |
| V. cholerae ViuP | Vibriobactin | 19 | 35 | 70 |
| VctD (VCA0228) | ||||
| B. subtilis Yc1N | Unknown | 43 | 72 | 36 |
| C. coli CeuB | Enterobactin | 42 | 69 | 50 |
| V. anguillarum FatD | Anguibactin | 35 | 66 | 35 |
| V. cholerae ViuD | Vibriobactin | 24 | 43 | 70 |
| VctG (VCA0229) | ||||
| B. subtilis Yc1O | Unknown | 48 | 69 | 36 |
| C. coli CeuC | Enterobactin | 40 | 65 | 50 |
| V. anguillarum FatC | Anguibactin | 26 | 57 | 35 |
| V. cholerae ViuG | Vibriobactin | 22 | 44 | 70 |
| VctC (VCA0230) | ||||
| B. subtilis Yc1P | Unknown | 51 | 75 | 36 |
| C. coli CeuD | Enterobactin | 43 | 67 | 50 |
| V. cholerae ViuC | Vibriobactin | 27 | 51 | 70 |
ORF numbering is according to the TIGR database (27).
TABLE 3.
IrgA and VctA promote utilization of enterobactin
| Indicator strain | Relevant phenotype | Zone of stimulation (mm) witha:
|
|||
|---|---|---|---|---|---|
| DH5α (enterobactin) | Lou15 (vibriobactin) | Heme | Ferrichrome | ||
| CA40130N | Parent | 17 | 26 | 18 | 24 |
| ARM316 | IrgA− | 14 | 24 | 17 | 22 |
| AGO310 | VctA− | 16 | 26 | 18 | 23 |
| ARM616/pWKS30 | VctA− IrgA− | 0 | 25 | 17 | 28 |
| ARM616/pCAT119 | VctA+ IrgA− | 16 | 28 | 16 | 26 |
| ARM616/pCAT121 | VctA− IrgA+ | 16 | 19 | 16 | 25 |
Cultures of the indicator strains were seeded into L agar containing 100 μg of EDDA per ml. E. coli strain DH5α (as a source of enterobactin), V. cholerae strain Lou15 (as a source of vibriobactin), heme, or ferrichrome was spotted onto the medium. The zone of growth was measured at 18 h after inoculation.
The second enterobactin receptor candidate gene, vctA, is located in a region containing other potential iron transport genes (Fig. 1C). In a BLAST search, VctA showed the highest homology scores with the Neisseria gonorrhoeae enterobactin receptor FetA and also with heme receptors from a variety of organisms (Table 2). A mutation in vctA was constructed by allelic exchange, and the mutant was tested for the ability to use enterobactin. As shown in Table 3, the vctA mutant strain, AGO310, used enterobactin as well as the parent strain did. Although VctA also has homology with heme receptors, AGO310 used heme efficiently (Table 3), and our studies of heme utilization in V. cholerae indicated that VctA is not one of the heme receptors (40).
The failure of both the irgA and vctA single mutants to show a defect in enterobactin utilization could be due to functional redundancy of these transport systems. To test this, we constructed an irgA vctA double mutant. Unlike either of the single mutants, the double mutant strain, ARM616, was completely defective in enterobactin utilization (Table 3). In complementation studies, a plasmid encoding either irgA (pCAT121) or vctA (pCAT119) was introduced into the double mutant strain. The ability to use enterobactin was restored when either the vctA or the irgA gene was supplied on a plasmid (Table 3). This is strong evidence that both IrgA and VctA transport enterobactin in V. cholerae.
In The Institute for Genomic Research (TIGR) genomic database for V. cholerae El Tor strain N16961, the vctA ORF (VCA0232) contains a frameshift mutation and should not encode an active protein (27). Our genetic data, however, suggested that vctA encodes a functional protein in CA40130N, a derivative of classical strain CA401. To resolve this apparent discrepancy, the sequence of the entire vctA gene from CA401 was determined. The CA401 vctA sequence contained an additional G residue not present in the published sequence. The vctA gene containing this additional residue is predicted to encode a full-length receptor protein. To determine whether a functional vctA gene is present in other V. cholerae strains, the region of vctA containing the additional G was sequenced from the classical strain O395 and from the El Tor strains Lou15 and N16961. In each of these strains, the G which restored the ORF was present, suggesting that a functional vctA gene is widely distributed among V. cholerae strains (data not shown). It is unclear why our sequence for N16961 vctA did not match the sequence in the database.
Characterization of a periplasmic binding protein-dependent ABC transport system linked to vctA.
vctA is linked to several genes that could also function in iron transport (Fig. 1C). Divergently transcribed from vctA is the ORF VCA0231, which has homology to the AraC/XylS family of transcriptional regulators. The function of this gene is not known. Upstream of VCA0231 are four genes with sequences that suggest that they encode a periplasmic binding protein-dependent ABC transport system. The vctPDGC genes have the highest homology to the enterobactin transport systems of N. gonorrhoeae (14) and Campylobacter coli (50) and to the anguibactin transport system of Vibrio anguillarum (1, 35) (Table 2). These homologies suggested that VctPDGC might function in the transport of enterobactin and possibly other catechol siderophores across the inner membrane. From these homologies it can be deduced that the vctP product is the periplasmic binding protein, the vctD and vctG products function as the inner membrane permease, and the vctC product is the ATPase for the system. A second set of genes, viuPDGC, had previously been identified as an ABC transport system for vibriobactin and enterobactin in V. cholerae (Fig. 1B) (70). Mutations in viuP or viuG reduced, but did not eliminate, the transport of vibriobactin and enterobactin, indicating the presence of an additional system for the transport of catechol siderophores across the inner membrane.
To determine whether VctPDGC constitute this second catechol siderophore transport system, a chromosomal mutation was created by allelic exchange, in which the 3′ region of vctP through the 5′ region of vctD was replaced by a kanamycin cassette. This mutation was created both in a wild-type Lou15 background to make the single vctPD mutant strain EWV108 and in the viuP mutant strain EWV103 to make a vctPD viuP double mutant, strain EWV109. As shown in Table 4, the vctPD mutant had no significant defect in either enterobactin or vibriobactin utilization, whereas the utilization of vibriobactin was reduced, but not abolished, in the viuP mutant. However, the vctPD viuP double mutant was completely defective in the utilization of both siderophores. When a plasmid clone containing either the vctPDGC transport genes (pCAT120) or the viuPDGC genes (pVIB147) was introduced into the double mutant strain, the utilization of both siderophores was restored. This indicates that both systems can function in the transport of both vibriobactin and enterobactin.
TABLE 4.
ViuPDGC and VctPDGC promote utilization of enterobactin and vibriobactin
| Indicator strain | Relevant phenotype | Zone of stimulation (mm) witha:
|
||
|---|---|---|---|---|
| DH5α (enterobactin) | Lou15 (vibriobactin) | Ferrichrome | ||
| Lou15 | Wild type | 18 | 24 | 28 |
| EWV103/pWKS30 | ViuP− | 16 | 12 | 18 |
| EWV108/pWKS30 | VctPD− | 17 | 21 | 28 |
| EWV109/pWKS30 | ViuP− VctPD− | 0 | 0 | 17 |
| EWV109/pCAT120 | ViuP− VctPD+ | 13 | 20 | 22 |
| EWV109/pVIB147 | ViuP+ VctPD− | 12 | 22 | 22 |
Cultures of the indicator strains were seeded into L agar containing 250 μg of EDDA per ml. E. coli strain DH5α (as a source of enterobactin), V. cholerae strain Lou15 (as a source of vibriobactin), or ferrichrome was spotted onto the medium. The zone of growth was measured at 18 h after inoculation.
As previously observed, the viuP mutant strain had a smaller zone of growth around both vibriobactin and ferrichrome than either the wild type or the vctPD mutant (Table 4). Since all of the Lou15-derived strains in this experiment produce vibriobactin, a low level of vibriobactin is present throughout the plate. This vibriobactin may withhold iron from the viuP mutant, which uses vibriobactin with reduced efficiency. Thus, the viuP mutant is likely to be somewhat more iron starved than the wild-type parent, resulting in the smaller zones of growth stimulation around vibriobactin and ferrichrome observed with this mutant.
Virulence assays of irgA mutants.
It has been reported that the irgA mutant MBG40 has reduced virulence in animal models. However, it is unclear why loss of one of two enterobactin receptors would attenuate virulence. To address this question, we used allelic exchange to construct a new, defined irgA mutation in the classical V. cholerae strain O395, the parent strain of MBG40. The presence of the cam cassette within irgA in mutant strain ARM516 was confirmed by PCR and by Southern blotting (data not shown). To further characterize the defect in this strain, outer membrane fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. A protein band with a molecular weight expected for mature IrgA was observed in Coomassie blue-stained fractions from the wild-type parent O395 but not in those from the irgA mutant strain ARM516. The presence of this protein band was restored when irgA was supplied to ARM516 on the plasmid pCAT121 (data not shown). As expected, ARM516 used enterobactin in a bioassay (data not shown), and this is likely due to the presence of a functional vctA gene in this strain.
The ability of ARM516 to compete with its wild-type parental strain O395 in an infant mouse assay was determined (Table 5). In these experiments, infant mice were given an intragastric inoculation of an equal number of wild-type and mutant bacteria, and the ratio of mutant to wild-type bacteria recovered 24 h later from the intestine was determined by plating on selective media. The competitive index of ARM516 was less than 1, but this reduction in competitive index was not statistically significant. In the same experiment, the previously characterized irgA mutant MBG40 had the very low competitive index of 0.12 (Table 5). This is in agreement with the competitive index of 0.11 previously reported for this strain (24). Interestingly, when the irgA gene was supplied on a plasmid, the ability of MBG40 to compete with the wild-type parent was not restored. This suggests that the defect in MBG40 is not simply the lack of a functional irgA gene.
TABLE 5.
In vivo competition assays of wild-type and irgA mutants of V. cholerae
| Strain | Competitive indexa
|
No. of mice | |
|---|---|---|---|
| Mean (SD) | Range | ||
| ARM516 | 0.71 (0.46) | 0.51-1.21 | 8 |
| MBG40 | 0.12 (0.11) | <0.01-0.38 | 10 |
| MBG40/pCAT121 | 0.07 (0.02) | 0.04-0.09 | 4 |
The competitive index of the indicated strain was determined relative to the wild-type parent O395 in an infant mouse model. The competitive index is the output ratio adjusted for the input ratio of the two competing strains, as described in Materials and Methods.
MBG40 has also been reported to have reduced lethality in a mouse model (24). To test whether the newly constructed irgA mutant, ARM516, has a significant reduction in virulence, its LD50 in infant mice was determined as described in Materials and Methods. The LD50 of ARM516 (8.4 × 105) was nearly identical to that of its wild-type parent O395 (1.0 × 106), indicating that ARM516 is not attenuated for virulence. Thus, it appears that the virulence defect observed for MBG40 is specific to that strain and not a general property of irgA mutants.
DISCUSSION
We had previously proposed a model for catechol siderophore transport in V. cholerae in which ferri-vibriobactin and ferri-enterobactin are each transported by a specific outer membrane receptor (70). Both of these siderophores could then be transported across the inner membrane by either the ViuPDGC or VctPDGC system. Based on the data presented in this work, we have modified our model to show that there are two enterobactin receptor genes, and these are identified as irgA (VC0475) and vctA (VCA0232) (Fig. 2). An additional system for the transport of catechol siderophores was also identified. These genes, vctPDGC, are closely linked to the vctA gene (VCA0227-230) (Fig. 1C). The observation that the outer membrane receptors are specific for their ligands while the inner membrane permeases transport a variety of structurally related ligands is reminiscent of the transport of hydroxamate siderophores. In E. coli, each hydroxamate siderophore is transported by a specific outer membrane receptor, whereas all are transported across the inner membrane by a single system, FhuBCD (8).
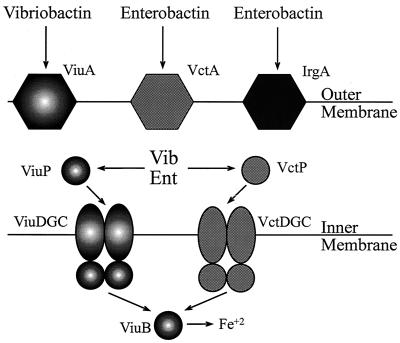
FIG. 2.
A model for catechol siderophore transport in V. cholerae. Enterobactin can be transported across the outer membrane by either of the two outer membrane receptors, IrgA (VC0475) or VctA (VCA0232), while the endogenous siderophore vibriobactin is transported by ViuA (VC2211). Following entry into the periplasm, both siderophores can be transported across the inner membrane by either the ViuPDGC (VC0776 to VC0779) or the VctPDGC (VCA0227 to VCA0230) system. The iron is then removed from the siderophore by ViuB (VC2210), which has been shown to remove the iron from both enterobactin and vibriobactin (9).
Most of the essential V. cholerae genes are encoded on the large chromosome, while the function of many of the genes encoded on the smaller chromosome is unknown. In addition, significant portions of the small chromosome appear to have been acquired by horizontal gene transfer (27). IrgA and ViuPDGC are encoded on the large chromosome, and both of these systems have high homology with the E. coli Fep system for enterobactin transport. In contrast, the VctA and VctPDGC genes are encoded on the small chromosome and have homology with enterobactin transport proteins from a diverse group of organisms. These data suggest that irgA and viuPDGC may have evolved from ancestral Vibrio fep-like genes, while the vctA and vctPDGC genes may have been acquired more recently. The vct genes are closely linked to a region encoding several secreted proteins, including an iron-regulated hemolysin, a lipase, and an extracellular protease (45) (Fig. 1D). It was previously proposed that this region could be part of a pathogenicity island promoting host tissue damage and thus aiding in the acquisition of iron and other nutrients by V. cholerae (45). The observation that this region is adjacent to siderophore transport genes supports this model.
Enterobactin is the prototype catechol siderophore produced by a group of closely related members of the Enterobacteriaceae, including E. coli, Salmonella, Klebsiella, and some strains of Shigella. It was recently shown that enterobactin is also produced by the gram-positive bacteria Streptomyces spp., suggesting that enterobactin synthesis is more widely distributed phylogenetically than has been previously recognized (19). Many organisms transport siderophores that they do not make, and this may reflect the relative energetic expense of making a siderophore. Enterobactin transport genes have been identified in a number of gram-negative bacteria that do not make this siderophore, including N. gonorrhoeae (14), Bordetella spp. (5), Campylobacter spp. (47, 50), Pseudomonas aeruginosa (16), and Yersinia enterocolitica (56). In addition, enterobactin transport was observed in a variety of other gram-negative organisms in a survey (55). Although some of these organisms may encounter enterobactin in the gut, the significance of enterobactin utilization to their survival in the host or in the environment is not known for most of these organisms.
It is believed that acquisition of iron is important for virulence, and in some bacterial pathogens, loss of specific iron transport systems correlates with reduced virulence in animal models. However, for many organisms, including V. cholerae, it has been difficult to determine the role of specific iron transport systems in virulence. A general requirement for high-affinity iron transport systems in virulence can be inferred from results of virulence studies using tonB mutants. V. cholerae has two TonB systems (44), which are partially redundant in their transport functions (57). Strains carrying mutations in either of these tonB systems have reduced colonization of infant mice as observed in a competition assay, and a strain carrying mutations in both tonB systems has an even greater reduction in its competitive index (57). These data suggest that high-affinity iron transport is needed for optimal colonization in this assay; however, no single iron source has been identified as being required for virulence. Strains defective in vibriobactin biosynthesis (59), vibriobactin transport (30), or heme transport (30, 40, 64) are only weakly affected for virulence.
The irgA mutant stain MBG40 is more attenuated for virulence than any of the strains carrying mutations in a single iron transport gene (64). However, lack of an identified function for IrgA has made it difficult to understand how it might contribute to the virulence of V. cholerae. This has generated debate as to whether IrgA is the receptor for an important, but unidentified, iron source or whether it might have a function unrelated to iron acquisition. In this work we show that IrgA is an outer membrane receptor for enterobactin. The irgA single mutants tested in this study used enterobactin as efficiently as their wild-type parent in vitro, presumably due to the presence of the other enterobactin receptor gene, vctA. However, even if IrgA is needed for efficient enterobactin utilization in vivo, it seems unlikely that reduced enterobactin utilization would severely attenuate virulence. V. cholerae colonizes the lower portions of the small intestine (3, 4), while E. coli is present primarily in the large intestine. Thus, enterobactin is probably not available in significant quantities to V. cholerae during colonization of its host. V. cholerae may encounter enterobactin as it is shed through the large intestine, but this would not contribute to the competitive index in the infant mouse model.
To resolve this question, we constructed a different irgA mutation in O395, the parental strain of MBG40. This irgA mutant strain, ARM516, was not significantly defective in its ability to compete with the wild-type parental strain O395 in an infant mouse model (Table 5), suggesting that it is able to colonize and grow at wild-type levels within its host. In addition, the LD50 of ARM516 in an infant mouse model was similar to that of the wild-type strain O395. In contrast, MBG40 did not efficiently compete with the parental strain, consistent with the previous characterization of this mutant. The ability of MBG40 to compete with the parental strain was not restored when irgA was supplied on a plasmid, providing further evidence that the virulence defect in this strain is not due to loss of IrgA function. From these data we conclude that the virulence defect of MBG40 is specific to that strain. The virulence defect in MBG40 is unlikely to be due to polarity of the TnphoA insertion, because a previous study demonstrated that the gene downstream of irgA, which encodes erythrose-4-phosphate dehydrogenase, is not required for virulence (13). It is possible that this defect is the result of toxicity of the irgA::phoA fusion, although this is unlikely since MBG40 grows normally in vitro in low-iron media in which expression of the fusion protein is maximally induced (21). Alternatively, MBG40 may contain a mutation in addition to the irgA::TnphoA insertion. It was shown by Southern hybridization that MBG40 contains only a single TnphoA insertion, (24), implying that this mutation is one not detected by that method. The nature of the colonization defect in MBG40 is not known, but it is of interest to identify the defect, since it appears to have a significant effect on colonization and virulence.
Acknowledgments
This work was supported by a grant from the Foundation for Research and National Institutes of Health grant AI 50669 to S.M.P. and by National Institutes of Health grant AI25096 to R.K.T.
We thank Stephen Calderwood for strains and helpful discussions and Chris Tinkle, Bonnie Reus, and Ana-Maria Valle for technical assistance. We are grateful to Melissa Mann and Mary Lozano for assistance with the animal studies and to Laura Runyen-Janecky for critical reading of the manuscript.
Editor: J. T. Barbieri
REFERENCES
- 1.Actis, L. A., M. E. Tolmasky, D. H. Farrell, and J. H. Crosa. 1988. Genetic and molecular characterization of essential components of the Vibrio anguillarum plasmid-mediated iron-transport system. J. Biol. Chem. 263:2853-2860. [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Angelichio, M. J., J. Spector, M. K. Waldor, and A. Camilli. 1999. Vibrio cholerae intestinal population dynamics in the suckling mouse model of infection. Infect. Immun. 67:3733-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baselski, V. S., and C. D. Parker. 1978. Intestinal distribution of Vibrio cholerae in orally infected infant mice: kinetics of recovery of radiolabel and viable cells. Infect. Immun. 21:518-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beall, B., and G. N. Sanden. 1995. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology 141:3193-3205. [DOI] [PubMed] [Google Scholar]
- 6.Beucher, M., and P. F. Sparling. 1995. Cloning, sequencing, and characterization of the gene encoding FrpB, a major iron-regulated, outer membrane protein of Neisseria gonorrhoeae. J. Bacteriol. 177:2041-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braun, V. 1995. Energy-coupled transport and signal transduction through the gram-negative outer membrane via TonB-ExbB-ExbD-dependent receptor proteins. FEMS Microbiol. Rev. 16:295-307. [DOI] [PubMed] [Google Scholar]
- 8.Braun, V., K. Hantke, and W. Koster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation, p. 67-145. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems, vol. 35. Marcel Dekker, New York, N.Y. [PubMed]
- 9.Butterton, J. R., and S. B. Calderwood. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butterton, J. R., M. H. Choi, P. I. Watnick, P. A. Carroll, and S. B. Calderwood. 2000. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J. Bacteriol. 182:1731-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butterton, J. R., J. A. Stoebner, S. M. Payne, and S. B. Calderwood. 1992. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J. Bacteriol. 174:3729-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byers, B. R., and J. E. Arceneaux. 1998. Microbial iron transport: iron acquisition by pathogenic microorganisms. Metal Ions Biol. Syst. 35:37-66. [PubMed] [Google Scholar]
- 13.Carroll, P. A., G. Zhao, S. A. Boyko, M. E. Winkler, and S. B. Calderwood. 1997. Identification, sequencing, and enzymatic activity of the erythrose-4-phosphate dehydrogenase gene of Vibrio cholerae. J. Bacteriol. 179:293-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Carson, S. D. B., P. E. Klebba, S. M. C. Newton, and P. F. Sparling. 1999. Ferric enterobactin binding and utilization by Neisseria gonorrhoeae. J. Bacteriol. 181:2895-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dean, C. R., and K. Poole. 1993. Cloning and characterization of the ferric enterobactin receptor gene (pfeA) of Pseudomonas aeruginosa. J. Bacteriol. 175:317-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Earhart, C. F. 1996. Uptake and metabolism of iron and molybdenum, p. 1075-1090. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed., vol. 1. ASM Press, Washington, D.C.
- 18.Faruque, S. M., M. J. Albert, and J. J. Mekalanos. 1998. Epidemiology, genetics and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev. 62:1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fiedler, H.-P., P. Krastel, J. Müller, K. Gebhardt, and A. Zeeck. 2001. Enterobactin: the characteristic catecholate siderophore of Enterobacteriaceae is produced by Streptomyces species. FEMS Microbiol. Lett. 196:147-151. [DOI] [PubMed] [Google Scholar]
- 20.Gardner, E. W., S. T. Lyles, and C. E. Langkford. 1964. A comparison of virulence in Vibrio cholerae strains for embryonated egg. J. Infect. Dis. 114:412.. [DOI] [PubMed] [Google Scholar]
- 21.Goldberg, M. B., S. A. Boyko, J. R. Buttertion, J. A. Stoebner, S. M. Payne, and S. B. Calderwood. 1992. Characterization of a Vibrio cholerae virulence factor homologous to the family of TonB-dependent proteins. Mol. Microbiol. 6:2407-2418. [DOI] [PubMed] [Google Scholar]
- 22.Goldberg, M. B., S. A. Boyko, and S. B. Calderwood. 1991. Positive transcriptional regulation of an iron-regulated virulence gene in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:1125-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldberg, M. B., S. A. Boyko, and S. B. Calderwood. 1990. Transcriptional regulation by iron of a Vibrio cholerae virulence gene and homology of the gene to the Escherichia coli fur system. J. Bacteriol. 172:6863-6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goldberg, M. B., V. J. DiRita, and S. B. Calderwood. 1990. Identification of an iron-regulated virulence determinant in Vibrio cholerae, using TnphoA mutagenesis. Infect. Immun. 58:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griffiths, G. L., S. P. Sigel, S. M. Payne, and J. B. Neilands. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem. 259:383-385. [PubMed] [Google Scholar]
- 26.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 27.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson, D. P., and S. M. Payne. 1994. Characterization of the Vibrio cholerae outer membrane heme transport protein HutA: sequence of the gene, regulation of expression, and homology to the family of TonB-dependent proteins. J. Bacteriol. 176:3269-3277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson, D. P., and S. M. Payne. 1993. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol. Microbiol. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 30.Henderson, D. P., and S. M. Payne. 1994. Vibrio cholerae iron transport systems: role of heme and siderophore iron transport in virulence and identification of a gene associated with multiple iron transport systems. Infect. Immun. 62:5120-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henderson, D. P., E. E. Wyckoff, C. E. Rashidi, H. Verlei, and A. L. Oldham. 2001. Characterization of the Plesiomonas shigelloides genes encoding the utilization of iron from heme. J. Bacteriol. 183:2715-2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaper, J. B., J. G. Morris, and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522-15530. [DOI] [PubMed] [Google Scholar]
- 34.Keating, T. A., C. G. Marshall, and C. T. Walsh. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513-15521. [DOI] [PubMed] [Google Scholar]
- 35.Köster, W. L., L. A. Actis, L. S. Waldbeser, M. E. Tolmasky, and J. H. Crosa. 1991. Molecular characterization of the iron transport system mediated by the pJM1 plasmid in Vibrio anguillarum 775. J. Biol. Chem. 266:23829-23833. [PubMed] [Google Scholar]
- 36.Kunst, F., N. Ogasawara, I. Moszer, A. M. Albertini, G. Alloni, V. Azevedo, M. G. Bertero, P. Bessieres, A. Bolotin, S. Borchert, R. Borriss, L. Boursier, A. Brans, M. Braun, S. C. Brignell, S. Bron, S. Brouillet, C. V. Bruschi, B. Caldwell, V. Capuano, N. M. Carter, S. K. Choi, J. J. Codani, I. F. Connerton, A. Danchin, et al. 1997. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390:249-256. [DOI] [PubMed] [Google Scholar]
- 37.Lundrigan, M. D., and R. J. Kadner. 1986. Nucleotide sequence of the gene for the ferrienterochelin receptor FepA in Escherichia coli. J. Biol. Chem. 261:10797-10801. [PubMed] [Google Scholar]
- 38.Marshall, C. G., M. D. Burkart, T. A. Keating, and C. T. Walsh. 2001. Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochemistry 40:10655-10663. [DOI] [PubMed] [Google Scholar]
- 39.Mekalanos, J. J., D. J. Swartz, G. D. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 40.Mey, A. R., and S. M. Payne. 2001. Haem utilization in Vibrio cholerae involves multiple TonB-dependent haem receptors. Mol. Microbiol. 42:835-849. [DOI] [PubMed] [Google Scholar]
- 41.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mills, M., and S. M. Payne. 1997. Analysis of shuA, the gene encoding the heme receptor of Shigella dysenteriae, and analysis of invasion and intracellular multiplication of a shuA mutant. Infect. Immun. 65:5358-5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nau, C. D., and J. Konisky. 1989. Evolutionary relationship between the TonB-dependent outer membrane transport proteins: nucleotide and amino acid sequences of the Escherichia coli colicin I receptor gene. J. Bacteriol. 171:1041-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Occhino, D. A., E. E. Wyckoff, D. P. Henderson, T. J. Wrona, and S. M. Payne. 1998. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol. Microbiol. 29:1493-1507. [DOI] [PubMed] [Google Scholar]
- 45.Ogierman, M. A., A. Fallarino, T. Riess, S. G. Williams, S. R. Attridge, and P. A. Manning. 1997. Characterization of the Vibrio cholerae El Tor lipase operon lipAB and a protease gene downstream of the hly region. J. Bacteriol. 179:7072-7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olson, S. A. 1994. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol. Biol. 25:195-201. [DOI] [PubMed] [Google Scholar]
- 47.Park, S. F., and P. T. Richardson. 1995. Molecular characterization of a Campylobacter jejuni lipoprotein with homology to periplasmic siderophore-binding protein. J. Bacteriol. 177:2259-2264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 49.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 50.Richardson, P. T., and S. F. Park. 1995. Enterochelin acquisition in Campylobacter coli: characterization of components of a binding-protein-dependent transport system. Microbiology 141:3181-3191. [DOI] [PubMed] [Google Scholar]
- 51.Rogers, H. J. 1973. Iron-binding catechols and virulence in Escherichia coli. Infect. Immun. 7:445-456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogers, M. B., J. A. Sexton, G. J. DeCastro, and S. B. Calderwood. 2000. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J. Bacteriol. 182:2350-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Runyen-Janecky, L. J., M. Hong, and S. M. Payne. 1999. Virulence plasmid-encoded impCAB operon enhances survival and induced mutagenesis in Shigella flexneri after exposure to UV radiation. Infect. Immun. 67:1415-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Russo, T. A., U. B. Carlino, and J. R. Johnson. 2001. Identification of a new iron-regulated virulence gene, ireA, in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 69:6209-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rutz, J. M., T. Abdullah, S. P. Singh, V. I. Kalve, and P. E. Klebba. 1991. Evolution of the ferric enterobactin receptor in gram-negative bacteria. J. Bacteriol. 173:5964-5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schubert, S., D. Fischer, and J. Heesemann. 1999. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J. Bacteriol. 181:6387-6395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seliger, S. S., A. R. Mey, A.-M. Valle, and S. M. Payne. 2001. The two TonB systems in Vibrio cholerae: redundant and specific functions. Mol. Microbiol. 39:801-812. [DOI] [PubMed] [Google Scholar]
- 58.Sigel, S. P., and S. M. Payne. 1982. Effect of iron limitation on growth, siderophore production and expression of outer membrane proteins of Vibrio cholerae. J. Bacteriol. 150:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sigel, S. P., J. A. Stoebner, and S. M. Payne. 1985. Iron-vibriobactin transport system is not required for virulence of Vibrio cholerae. Infect. Immun. 47:360-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoebner, J. A., J. R. Butterton, S. B. Calderwood, and S. M. Payne. 1992. Identification of the vibriobactin receptor of Vibrio cholerae. J. Bacteriol. 174:3270-3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoebner, J. A., and S. M. Payne. 1988. Iron-regulated hemolysin production and utilization of heme and hemoglobin by Vibrio cholerae. Infect. Immun. 56:2891-2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stojiljkovic, I., and K. Hantke. 1992. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in gram-negative bacteria. EMBO J. 11:4359-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tashima, K. T., P. A. Carroll, M. B. Rogers, and S. B. Calderwood. 1996. Relative importance of three iron-regulated outer membrane proteins for in vivo growth of Vibrio cholerae. Infect. Immun. 64:1756-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang, R. F., and S. R. Kushner. 1991. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene 100:195-199. [PubMed] [Google Scholar]
- 67.Wyckoff, E. E., D. Duncan, A. G. Torres, M. Mills, K. Maase, and S. M. Payne. 1998. Structure of the Shigella dysenteriae haem transport locus and its phylogenetic distribution in enteric bacteria. Mol. Microbiol. 28:1139-1152. [DOI] [PubMed] [Google Scholar]
- 68.Wyckoff, E. E., S. L. Smith, and S. M. Payne. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J. Bacteriol. 183:1830-1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wyckoff, E. E., J. A. Stoebner, K. E. Reed, and S. M. Payne. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J. Bacteriol. 179:7055-7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wyckoff, E. E., A.-M. Valle, S. L. Smith, and S. M. Payne. 1999. A multifunctional ABC transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J. Bacteriol. 181:7588-7596. [DOI] [PMC free article] [PubMed] [Google Scholar]