Abstract
Chlamydia pneumoniae is a common intracellular human pathogen that has been associated with several severe pathological conditions, including coronary heart disease and atherosclerosis. There is no vaccine against C. pneumoniae infection, but CD8+ T cells have been shown to be crucial for protection during experimental infection. However, the effector functions and epitope specificity of the protective CD8+ T cell remain unknown. The aim of this study was to identify C. pneumoniae-derived mouse CD8 epitopes by using a recent epitope prediction method. Of four C. pneumoniae proteins (the major outer membrane protein, outer membrane protein 2, polymorphic outer membrane protein 5, and heat shock protein 60), 53 potential CD8+ T-cell epitopes were predicted by H-2 class I binding algorithms. Nineteen of the 53 peptides were identified as CD8 epitopes by testing for induction of a cytotoxic response after immunization. To test whether the predicted epitopes are naturally processed and presented by C. pneumoniae-infected cells, we generated a panel of seven peptide-specific cytotoxic T lymphocyte lines that were subsequently tested for recognition of C. pneumoniae-infected target cells. By using this strategy, we were able to identify three C. pneumoniae CD8 epitopes that were, indeed, processed and presented on infected cells. Identification of these natural CD8 epitopes provides tools for characterization of CD8+ T-cell function in vivo and generation of epitope-specific prevention strategies.
Chlamydia pneumoniae is a common human respiratory pathogen belonging to the Chlamydia genus of obligatorily intracellular bacteria (6). Like Chlamydia trachomatis and Chlamydia psittaci, C. pneumoniae has been shown to have a tendency to develop persistent infections in vitro (15) and in vivo (10, 13, 14). An increasing body of data suggests an association between persistent C. pneumoniae infection and severe sequelae, such as coronary heart disease and atherosclerosis. In addition to the presence of C. pneumoniae in the atheromatous plaques (reviewed in reference 26), the data include evidence from seroepidemiological studies (16), small-scale intervention studies (7, 8), and animal studies (1). Although the pathogenesis of persistent chlamydial infection is not well understood, the consequences are considered to be immunopathologically mediated (29). Because of the putative severe sequelae of C. pneumoniae, as well as other chlamydial infections, vaccination or other methods of prevention would be highly desirable but, at the same time, caution is needed in order not to exacerbate the pathology. Therefore, a specifically targeted vaccine that stimulates only a protective type of immune response is an attractive approach.
Establishment of a C. pneumoniae-infected mouse model has enabled characterization of infection kinetics and immune response in vivo. Primary infection initiated by intranasal inoculation of C. pneumoniae is self-restricted and leads to a partially protective acquired immunity, detected as faster clearance of bacteria from the lungs during reinfection (17). The mouse model has demonstrated the essential role of T cells in protection, as thymusless mice are incapable of clearing the infection (18). Of the two major T-cell subsets, the CD8+ cells play a major role in bacterial clearance. During primary infection, genetically altered mice lacking CD8+ T cells are more susceptible to infection than are immunocompetent wild-type mice or mice lacking CD4+ T cells (21). Furthermore, the acquired protection detected in reinfection is abolished if mice are treated with monoclonal antibodies to deplete CD8+ T cells (18). Besides the mouse model, activation of CD8+ T cells is also detected in the early phase of C. pneumoniae infection in humans (9).
Although CD8+ T cells seem to be the main effector cells in protection, the protective effector mechanisms are not well known. Knockout mice deficient in perforin or tumor necrosis factor (TNF) receptor p55 do not have an altered course of infection (21), but gamma interferon (IFN-γ) is important for clearance, at least during the early phases of primary infection (5, 21) and reinfection (28). Furthermore, the epitope specificity of the protective CD8+ T cells has not been identified. In the present study, we used the genomic sequence data of C. pneumoniae (12) and a recent epitope prediction method (20) for identification of CD8 epitopes from four C. pneumoniae proteins: the major outer membrane protein (MOMP), cysteine-rich outer membrane protein 2 (Omp2), polymorphic outer membrane protein 5 (Pomp5), and the 60-kDa heat shock protein (Hsp60). The CD8 epitopes identified may serve as tools in a more specific characterization of the CD8 response in vivo and in the development of epitope-specific approaches for prevention of C. pneumoniae infection.
MATERIALS AND METHODS
Mice.
Female inbred BALB/c (H-2d) and C57BL/6 (H-2b) mice were obtained from Bomholtgård Breeding and Research Center Ltd., Ry, Denmark, and used in experiments when 7 to 9 weeks old. The mice were given food and water ad libitum and kept in ventilated containers (Scantainer; Scanbur A/S, Køge, Denmark). The study was approved by the institutional animal care and use committee, which acts under the provincial board.
Cell lines and culture medium.
The cell lines used as target cells in this study were P815 (H-2d, derived from a DBA/2 mouse mastocytoma), BALB/3T3 (H-2d, derived from a BALB/c mouse embryo), RMA (H-2b, derived from a T-cell lymphoma), RMA-S (from a transporter associated with antigen processing [TAP]-deficient T-cell lymphoma), and thymic stromal cell line 1308.1 (H-2b), which was kindly provided by B. Knowles (3). All of these cell lines were maintained in complete Dulbecco modified Eagle medium (cDMEM; Gibco BRL, Paisley, Scotland) containing 10% heat-inactivated fetal calf serum, 50 μM 2-mercaptoethanol, 10 μg of streptomycin per ml, and 10 mM HEPES (all reagents were purchased from Sigma, St. Louis, Mo.) at 37°C in 7.5% CO2. Cytotoxic T-lymphocyte (CTL) lines were maintained in MEM alpha (αMEM; Gibco) supplemented with 10% fetal calf serum, 50 μM 2-mercaptoethanol, antibiotics (20 U of penicillin per ml and 20 μg of streptomycin per ml), 25 mM α-methylmannoside (Sigma), 5% concanavalin A (ConA) supernatant (supernatant from ConA-stimulated rat spleen cells), and 1 ng of recombinant murine interleukin-2 (R&D Systems, Abingdon, England) per ml. Proliferation and cytokine assays were performed in the same medium, except without α-methylmannoside, ConA supernatant, and recombinant interleukin-2.
In vitro infection of cell lines with C. pneumoniae.
Infected BALB/3T3 and 1308.1 cells were used as targets in CTL and cytokine assays. The cells were seeded into 24-well tissue culture plates at a density of 105 to 106 cells per well in cDMEM and incubated overnight. On the following day, 1 × 106 to 5 × 106 inclusion-forming units of C. pneumoniae isolate Kajaani 6 in 100 μl of sucrose-phosphate-glutamate solution was added to each well. To facilitate infection, the plates were centrifuged at 500 × g for 1 h, after which the inoculum was removed and replaced with 1 ml of cDMEM with 0.5 μg of cycloheximide (Sigma) per ml. Control cells were treated the same way as infected cells by using sucrose-phosphate-glutamate with UV-inactivated C. pneumoniae or without bacteria. After incubation for 48 h at 35°C in 5% CO2, the cells were trypsinized from the plates, rinsed twice with complete medium without cycloheximide, and used as target cells in cytokine assays. For CTL assays, similarly infected cells were labeled with 51Cr.
Peptide prediction.
All potential CTL epitopes were predicted and synthesized at the University of Tübingen as previously described (20). Briefly, the knowledge of the motifs for major histocompatibility complex (MHC) class I alleles was used to predict CD8 epitopes on the basis of the amino acid sequence of a protein. Peptides possessing known H-2d- and H-2b-binding anchor residues at positions 2 and 9 were identified from the published amino acid sequences of four C. pneumoniae proteins: the MOMP, Omp2, Pomp5, and Hsp60 (http://chlamydia-www.berkeley.edu:4231). Synthesis of 54 peptides with the highest expected binding scores was performed by applying 9-fluorenylmethoxy carbonyl chemistry, followed by high-performance liquid chromatography purification and mass spectroscopy analysis. Lyophilized peptides were diluted to 1 mg/ml with 5% dimethyl sulfoxide-phosphate-buffered saline, stored at −20°C, and used at the desired final concentration. In addition, previously described CTL epitopes of ovalbumin (SIINFEKL) (22) and E. coli β-galactosidase (TPHPARIGL) (4) of the H-2Ld and H-2Kb haplotypes, respectively, were synthesized and used as controls for induction and stimulation of CTL and target cell lysis.
Peptide-binding assay.
Peptides possessing H-2b-binding motifs were evaluated for the ability to increase the expression of MHC class I on the surface of TAP-deficient mutant cell line RMA-S. MHC class I molecules of TAP-deficient cells are labile when no peptide is added, but the binding of exogenous peptide to unstable MHC class I molecules at the cell surface stabilizes the complex and this can be measured by flow cytometric analysis. In the MHC class I stabilization assay, RMA-S was incubated with different peptide concentrations overnight. The cells were then washed and stained with an MHC class I-specific monoclonal antibody, followed by fluorescein isothiocyanate (FITC)-conjugated anti-mouse antibody. Intensity of fluorescence was analyzed by flow cytometry.
Peptide immunization.
Mice were immunized subcutaneously in the neck with 50 μg of synthetic C. pneumoniae-specific peptide and 25 μg of a known helper epitope from ovalbumin (Ad; ISQAVHAAHAEINEAGR) and 25 μg of library helper epitopes (no. 8047; Ed library, obtained from the University of Tübingen). Peptides were emulsified in TiterMax adjuvant (Sigma) at 1:1 (vol/vol). After 2 weeks, the mice were boosted once with the same dose. One week after the second injections, mice were sacrificed, spleens were removed and homogenized, and single-cell suspensions were prepared without depletion of red blood cells.
In vitro stimulations and maintenance of peptide-specific CTL lines.
After peptide immunization, approximately 5 × 107 splenocytes were suspended in CTL medium in 25-cm2 upright-standing tissue culture flasks (Medi-Cult). Peptide (300 ng/ml) was added to the culture. Cytotoxic activity of cultures against peptide-pulsed target cells was tested after 6 days with a standard chromium release assay. Subsequently, half of the responding cells were restimulated weekly by adding one-fourth of the freshly isolated irradiated (2,000 rads) splenocytes from a syngeneic spleen and peptide (100 ng/ml). Irradiated spleen cell suspensions used as feeder cells (peptide-presenting cells) had been previously depleted of red blood cells by treatment with hemolytic Gey's solution. Cultures were incubated at 37°C in 7.5% CO2 in a total volume of 10 ml of CTL medium.
Cytotoxicity assay.
CTL activity was determined by a standard 5-h chromium release assay. Briefly, target cells were labeled with 50 μCi of Na51Cr solution (Nycomed, Amersham, Buckinghamshire, England) for 1 h at 37°C. Labeled target cells were washed three times, resuspended in complete medium, and seeded at a final concentration of 5 × 104/ml. A 100-μl volume of target cells was plated in medium with or without peptide (1 μg/ml) supplementation in the wells of 96-well round-bottom plates (Nunclon; Nalge Nunc) that contained serial dilutions of effector cells in 100-μl aliquots. For experiments in which preincubation of effector cells with 20 μg of anti-CD4 or -CD8 monoclonal antibodies was required, the pretreatment was performed at 37°C for 45 min before the addition of target cells. After incubation for 5 h or overnight at 37°C in a 7.5% CO2 atmosphere, 50 μl of cell-free supernatant from each well was collected and radioactivity was counted with a beta counter (Wallac1450; MicroBeta, Turku, Finland). Spontaneous release from wells containing target cells alone was measured. Maximum release was induced by 1% Triton X-100 (Sigma) treatment. Specific CTL lysis was calculated as follows: percent specific lysis = 100 × [(experimental release − spontaneous release)/(maximum release − spontaneous release)]. All of the data reported are means of duplicate samples, and spontaneous release was typically 4 to 10% of the maximal release.
Proliferation assays.
Lymphocyte proliferative responses to specific peptides were determined by measuring [3H]thymidine (Amersham, Aylesbury, United Kingdom) incorporation. We incubated 2 × 105 CTLs/well in complete αMEM with a peptide concentration of 1 μg/ml and with 2 × 105 irradiated syngeneic splenocytes on 96-well plates. Control wells received antigen-presenting cells and medium without peptide (background control). The cells were incubated at 37°C in an atmosphere of 7.5% CO2. On day 2, the wells were pulsed with 1 μCi of [3H]thymidine and harvested 16 to 18 h later. All assays were performed in quadruplicate. The incorporated activity was measured by standard scintillation counting. The proliferation index (PI) was calculated as follows: PI = (peptide-induced proliferation − background proliferation)/background proliferation.
Cytokine assays.
The amounts of TNF-α and IFN-γ produced by CTL lines stimulated with a specific peptide and irradiated feeder cells or C. pneumoniae-infected target cells were determined by enzyme-linked immunosorbent assay (ELISA). When the CTLs were stimulated with the peptide, cytokine samples were collected from proliferation plates before addition of [3H]thymidine on day 2. The supernatants (50 μl from each well) were frozen and analyzed later. When the lines were stimulated with infected targets, CTLs were plated into 24-well plates at 3 × 105 per well. C. pneumoniae-infected 3T3 or 1308.1 cells were added at 3 × 105 cells per well, and the final volume was adjusted to 700 μl with complete αMΕM without penicillin. Control wells contained uninfected target cells. The cells were incubated at 37°C in an atmosphere of 7.5% CO2 for 48 h, after which the supernatants were collected. The supernatants were analyzed for IFN-γ and TNF-α with DuoSet mouse kits (R&D Systems, Minneapolis, Minn.). IFN-γ and TNF-α detection limits were typically 50 and 24 pg/ml, respectively.
Flow cytometric analysis.
The phenotype of cultured CTL lines was determined by immunofluorescence staining and flow cytometric analysis. Before immunostaining, viable CTLs were separated from irradiated feeder cells by Ficoll-Hypaque (Pharmacia Biotech AB, Uppsala, Sweden) centrifugation. CTLs were stained with 5 μl of FITC-conjugated anti-CD8 and phycoerythrin-conjugated anti-CD4 (all purchased from Caltag, Burlingame, Calif.) antibodies for 0.5 h. After incubation, the cells were washed with phosphate-buffered saline and fixed with 1% paraformaldehyde (Sigma). Unstained cells and cells labeled with fluorochrome-conjugated irrelevant rat immunoglobulin G (IgG) antibodies were used as controls and for adjustment of the flow cytometer (FACScan; Becton-Dickinson, San Jose, Calif.), and data were typically collected from 10,000 events.
RESULTS
Epitope prediction.
Peptide sequences binding to MHC class I of either the H-2d or H-2b haplotype were predicted by computer-based algorithms as potential CD8 epitopes. Fifty-three peptides (19 from the MOMP, 11 from Pomp5, 15 from Hsp60, and 8 from Omp2) with the highest expected binding capacities (highest prediction scores) were synthesized (20).
MHC-binding capacities of peptides.
The binding capacities of the synthetic peptides were evaluated by an MHC class I stabilization assay before the immunizations. The ability of peptides possessing a proposed H-2b motif to stabilize the dimeric complex of heavy chain and β2-microglobulin on the surface of the TAP-deficient cell line RMA-S was determined by flow cytometric analysis. Seventy-five percent of the synthetic H-2b peptides tested were able to bind to the RMA-S cell line in vitro. Peptides containing the H-2d-binding motif were used in immunizations without performing peptide-binding assays because a suitable TAP-deficient cell line for peptides that bind to H-2d class I molecules was not available.
Synthetic peptides induce a CTL response in vivo.
A total of 53 predicted CD8 epitopes from C. pneumoniae MOMP, Omp2, Pomp5, and Hsp60 were tested for the ability to induce a CTL response in vivo. This was achieved by subcutaneous immunization of mice with a mixture of peptides in adjuvant, in vitro restimulation of isolated splenocytes with the corresponding peptide, and cytotoxic response detection in a 51Cr release assay. Nineteen of the 53 peptides were shown to be capable of stimulating a CTL response following peptide immunizations. However, we were able to establish and maintain long-term peptide-specific CTL cultures against only seven of the peptides: 7087 (H-2Kb) and 7093 (H-2Db) from the MOMP, 8482 (H-2Dd) and 8511 (H-2Kd) from Omp2, 9165 (H-2Db) from Pomp5, and 7233 (H-2Dd), and 7239 (H-2Dd) from Hsp60 (Table 1). All of the H-2b peptides (7087, 7093, and 9165) that were capable of inducing long-term CTL lines in vivo were also able to bind class I molecules in the binding assay. The MHC binding of these peptides is shown in Table 1.
TABLE 1.
List of peptides that allowed generation of long-term CTL lines
| Protein | Peptide name | Sequence and position | Binding motif | Prediction scorea | Expt binding to MHC class Ib |
|---|---|---|---|---|---|
| MOMP | 7087 | 67 GDYVFDRI 74 | H-2Kb | 22 | +++ |
| MOMP | 7093 | 319 SLLGNATAL 327 | H-2Db | 27 | + |
| Pomp5 | 9165 | 283 QAVANGGAI 291 | H-2Db | 25 | +++ |
| Omp2 | 8482 | 59 RGAFCDKEF 67 | H-2Dd | 18 | ND |
| Omp2 | 8511 | 85 CYGRLYSVKV 94 | H-2Kd | 18 | ND |
| Hsp60 | 7233 | 7 KYNEEARKKI 16 | H-2Kd | 24 | ND |
| Hsp60 | 7239 | 32 GPKGRHVVI 40 | H-2Ld | 18 | ND |
| Ovalbuminc | 7548 | 257 SIINFEKL 264 | H-2Kb | 25 | +++ |
| β-Galactosidasec | 9339 | 876 TPHPARIGL 884 | H-2Ld | 25 | ND |
The binding capacity of the peptides was scored according to the presence of anchor amino acids and other amino acids frequently occurring in natural ligands (20).
Peptides possessing H-2b-binding anchor residues were tested for the ability to bind to class I molecules in vitro as described in Materials and Methods. ND, not determined; +++, strong stabilization of MHC assembly by peptide; +, stabilization of MHC assembly by peptide.
Peptide specificity of CTL lines.
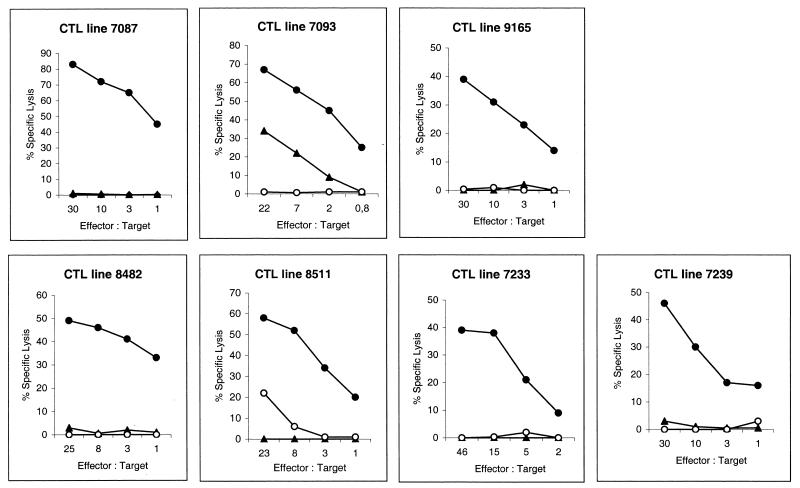
The specificity of the CTL lines was further studied by analyzing their in vitro responses to specific peptides. Figure 1 shows the percentage of specific lysis of the seven long-term CTL lines at effector/target (E/T) cell ratios of 30, 10, 3, and 1 against peptide-pulsed and unpulsed target cells. The lysis of target cells pulsed with a specific peptide was significantly higher (30 to 80%) than that of target cells pulsed with unspecific peptides (0 to 30%) or that of unpulsed targets (0 to 3%) at an E/T cell ratio of 30, indicating the specificity of CTL lines for a particular epitope. The lysis of peptide-pulsed target cells by control CTL lines (CTL lines specific for peptides from ovalbumin and β-galactosidase) was 30 to 60% at an E/T cell ratio of 30. The affinity of the CTL lines was assessed by titrating down the concentration of the peptide used to coat the target cells. All seven of the peptide-induced CTL lines were efficient at lysing target cells coated with picogram amounts of peptide, indicating that the affinities of these CTL lines were high (data not shown).
FIG. 1.
Peptide specificity of the seven long-term CTL lines. CTL lines were enriched from splenocytes of BALB/c (H-2d) and C57BL/6 (H-2b) mice that had been subcutaneously immunized with a mixture of TiterMax adjuvant and C. pneumoniae-derived peptide. The splenocytes were restimulated in vitro weekly with the corresponding peptide and irradiated autologous splenocytes. The cytotoxic activity of the CTL lines was tested with a 5-h 51Cr release assay on target cells alone (○), target cells coated with corresponding peptide (•), or target cells coated with another peptide specific for the same H-2 allele from the same protein (▴). P815 (for H-2d effectors) and RMA (for H-2b effectors) cells were used as target cells. The E/T cell ratio describes the number of CTLs (effectors) in comparison with the number of target cells in each well. Percent specific lysis was calculated as follows: % specific lysis = 100 × [(experimental release − spontaneous release)/(maximum release − spontaneous release)].
In addition to a cytotoxic response, significant proliferation was observed following culturing with the specific peptide, with PIs ranging from 3 to 98 (Table 2), while the CTL lines failed to proliferate following culturing with unspecific peptides (data not shown). Stimulation of the CTL lines with specific peptides also led to significant TNF-α and IFN-γ production (Table 2). CTL lines produced 230 to 11,000 pg of TNF-α per ml and 4,000 to 100,000 pg of IFN-γ per ml in the presence of the specific peptides (0.5 to 1 μg/ml), whereas undetectable amounts of TNF-α and IFN-γ were produced in the absence of the peptides.
TABLE 2.
Induction of proliferation and secretion of TNF-α and IFN-γ by peptide-specific CTL lines in response to stimulation with C. pneumoniae-derived peptides
| CTL line specific for peptide | PIa | TNF-α concn (pg/ml)b | IFN-γ concn (pg/ml) |
|---|---|---|---|
| 7087 (MOMP) | 3 | 230 | 11,000 |
| 7093 (MOMP) | 69 | 8,700 | 33,000 |
| 8482 (Omp2) | 52 | 1,400 | 8,000 |
| 8511 (Omp2) | 7 | 11,000 | 118,000 |
| 9165 (Pomp5) | 88 | 2,400 | 23,000 |
| 7233 (Hsp60) | 13 | 4,600 | 23,000 |
| 7239 (Hsp60) | 3 | 240 | 8,000 |
| 7548 (Ovalbumin) | 92 | 4,100 | 13,000 |
| 9339 (β-galactosidase) | 98 | 6,200 | 4,000 |
Three-day proliferation assays based on the incorporation of [3H]thymidine during the last 16 to 18 h were performed at an E/T cell ratio of 1 and a peptide concentration of 1 μg/ml. The PI was calculated as follows: PI = (peptide-induced proliferation − background proliferation)/background proliferation.
Secretion of cytokines was determined by ELISA in samples of cell culture supernatants taken from the proliferation assay wells 48 h after peptide stimulation.
Cytolysis is mediated by CD8+ T lymphocytes.
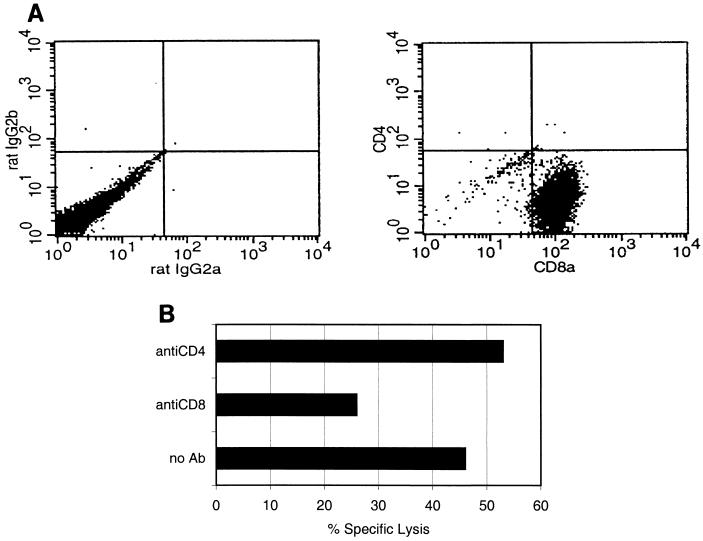
To identify the population of cells responsible for peptide-specific cytotoxicity, the phenotype of the long-term CTL lines was confirmed by direct immunostaining with fluorochrome-labeled anti-CD4 and anti-CD8 antibodies and flow cytometric analysis. After multiple rounds of restimulation, an enriched population of peptide-specific CTLs was obtained, >95% of all viable cells being CD8+ cells. A representative flow cytometric analysis of CTL line 8511 is shown in Fig. 2A. Similar results were obtained with the other six long-term CTL lines. To further confirm the phenotype of the effector cells, CD8- and CD4-expressing lymphocytes were neutralized from stimulated cultures with monoclonal antibodies specific for murine CD4 and CD8 molecules. The neutralized cells were tested for the ability to lyse peptide-pulsed target cells. Only neutralization of CD8+ effector cells reduced the amount of specific lysis of the target cells. In contrast, neutralization of CD4+ cells did not affect the cytotoxic activity, suggesting that cytolysis was due to CD8+ lymphocytes. Figure 2B shows CTL line 8511 neutralized for CD8 or CD4 molecules before the CTL assay. Reduction of cytotoxic activity after neutralization of CD8+ cells was also detected with the other six CTL lines (inhibition varied from 10 to 60%).
FIG. 2.
(A) Expression of CD4 and CD8 molecules on a CTL line specific for peptide 8511. After enrichment by Ficoll-Paque gradient centrifugation, the viable cells were stained with either FITC-conjugated anti-mouse CD8 and phycoerythrin-conjugated anti-mouse CD4 antibodies or with irrelevant anti-mouse antibodies (rat IgG2a and rat IgG2b). The labeled cells were fixed with 1% paraformaldehyde, and 10,000 events were subjected to flow cytometric analysis. Similar results (>95% of the cells were CD8+) were also obtained with the CTL lines specific for the other peptides. (B) Neutralization of CD8+ cells reduced the cytotoxic activity of CTL line 8511 compared to that of an untreated culture. CTL effector cells were preincubated with 20 μg of anti-CD4 or -CD8 monoclonal antibodies (Ab) for 45 min before the addition of 51Cr-labeled peptide-pulsed target cells. The E/T cell ratio was 40. A similar reduction of cytotoxic activity after neutralization of CD8+ cells was also detected with other CTL lines, inhibition varying from 10 to 60%.
CTL lines respond to infected target cells.
To test whether peptides recognized by our CTL lines are produced and presented through the natural class I processing pathway, we tested the ability of the CTL lines to respond to target cells infected with C. pneumoniae. In addition to cytotoxic activity, cytokine (TNF-α and IFN-γ) production was measured.
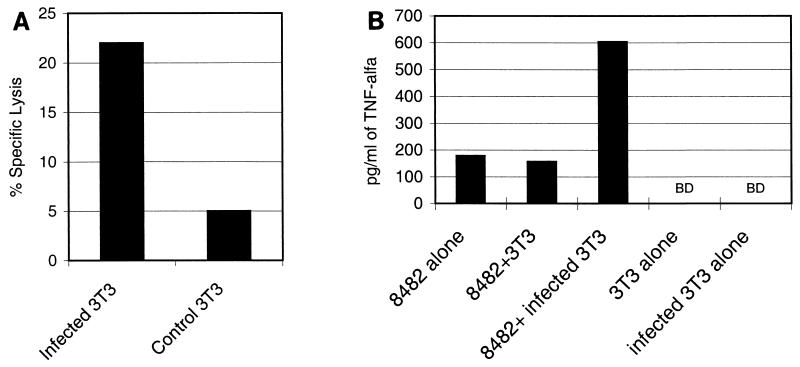
To test the CTL lines isolated from BALB/c mice (H-2d), BALB/3T3 cells were infected with C. pneumoniae. In these experiments, addition of cycloheximide was essential for detectable infection of the mouse cell lines. Without this eucaryotic protein inhibitor, no intracellular inclusions could be seen. Before using infected cells as target cells, cycloheximide was washed away and the ability of target cells to proliferate was confirmed by measuring [3H]thymidine incorporation (data not shown). In our experiments, 60 to 90% of the BALB/3T3 cells were infected (contained inclusions). Figure 3A shows a moderate cytotoxic response against BALB/3T3 cells infected with C. pneumoniae (multiplicity of infection of 5) by the CTL line specific for peptide 8482 from Omp2. No lysis was observed when the target cells were treated with equal numbers of UV-inactivated bacteria or without bacteria. CTL line 8482 also responded to infected BALB/3T3 cells with increased TNF-α production (Fig. 3B). Furthermore, CTL line 8511, which is specific for another Omp2-derived peptide, showed increased cytotoxicity but no TNF-α or IFN-γ production in response to infected target cells (Table 3). The other two H-2d CTL lines did not show any recognition of infected cells (Table 3).
FIG. 3.
(A) Lysis of C. pneumoniae-infected target cells by CTL line 8482 in a 10-h 51Cr release assay. The CTL line was stimulated with splenocytes of a mouse immunized with Omp2-derived peptide 8482. BALB/3T3 target cells were infected in vitro with C. pneumoniae and labeled with 51Cr. Uninfected control cells were treated similarly without addition of C. pneumoniae. An E/T cell ratio of 10 was used. (B) The TNF-α production of CTL line 8482 in the presence of infected BALB/3T3 cells was significantly higher than the background level or the response to uninfected target cells. We stimulated 3 × 105 CTLs with the same number of infected target cells (E/T cell ratio of 1) on 24-well plate for 48 h, after which the supernatants were collected and analyzed by ELISA. BD, below detection limit.
TABLE 3.
Summary of cytotoxic activity and TNF-α and IFN-γ secretion detected in seven peptide-specific CTL lines in response to C. pneumoniae-infected target cells
| CTL line specific for peptide | Infected target cella | % Specific lysis (uninfected target cells)b | Concn of TNF-α (pg/ml) (uninfected target cells)c | Concn of IFN-γ (pg/ml) (uninfected target cells)c |
|---|---|---|---|---|
| 7087 (MOMP) | 1,308.1 | 8 (0) | 38 (26) | 1,594 (171) |
| 7093 (MOMP) | 1,308.1 | 3 (8) | 36 (40) | 6,116 (4,861) |
| 9165 (Pomp5) | 1,308.1 | 0 (0) | ND | ND |
| 8482 (Omp2) | BALB/3T3 | 22 (5) | 604 (157) | 2,722 (2,298) |
| 8511 (Omp2) | BALB/3T3 | 20 (2) | 21 (18) | 2,141 (2,271) |
| 7233 (Hsp60) | BALB/3T3 | 10 (5) | 11 (9) | 150 (210) |
| 7239 (Hsp60) | BALB/3T3 | 14 (8) | 12 (12) | 180 (260) |
The infection frequency obtained with 1308 cells was 50 to 70%, whereas that obtained with BALB/3T3 cells was ∼70%.
Ten-hour chromium release assays were performed at an E/T ratio of 10. Lysis of in vitro-infected BALB/3T3 target cells was compared to lysis of uninfected targets.
Production of TNF-α and IFN-γ by the peptide-specific CTL lines stimulated for 48 h with infected or uninfected target cells at an E/T ratio of 1 was determined in cell culture supernatants by ELISA. ND, not determined.
For testing of CTL lines isolated from C57BL/6 (H-2b) mice, 1308.1 cells were infected with C. pneumoniae (multiplicity of infection of 10). As with BALB/3T3 cells, addition of cycloheximide was found to be essential for initiation of the infection but it was washed away before coculture with effector cells. The infection frequency of 1308.1 cells varied from 50 to 70%. None of the H-2b CTL lines tested were capable of specific lysis of infected targets in an overnight chromium release assay (Table 3). However, infected 1308.1 cells stimulated the CTL line specific for peptide 7087 from the MOMP to secrete more than six times the amount of IFN-γ secreted after stimulation by uninfected control cells (Table 3). When the infected target cells were sensitized with the specific peptide, all of the CTL lines tested killed them efficiently (data not shown). Moreover, control CTL lines of both the H-2d and H-2b haplotypes that recognized nonchlamydial peptides did not respond to C. pneumoniae-infected cells (data not shown).
DISCUSSION
The importance of CD8+ T cells for protection against infection by many intracellular bacteria is well recognized (23). CD8+ T cells control intracellular infections in two general ways, by direct lysis of infected cells and by secretion of cytokines such as IFN-γ and TNF-α (24). Identification of specific CD8 epitopes underlying this protection is important for several reasons. First, efficient methods for analyzing CD8+ T-cell responses after infection or immunization require prior knowledge of epitope specificity. Second, identification of pathogen-specific protective CD8 epitopes enables the development of novel and highly specific preventive and therapeutic approaches.
Here we report a strategy for the identification of C. pneumoniae-derived CD8 epitopes in which the known nucleotide sequence of C. pneumoniae proteins (http://chlamydia-www.berkeley.edu:4231) and known MHC class I peptide motifs (20) were used as tools. We screened the amino acid sequences of four C. pneumoniae genes (coding for the MOMP, Omp2, Hsp60, and Pomp5) for the presence of peptides (8 to 10 amino acids) with known MHC class I anchor residues. By using computer-based H-2d and H-2b class I molecules algorithms, we identified and synthesized a total of 53 peptides (33 H-2d and 20 H-2b). When the peptides were used to immunize mice, a peptide-specific CTL response was induced for 19 of the 53 peptides. We were able to establish long-term CTL lines against seven of these CD8 epitopes, and they were characterized further. Besides the observed cytolytic activity, activation of the CTL lines with a specific peptide led to proliferation and production of IFN-γ and TNF-α cytokines.
In order to determine if the identified CD8 epitopes were present on C. pneumoniae-infected cells, we analyzed the ability of in vitro-infected target cells to activate the peptide-specific CTL lines. Three of the seven CTL lines were activated in the presence of infected target cells. The activation was detected as different types of responses in different CTL lines. Cell line 8482 showed an increased cytotoxic response and increased TNF-α production, whereas cell line 8511 also showed an increased cytotoxic response but did not secrete TNF-α or IFN-γ. Furthermore, cell line 7087 showed increased IFN-γ production but no cytotoxic response was detected. The lack of a cytotoxic response in cell line 7087 may be due to the insensitivity of the 51Cr release assay since very low levels of specific lysis were observed against both infected and uninfected cells (8 and 0%, respectively). The TNF-α and IFN-γ response levels varied among the different CTL lines also when they were stimulated with target cells loaded with specific peptides. Thus, this variation may reflect phenotype or activation stage differences between the CTL lines.
Cytotoxic response and cytokine secretion levels in response to infected cells were much lower than those of cells pulsed with a specific peptide. There are several possible explanations for this. First, the target cell infection frequency was approximately 50 to 70% and this results, in practice, in lower numbers of target cells than in the epitope-pulsing technique. Second, the infected cells most likely present multiple epitopes from the infecting bacteria on the MHC class I molecules, thus reducing the effective density of the specific peptide recognized by this CTL line. Third, the fact that a eucaryotic protein inhibitor, cycloheximide, was required to initiate C. pneumoniae infection in the mouse cell lines may have resulted in lower MHC class I expression on the surface of the infected target cells. However, cycloheximide did not cause an irreversible protein synthesis block since proliferative activity was shown to be restored after cycloheximide removal. Finally, C. trachomatis infection has been shown to downregulate MHC class I and II expression (31, 32) through production of a chlamydial protease-like activity factor that degrades host transcription factors required for MHC expression (30). Because of the chlamydial protease-like activity factor sequence homology between the two Chlamydia strains, it is reasonable to assume that the same mechanism may be active in C. pneumoniae infection as well. Indeed, MHC class I downregulation was recently observed during C. pneumoniae infection of human monocytes (2). However, our observation that the infected target cells were efficiently presenting peptide provided from outside to CTL lines of corresponding specificity speaks against complete inhibition of MHC class I expression.
On the basis of these data, we concluded that from four C. pneumoniae antigens, we identified 19 immunogenic C. pneumoniae-derived CD8 epitopes. By using long-term CTL lines against seven CD8 epitopes, we were able to further identify three natural epitopes presented on infected target cells (8482 and 8511 from Omp2 and 7087 from MOMP). Identification of the CD8 epitopes enables us to proceed to studying their role in infection and protective immunity in vivo. Furthermore, the strategy described here for identification of H-2d- and H-2b-specific CD8 epitopes can be used for the identification of human CD8 epitopes with the help of HLA class I algorithms and transgenic mice expressing HLA class I molecules instead of H-2 molecules.
The results of this study could eventually contribute to the development of a vaccine against C. pneumoniae. Starting from the demonstration of the important role of CD8+ cells in acquired immunity (18), DNA vaccines expressing C. pneumoniae proteins have been tested for protection in mice (19, 25). The results have indeed shown moderate protection but to a degree less than that seen after infection. It has been reported that stronger epitope-specific CTL responses can be obtained by immunization with DNA minigenes containing CD8 epitopes than by immunization with DNA encoding the whole protein antigen (11). Furthermore, CD8 epitopes may be dominant epitopes, inducing a response upon immunization with whole antigens, or subdominant, inducing no response when a whole antigen is used for immunization (27). We have detected a CTL response against only one of the identified natural CD8 epitopes following DNA immunization with constructs encoding whole antigens, thus suggesting that the other two are indeed subdominant (our unpublished data). The presence of subdominant CD8 epitopes might, in part, explain the moderate level of protection obtained so far by DNA immunization and speak in favor of an epitope immunization strategy. Identification of specific CD8 epitopes will enable the development of multiepitope vaccines against C. pneumoniae infection, first to be tested in the mouse model with the appropriate mouse-specific epitopes and then to be extended to a cocktail of epitopes specific for the major HLA class I alleles.
Acknowledgments
We thank P. H. Mäkelä for advice and comments on the manuscript and Irene Viinikangas, Leena Erkkilä, and Anu Haveri for technical assistance.
This work was partially supported by the Biotechnology Program of the Commission of the European Union (contract BIO-CT96-0152), the Finnish Cultural Foundation, and Spectrum Medical Sciences (Helsinki, Finland).
Editor: J. D. Clements
REFERENCES
- 1.Campbell, L. A., M. Rosenfeld, and C.-C. Kuo. 2000. The role of Chlamydia pneumoniae in atherosclerosis—recent evidence from animal models. Trends Microbiol. 8:255-257. [DOI] [PubMed] [Google Scholar]
- 2.Caspar-Bauguil, S., B. Puissant, D. Nazzal, J.-C. Lefèvre, M. Thomsen, R. Salvayre, and H. Benoist. 2000. Chlamydia pneumoniae induces interleukin-10 production that down-regulates major histocompatibility complex class I expression. J. Infect. Dis. 182:1394-1401. [DOI] [PubMed] [Google Scholar]
- 3.Faas, S. J., J. L. Rothstein, B. L. Kreider, G. Rovera, and B. B. Knowles. 1993. Phenotypically diverse mouse thymic stromal cell lines which induce proliferation and differentiation of hematopoietic cells. Eur. J. Immunol. 23:1201-1214. [DOI] [PubMed] [Google Scholar]
- 4.Gavin, M. A., M. J. Gilbert, S. R. Riddell, P. D. Greenberg, and M. J. Bevan. 1993. Alkali hydrolysis of recombinant proteins allows for the rapid identification of class I MHC-restricted CTL epitopes. J. Immunol. 151:3971-3980. [PubMed] [Google Scholar]
- 5.Geng, Y., K. Berencsi, Z. Gyulai, T. Valyi-Nagy, E. Gonczol, and G. Trinchieri. 2000. Roles of interleukin-12 and gamma interferon in murine Chlamydia pneumoniae infection. Infect. Immun. 68:2245-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grayston, J. T., L. A. Campbell, C.-C. Kuo, C. H. Mordhorst, P. Saikku, D. H. Thom, and S.-P. Wang. 1990. A new respiratory tract pathogen: Chlamydia pneumoniae strain TWAR. J. Infect. Dis. 161:618-625. [DOI] [PubMed] [Google Scholar]
- 7.Gupta, S., E. W. Leatham, D. Carrington, M. A. Mendall, J. C. Kaski, and A. J. Camm. 1997. Elevated Chlamydia pneumoniae antibodies, cardiovascular events, and azithromycin in male survivors of myocardial infarction. Circulation 96:404-407. [DOI] [PubMed] [Google Scholar]
- 8.Gurfinkel, E., G. Bozovich, A. Daroca, E. Beck, B. Mautner, and The ROXIS Pilot Study Group. 1997. Randomised trial of roxithromycin in non-Q-wave coronary syndromes: ROXIS pilot study. Lancet 350:404-407. [DOI] [PubMed] [Google Scholar]
- 9.Halme, S., J. Latvala, R. Karttunen, I. Palatsi, P. Saikku, and H.-M. Surcel. 2000. Cell-mediated immune response during primary Chlamydia pneumoniae infection. Infect. Immun. 68:7156-7158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerschlag, M. R., K. Chirgwin, P. M. Roblin, M. Gelling, W. Dumornay, L. Mandel, P. Smith, and J. Schachter. 1992. Persistent infection with Chlamydia pneumoniae following acute respiratory illness. Clin. Infect. Dis. 14:178-182. [DOI] [PubMed] [Google Scholar]
- 11.Ishioka, G. Y., J. Fikes, G. Hermanson, B. Livingston, C. Crimi, M. Qin, M.-F. del Guercio, C. Oseroff, C. Dahlberg, J. Alexander, R. W. Chesnut, and A. Sette. 1999. Utilization of MHC class I transgenic mice for development of minigene DNA vaccines encoding multiple HLA-restricted CTL epitopes. J. Immunol. 162:3915-3925. [PubMed] [Google Scholar]
- 12.Kalman, S., W. Mitchell, R. Marathe, C. Lammel, J. Fan, R. W. Hyman, L. Olinger, J. Grimwood, R. W. Davis, and R. S. Stephens. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat. Genet. 21:385-389. [DOI] [PubMed] [Google Scholar]
- 13.Laitinen, K., A. L. Laurila, M. Leinonen, and P. Saikku. 1996. Reactivation of Chlamydia pneumoniae infection in mice by cortisone treatment. Infect. Immun. 65:4832-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malinverni, R., C.-C. Kuo, L. A. Campbell, and J. T. Grayston. 1995. Reactivation of Chlamydia pneumoniae lung infection in mice by cortisone. J. Infect. Dis. 172:593-594. [DOI] [PubMed] [Google Scholar]
- 15.Mehta, S. J., R. D. Miller, J. A. Ramirez, and J. T. Summersgill. 1998. Inhibition of Chlamydia pneumoniae replication in HEp-2 cells by interferon-γ: role of tryptophan catabolism. J. Infect. Dis. 177:1326-1331. [DOI] [PubMed] [Google Scholar]
- 16.Orfila, J. J. 1998. Seroepidemiological evidence for an association between Chlamydia pneumoniae and atherosclerosis. Atherosclerosis 140:S11-S15. [DOI] [PubMed] [Google Scholar]
- 17.Penttilä, J. M., M. Anttila, M. Puolakkainen, A. Laurila, K. Varkila, M. Sarvas, P. H. Mäkelä, and N. Rautonen. 1998. Local immune responses to Chlamydia pneumoniae in the lungs of BALB/c mice during primary infection and reinfection. Infect. Immun. 66:5113-5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Penttilä, J. M., M. Anttila, K. Varkila, M. Puolakkainen, M. Sarvas, P. H. Mäkelä, and N. Rautonen. 1999. Depletion of CD8+ cells abolishes memory in acquired immunity against Chlamydia pneumoniae in BALB/c mice. Immunology 97:490-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Penttilä, T., J. M. Vuola, V. Puurula, M. Anttila, M. Sarvas, N. Rautonen, P. H. Mäkelä, and M. Puolakkainen. 2000. Immunity to Chlamydia pneumoniae induced by vaccination with DNA vectors expressing a cytoplasmic protein (Hsp60) or outer membrane proteins (MOMP and Omp2). Vaccine 19:1256-1265. [DOI] [PubMed] [Google Scholar]
- 20.Rammensee, H.-G., J. Bachmann, N. P. N. Emmerich, O. A. Bachor, and S. Stevanovic. 1999. SYFPEITHI: database for MHC ligands and peptide motifs. Immunogenetics 50:213-219. [DOI] [PubMed] [Google Scholar]
- 21.Rottenberg, M. E., A. C. Gigliotti Rothfuchs, D. Gigliotti, C. Svanholm, L. Bandholtz, and H. Wigzell. 1999. Role of innate and adaptive immunity in the outcome of primary infection with Chlamydia pneumoniae, as analyzed in genetically modified mice. J. Immunol. 162:2829-2836. [PubMed] [Google Scholar]
- 22.Rotzschke, O., K. Falk, S. Stevanovic, G. Jung, P. Walden, and H.-G. Rammensee. 1991. Exact prediction of a natural T cell epitope. Eur. J. Immunol. 21:2891-2894. [DOI] [PubMed] [Google Scholar]
- 23.Schaible, U. E., H. L. Collins, and S. H. E. Kaufmann. 1999. Confrontation between intracellular bacteria and the immune system. Adv. Immunol. 71:267-377. [DOI] [PubMed] [Google Scholar]
- 24.Slifka, M. K., and J. L. Whitton. 2000. Antigen-specific regulation of T cell-mediated cytokine production. Immunity 12:451-457. [DOI] [PubMed] [Google Scholar]
- 25.Svanholm, C., L. Bandholtz, E. Castaños-Velez, H. Wigzell, and M. E. Rottenberg. 2000. Protective DNA immunization against Chlamydia pneumoniae. Scand. J. Immunol. 51:345-353. [DOI] [PubMed] [Google Scholar]
- 26.Taylor-Robinson, D., and B. J. Thomas. 1998. Chlamydia pneumoniae in arteries: the facts, their interpretation, and future studies. J. Clin. Pathol. 51:793-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vitiello, A., A. Sette, L. Yuan, P. Farness, S. Southwood, J. Sidney, R. W. Chesnut, H. M. Grey, and B. Livingston. 1997. Comparison of cytotoxic T lymphocyte responses induced by peptide or DNA immunization: implications on immunogenicity and immunodominance. Eur. J. Immunol. 27:671-678. [DOI] [PubMed] [Google Scholar]
- 28.Vuola, J. M., V. Puurula, M. Anttila, P. H. Mäkelä, and N. Rautonen. 2000. Acquired immunity to Chlamydia pneumoniae is dependent on gamma interferon in two mouse strains that initially differ in this respect after primary challenge. Infect. Immun. 68:960-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ward, M. E. 1995. The immunobiology and immunopathology of chlamydial infections. APMIS 103:769-796. [DOI] [PubMed] [Google Scholar]
- 30.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon γ-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 189:1931-1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon γ-inducible major histocompatibility complex class I expression in Chlamydia-infected cells. J. Exp. Med. 191:1525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]