Abstract
Selective capture of transcribed sequences (SCOTS) has been employed to identify 54 cDNA molecules that represent 46 genes that are expressed by Mycobacterium avium during growth in human macrophages. Some cDNA molecules correspond to genes that are apparently expressed 48 h after infection of macrophages, while others correspond to genes expressed 110 h after infection, and still others correspond to genes expressed throughout the course of infection in our model system. Genes expressed by M. avium during growth in macrophages include genes encoding enzymes of several biosynthetic pathways (pyrimidines, mycobactin, and polyketides); genes that encode enzymes involved in intermediary metabolism, energy metabolism (tricarboxylic acid cycle, glyoxalate shunt), and nitrogen metabolism; and genes that encode regulatory proteins. A number of genes of unknown function were also identified, including genes that code for proteins similar to members of the PPE family of proteins of Mycobacterium tuberculosis and proteins similar to those encoded by the M. tuberculosis mce genes, which have been previously associated with mycobacterial virulence. The SCOTS technique, followed by enrichment for cDNA molecules that are up-regulated or are uniquely expressed by M. avium during growth in human macrophages (compared to growth in laboratory broth culture), allows recovery and identification of a greater diversity of cDNA molecules than does subtractive hybridization between cDNA mixtures from macrophage-grown and broth-grown M. avium. Data are presented demonstrating the reproducibility of recovery of a subset of cDNA molecules from cDNA mixtures purified by SCOTS on several different occasions. These results further demonstrate the beneficial utility of the SCOTS technique for identifying genes whose products are needed for successful survival and growth by an organism in a specific environment.
Mycobacterium avium is a ubiquitous, saprophytic mycobacterium commonly found in soil and water (13, 20, 27). Prior to the AIDS epidemic, M. avium was rarely identified as a pathogen in humans. However, during the early years of the AIDS epidemic, M. avium became one of the most significant opportunistic pathogens of severely immunocompromised human immunodeficiency virus (HIV)-infected individuals (those whose CD4+ T-cell counts were ≤100/ml of blood; 11, 26, 29). Implementation of highly active antiretroviral therapy (HAART) in the treatment of HIV-infected individuals has significantly improved the degree of immunocompetency of AIDS patients and has resulted in a concomitant decrease in the incidence of opportunistic infections in these individuals (1, 30, 38). However, newly diagnosed AIDS patients, those for whom HAART has been ineffective, and those who are unable to afford HAART remain at risk for opportunistic infections. Thus, we must not become complacent in thinking that M. avium is no longer a significant pathogen. Moreover, increasing numbers of non-HIV-infected individuals are being diagnosed with M. avium infections of the lung (14, 28).
Like other pathogenic mycobacteria (e.g., Mycobacterium tuberculosis and Mycobacterium leprae), M. avium is phagocytosed by macrophages following entry into the human body (5, 41, 42, 44). Uptake of pathogenic mycobacteria occurs via the complement receptors CR3b and CR1 (41, 42), the mannosyl-fucosyl receptor (5), the fibronectin receptor (5), and/or the αvβ3 integrin receptor (40, 44). The phagosomal compartments of nonactivated macrophages containing M. avium do not mature to phagolysosomes but retain the cellular markers associated with early endosomes (10, 16, 18, 21, 22, 43). Thus, M. avium, like M. tuberculosis, is not destroyed by nonactivated macrophages but is able to survive and multiply within the phagosomes of these cells. The mechanism(s) whereby pathogenic mycobacteria are able to preclude phagosomal maturation is not well understood, but the ability to survive and multiply within macrophages is a key component of mycobacterial pathogenesis.
Several years ago, we reasoned that identification of genes that are up-regulated for expression or that are uniquely expressed by mycobacteria when they are growing in macrophages compared to when they are growing in broth would enable us to gain insight into gene products that are important for mycobacterial survival and growth within macrophages. To identify such genes, we initially developed a cDNA subtractive hybridization technique and were successful in identifying a gene (mig) that appeared to be uniquely expressed by M. avium during growth in macrophages (39). More recently, we have developed an improved technique, called selective capture of transcribed sequences (SCOTS), which has enabled us to identify a number of genes that are up-regulated for expression or are only expressed by M. tuberculosis during growth in human macrophages (23). Here we describe studies comparing the cDNA subtractive hybridization (i.e., removal of unwanted cDNA molecules) and SCOTS (i.e., selective capture of desired cDNA molecules) techniques and report the identification of a number of genes expressed by M. avium at different times after infection of human macrophages.
MATERIALS AND METHODS
Bacterial strains.
M. avium strain 5-8, a serovar 4 strain originally isolated from an AIDS patient, was kindly provided by Anna Tsang (39).
Escherichia coli K-12 DH5α (35) was used as the host strain for construction and maintenance of all cDNA libraries prepared in the plasmid vector pBluescript II SK+ (Stratagene, Inc., La Jolla, Calif.).
E. coli LE 392 (35) was the host strain for all of the individual cosmid clones used in the experiments. Construction of the M. avium cosmid library was described by Plum and Clark-Curtiss (39).
Media.
M. avium was grown in Middlebrook 7H9 broth (Difco) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC; obtained from Microbios [Phoenix, Ariz.] or Remel, Inc. [Lenexa, Kans.]) and 0.05% Tween 80 (Sigma, St. Louis, Mo.).
E. coli strains were grown in Lennox broth (32), in Superbroth (33), or on Antibiotic Medium 2 agar (Difco). Superbroth consists of 32 g of Bacto-Tryptone (Difco), 20 g of yeast extract (Difco), and 5 g of NaCl per liter of water, with the pH adjusted to 7.0 by addition of 1 M NaOH (33).
Human macrophages were cultured in RPMI 1640 medium (Gibco BRL, Gaithersburg, Md.) supplemented with 4 mM l-glutamic acid, 25 mM HEPES, and 1% modified Eagle’s medium containing nonessential amino acids (supplemented RPMI 1640 medium). Autologous human serum was added to supplemented RPMI 1640 medium to a final concentration of 1 to 20% as indicated at different steps of the protocols described below and as previously described (23).
Cultivation and infection of macrophages.
Peripheral blood was collected from healthy, tuberculin skin test-negative volunteers, and mononuclear cells were isolated by passage through Ficoll gradients (Amersham Pharmacia Biotech, Piscataway, N.J.) in accordance with the manufacturer's instructions. Monocytes were cultivated in Teflon wells in supplemented RPMI 1640 medium plus 20% autologous serum and incubated at 37°C in 5% CO2 for 5 days to allow adherence. The adherent monocytes were removed from the Teflon wells, suspended in supplemented RPMI 1640 medium plus 20% autologous serum, introduced into 25-cm2 tissue culture flasks, and incubated at 37°C in 5% CO2 for 7 days to allow further maturation of the macrophages, as described previously (23). Mature, adherent macrophages were infected at a multiplicity of 0.5 to 1 bacterium per macrophage with a predominantly single-cell suspension of M. avium (10) that had been grown in Middlebrook 7H9 broth to mid-logarithmic phase. Infections were carried out by incubating macrophage monolayers (2 × 106 cells per 25-cm2 flask) and bacteria for 2 h at 37°C with 5% CO2 in supplemented RPMI 1640 medium with 10% autologous serum (23). After infection, the nonphagocytosed bacteria were removed by washing the monolayers with RPMI 1640 medium three times. Approximately 30% of the macrophages were infected, as determined by microscopic examination. The infected monolayers were maintained at 37°C with 5% CO2 in supplemented RPMI 1640 medium plus 1% autologous serum. The medium was changed at 16 h postinfection as described previously (23). The macrophage monolayers remained intact and adherent until approximately 96 h after infection. At that point, some of the macrophages began to detach from the surface of the flask and by 120 h after infection, approximately half of the macrophages had become detached. Infection experiments were not continued beyond 120 h. Of the intact, adherent macrophages, 40% were infected with M. avium. The number of bacilli per macrophage varied from 2 to 3 to more than 25. The infected monolayers were washed three times with RPMI 1640 medium just prior to harvesting to remove the nonadherent macrophages. Infected macrophages were harvested at 48, 110, or 120 h after infection.
The generation time of M. avium growing within macrophages, determined by CFU counting and microscopic enumeration of acid-fast bacilli (23), was approximately 20 h.
Isolation of nucleic acids.
Chromosomal DNA was isolated from broth-grown M. avium as described previously (9), except that the bacilli were disrupted in a Mini-Bead Beater (Bio-Spec, Inc., Bartlesville, Okla.), in tubes containing 0.1-mm silica-zirconium beads. Carbohydrates associated with M. avium chromosomal DNA were removed by mixing the nucleic acid solution with cetyltrimethylammonium bromide, followed by organic extraction and ethanol precipitation (45).
Total RNA from broth-grown bacteria or from M. avium-infected macrophages was isolated by mechanical disruption and organic extraction with hot guanidinium-thiocyanate-phenol-chloroform as described previously (8, 23).
cDNA synthesis.
Total RNA isolated from infected macrophages or from broth-grown M. avium was treated with DNase I (Ambion, Inc., Austin, Tex.) at 37°C for 30 min and converted to first-strand cDNA by random priming with Moloney murine leukemia virus reverse transcriptase (Superscript II; Gibco BRL) in accordance with the manufacturer's instructions. As described previously (23), primers with defined 5′ sequences and random nonamers at the 3′ ends were used for both first- and second-strand syntheses. Different terminal sequences were added to cDNA mixtures from broth-grown M. avium (PCR-K-9 primer: 5′-GACACTCTCGAGACATCA CCGG-3′), to cDNA mixtures from M. avium grown for 48 h in macrophages (Xba-PCR primer: 5′-TGCTCTAGACGTCCTGATGGTT-3′), and to cDNA mixtures from M. avium grown for 110 h in macrophages (Sal-PCR primer: 5′-ATATGTCGACTGAATTCCGTAGG-3′).
cDNA mixtures were amplified by PCR with 25 cycles of amplification (95°C for 30 s, 57°C [for cDNA from broth-grown M. avium] or 49°C [for macrophage-grown M. avium] for 90 s, and 72°C for 50 s). A portion of each cDNA mixture was frozen at −70°C as the stock cDNA mixture for each growth condition.
cDNA subtractive hybridization.
The procedure used for cDNA subtractive hybridization has been described in detail by Plum and Clark-Curtiss (39), except that rRNA was not removed from the total RNA preparation prior to conversion to cDNA and macrophages were grown differently. In these experiments, peripheral blood mononuclear cell (PBMC)-derived macrophages were infected as described above whereas Plum and Clark-Curtiss infected adherent macrophages 5 days after separation of monocytes from peripheral blood (39). Five days after infection, the infected macrophages were removed from the tissue culture flasks and lysed and the M. avium bacilli were recovered as described previously (39). Total bacterial RNA was isolated and converted to first-strand cDNA by random priming and addition of specific primers, as described above, with Superscript II reverse transcriptase and then to double-stranded cDNA (cDNA mixture 1 consisted of an unsubtracted cDNA mixture from macrophage-grown M. avium [Table 1]). cDNA prepared from M. avium grown in Middlebrook 7H9 broth-OADC-0.05% Tween 80 was denatured and biotinylated as described previously (39). The denatured, biotinylated cDNA mixture from broth-grown M. avium was mixed with cDNA from macrophage-grown M. avium at a 10:1 ratio, and three rounds of subtractive hybridization were carried out as described previously (39). Thus, cDNA molecules corresponding to genes expressed by M. avium under both growth conditions hybridized to one another and were removed from the mixture by reaction of the hybrids with streptavidin-coated magnetic beads as described previously (39). Samples of the nonhybridized cDNA mixtures (corresponding to genes that are up-regulated or are uniquely expressed by M. avium during growth in macrophages) obtained after each round of subtraction were radioactively labeled and used as probes in dot blot hybridization experiments with individual M. avium cosmid clones as described below.
TABLE 1.
cDNA Mixtures used in experiments
| cDNA mixture(s) | Description | Method of preparation |
|---|---|---|
| 1 | Unsubtracted cDNA mixture from macrophage-grown M. avium | Macrophages lysed 120 h after infection with M. avium; RNA isolated from bacteria separated from macrophages and then converted to cDNA |
| 2 | cDNA mixture from broth-grown M. avium | M. avium grown to mid-log phase in Middlebrook 7H9 broth-OADC-Tween 80; RNA isolated and converted to cDNA |
| 3 | Threefold-subtracted cDNA mixture | cDNA mixture 1 subjected to 3 rounds of subtraction with biotinylated cDNA mixture 2 |
| 4-1, 4-2, 4-3 | SCOTS-derived cDNA mixtures from broth-grown M. avium | cDNA mixtures prepared from M. avium grown in Middlebrook 7H9 broth-OADC-Tween 80 and subjected to 3 rounds of SCOTS on 3 separate occasions, 0 (4-1), 6 (4-2), and 11 (4-3) mo after original cDNA stock mixture was prepared |
| 5-1, 5-2, 5-3 | SCOTS-derived cDNA mixtures from M. avium grown for 48 h in human macrophages | cDNA mixtures prepared from macrophages infected for 48 h with M. avium; M. avium cDNA mixtures obtained by 3 rounds of SCOTS on 3 separate occasions, 0 (5-1), 6 (5-2), and 11 (5-3) mo after original stock cDNA mixture was prepared |
| 6-1, 6-2, 6-3 | SCOTS-derived cDNA mixtures from M. avium grown for 110 h in human macrophages | cDNA mixtures prepared from macrophages infected for 110 h with M. avium; M. avium cDNA mixtures obtained by 3 rounds of SCOTS on 3 separate occasions, 0 (6-1), 6 (6-2), and 11 (6-3) mo after original stock cDNA mixture was prepared |
| 7 | Enriched cDNA mixture from M. avium grown for 48 h in human macrophages | SCOTS-derived cDNA mixture from M. avium grown for 48 h in macrophages subjected to 3 rounds of enrichment for cDNA molecules corresponding to genes up-regulated for expression or uniquely expressed by M. avium during growth in macrophages; enrichment accomplished through competitive hybridization of cDNA mixtures 4-1 and 5-1 with biotinylated M. avium chromosomal DNA, followed by selective amplification of cDNA molecules from macrophage-grown M. avium |
| 8 | Enriched cDNA mixture from M. avium grown for 110 h in human macrophages | Same procedure as for cDNA mixture 7, except with cDNA mixture 6-1 and 4 rounds of enrichment |
SCOTS.
The SCOTS procedure used is a slightly modified form of that described by Graham and Clark-Curtiss (23). Chromosomal DNA from M. avium 5-8 was biotinylated as follows. Photoactivatable Biotin Acetate (PAB acetate; Clontech, Palo Alto, Calif.) was dissolved in sterile distilled water to a final concentration of 1 μg/μl. Twenty microliters of M. avium chromosomal DNA (at a concentration of 0.6 μg/μl in sterile distilled water) was added to a 0.6-ml thin-walled PCR tube, and an equal volume of PAB acetate was added. After the contents of the tube were mixed, the tube was left open and placed in crushed ice so that the tube was 2 to 3 cm below a 250-W incandescent light bulb. The mixture was photoactivated for 30 min while being kept on ice. A volume of fresh PAB acetate solution equal to the first volume of PAB acetate was added to the contents of the tube, and an additional 30-min photoactivation (with the tube kept in ice) was done to ensure biotinylation of the chromosomal DNA. The reaction mixture was then diluted fourfold with 10 mM Tris-1 mM EDTA (pH 9.0) buffer. The biotinylated DNA was removed from unincorporated PAB acetate by extraction with 2-butanol (three times), followed by ethanol precipitation of the DNA.
The biotinylated M. avium chromosomal DNA (12 μg) was mixed with 100 μg of pYA1403 plasmid DNA (M. avium rrnA DNA cloned into pBluescript II SK+). The mixture was sonicated for 10 s at an output setting of 4 with a W-380 Sonicator using a microprobe (Heat Systems-Ultrasonic, Inc., Farmingdale, N.Y.). After sonication, the mixture was precipitated and resuspended in 160 μl of 10 mM N-(2-hydroxyethyl)piperazine-N′-(3-propanesulfonic acid) (EPPS)-1 mM EDTA. The mixture was then divided into 20 samples of 8 μl each. Prepared in this way, the mixture contains sufficient biotinylated chromosomal DNA and rrnA DNA for three rounds of SCOTS purification and three rounds of enrichment for specific cDNA sequences (as described below).
For each round of SCOTS, an 8-μl sample of the mixture (containing 0.6 μg of M. avium chromosomal DNA and 5 μg of rrnA DNA) was denatured by incubation at 98°C for 3 min under mineral oil. Two microliters of 1 M NaCl was added to the mixture, which was then incubated at 77°C for 30 min. This step allowed the plasmid rrnA DNA to hybridize to the rrnA sites on the M. avium chromosomal DNA, thereby rendering these sites unavailable for hybridization with ribosomal DNA present in the cDNA mixtures.
In separate reaction mixtures, total amplified cDNA from either infected macrophages or from broth-grown bacteria in 8 μl of 10 mM EPPS-1 mM EDTA was boiled for 3 min, followed by the addition of 2 μl of 1 M NaCl. The denatured cDNA mixture was added to the biotinylated chromosomal DNA-rrnA prehybridized mixture, and hybridization continued at 77°C for 20 to 24 h. One hundred twenty micrograms of streptavidin-coated magnetic beads (Dynal M280) was washed in accordance with the manufacturer's instructions. The bacterial cDNA-chromosomal DNA hybrids were removed from the hybridization mixture by binding to 120 μg of streptavidin-coated magnetic beads in a solution of 1 ml of 1 M NaCl, 5 mM EPPS, and 0.5 mM EDTA. After the beads were washed three times with 20 mM NaCl-0.5% sodium dodecyl sulfate (SDS) (once at room temperature and twice at 65°C), the cDNA molecules were eluted from the chromosomal DNA bound to the beads with 100 μl of 0.25 M NaOH-0.1 M NaCl. The mixture was neutralized by the addition of 20 μl of 1 M Tris, pH 7.4. The cDNA molecules were precipitated with ethanol in the presence of 10 μg of glycogen (Roche), which served as a carrier, and were then PCR amplified as described above.
For each growth condition, in the first round of SCOTS, 10 separate samples of the cDNA mixtures were captured by hybridization to biotinylated, rRNA gene (rDNA)-blocked chromosomal DNA in parallel reactions. This was done to enhance the likelihood of recovering cDNA molecules corresponding to the full complement of transcripts present at the time of RNA preparation for each growth condition. The 10 amplified cDNA preparations were then combined (for each growth condition), denatured, and again hybridized to fresh aliquots of rDNA-blocked, biotinylated chromosomal DNA. At least three rounds of SCOTS were done with cDNA mixtures from each growth condition. Table 1 provides a brief description of each of the SCOTS-derived cDNA mixtures.
Enrichment for cDNA molecules from macrophage-grown M. avium and preparation of cDNA libraries.
To identify cDNA molecules that represent transcripts from genes that may be up-regulated for expression during growth of M. avium in macrophages, an additional step was included in the experiments (23). Preparations of cDNA mixtures from M. avium grown in macrophages that had been obtained by three rounds of SCOTS were added to biotinylated chromosomal DNA that had been prehybridized with both rDNA and a cDNA preparation from broth-grown M. avium (also obtained by three rounds of SCOTS). Hybridization proceeded for 24 h at 77°C, and the hybridized molecules were recovered by binding to streptavidin-coated beads as described above. Following elution of the cDNA molecules from the bound chromosomal DNA, the cDNA molecules derived from macrophage-grown M. avium were recovered by PCR amplification with primers specific for the macrophage-grown M. avium cDNA preparations. After three rounds of this enrichment procedure, the cDNA molecules from M. avium grown for 48 h in macrophages were cloned into the XbaI site of pBluescript II SK+ and the cDNA molecules from M. avium grown for 110 h in macrophages were cloned into the SalI site of pBluescript II SK+ to generate cDNA libraries. Table 1 provides a brief description of the SCOTS-derived and enriched cDNA mixtures.
Analysis of individual cDNA clones.
Individual cDNA clones were randomly chosen from the libraries prepared from cDNA mixtures from M. avium grown for 48 h in macrophages, from M. avium grown for 110 h in macrophages, and from broth-grown M. avium. The cDNA clones were used in Southern hybridizations as described below. The inserts of some cDNA clones from each library were PCR amplified and sequenced by using ABI Prism Big Dye primer cycling sequencing kits in accordance with the manufacturer's (PE Applied Biosystems, Branchburg, N.J.) instructions.
Database searches and DNA and protein similarity comparisons were carried out by using the BLAST algorithm from the National Center for Biotechnology Information at the National Library of Medicine. Databases included the M. tuberculosis H37Rv genome sequence (Sanger Centre, Hinxton, United Kingdom) and the M. avium genome sequence (The Institute for Genomic Research, Rockville, Md.) through the website at http://www.tigr.org.
Preparation of cDNA probes.
Samples of cDNA mixtures were prepared for use as probes in hybridization experiments. Radioactively labeled cDNA probes were prepared by random priming with incorporation of [32P]dCTP (New England Nuclear Corp., Boston, Mass.) with Hexanucleotide Mix Labeling kits from Roche Molecular Biochemicals (Indianapolis, Ind.) in accordance with the manufacturer's instructions. Digoxigenin-labeled probe mixtures were prepared with PCR DIG Probe Synthesis kits (Roche Molecular Biochemicals) to incorporate digoxigenin-11-dUTP during PCR amplification of the probe mixture in accordance with the manufacturer's instructions.
Dot blot hybridization to individual cosmid clones.
Three hundred sixty-eight individual cosmid clones from the pYA3060::M. avium library in E. coli LE392 (39) were each inoculated into 200 μl of Superbroth-60 μg of ampicillin/ml (final concentration) in 96-well microtiter plates. After overnight growth at 37°C, samples from each well were stamped onto sterile Nytran filters (8 by 11 cm; Schleicher & Schuell, Keene, N.H.) overlaid on petri dishes (150 by 50 mm) containing Antibiotic Medium 2 agar (Difco)-60 μg of ampicillin/ml (final concentration) with a Sigma Array Replica Plater (8 by 6 cm; Sigma). The agar plates and filters were incubated for approximately 9 h (until discrete spots of bacterial growth were visible on all of the filters). The bacteria were disrupted, and the DNA was denatured and fixed to the filters by standard procedures (35).
After the filters were processed and dried, they were prehybridized with 50 μg of denatured salmon sperm DNA per ml in 1 M NaCl-1% SDS-10% dextran sulfate at 80°C for 1.5 h. Radioactively labeled cDNA probe mixtures were denatured and added to the hybridization bottles containing the filters and the hybridization buffer. Hybridization continued at 80°C for approximately 24 h. The filters were washed briefly with 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at room temperature and then twice with 1× SSC-0.1% SDS for 20 min each time; these washes were done at 80°C. A final brief rinse with 0.1× SSC at room temperature completed the washing process. The filters were wrapped in clear plastic wrap and placed on a PhosphorImager (Molecular Dynamics, Houston, Tex.) screen overnight at room temperature. Successful hybridization was detected by PhosphorImager analysis (23).
Southern hybridization experiments.
Inserts of individual cDNA clones were amplified by PCR with primers specific for the terminal sequences and cloning sites (i.e., XbaI for 48-h cDNA, SalI for 110-h cDNA) of pBluescript II SK+ into which the cDNA molecules had been cloned. The amplified inserts were run on 1% agarose gels and then transferred to GeneScreen Plus filters (NEN Life Science Products, Boston, Mass.) in accordance with the manufacturer's instructions. When 32P-labeled cDNA mixtures were used as probes, hybridizations were done with 50 μg of denatured salmon sperm DNA per ml in 1 M NaCl-1% SDS-10% dextran sulfate at 80°C for 24 h.
When digoxigenin-labeled cDNA mixtures were used as probes, hybridizations were carried out in 5× SSC-0.1% (wt/vol) N-lauroyl sarcosine-0.02% SDS-50% formamide-2% Blocking Reagent (Roche Molecular Biochemicals) at 45°C for 24 h. Filters were processed in accordance with the manufacturer's instructions for hybridizations with digoxigenin-labeled probes (Roche Molecular Biochemicals). Detection of successful hybridization was done by detecting chemiluminescence by exposure to X-ray film as recommended by the manufacturer (Roche Molecular Biochemicals).
RESULTS
Reduction of the amount of rRNA cDNA in cDNA mixtures.
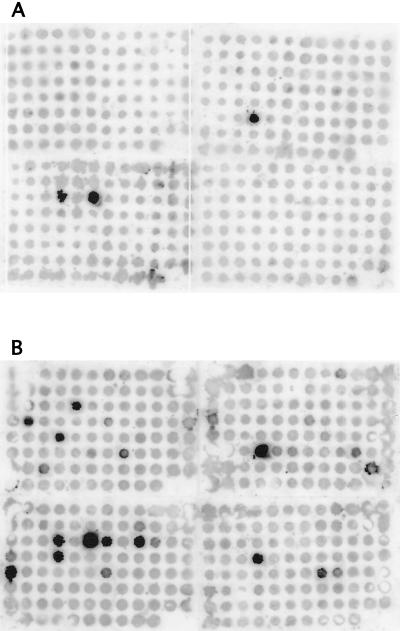
Figure 1 illustrates the importance of reducing the amount of cDNA corresponding to rRNA in cDNA mixtures. Cosmid DNA from 368 individual clones from the pYA3060::M. avium library (39) was blotted onto filters and hybridized with cDNA probe mixtures prepared from M. avium grown for 5 days (120 h) in human macrophages. For the hybridization in Fig. 1A, the probe was from the cDNA mixture before any subtractive hybridization had been done (cDNA mixture 1 [Table 1]). The unsubtracted probe mixture hybridized strongly with three cosmids (120, 122, and 248), each of which possesses the rRNA operon (previously determined by hybridization of the cloned M. avium rRNA operon as a probe with this set of cosmid clones; data not shown). Hybridization signals between the unsubtracted cDNA probe mixture and all of the other cosmid DNA dots were uniform and low in intensity. Figure 1B depicts the same set of cosmid clones hybridized with a probe prepared from the cDNA mixture from macrophage-grown M. avium after three rounds of subtractive hybridization with the cDNA mixture from broth-grown M. avium (cDNA mixture 3 [Table 1]). Strong hybridization signals were detected between the threefold-subtracted cDNA probe mixture and eight cosmids in addition to the cosmids with the rrnA operon, and moderately strong signals were detected between the cDNA probe mixture and five other cosmids.
FIG. 1.
Dot blot hybridizations between 368 individual M. avium cosmid clones and cDNA mixtures prepared from M. avium grown for 5 days in human macrophages. Hybridization and wash conditions are described in Materials and Methods. In panel A, the cDNA probe was prepared from an unsubtracted M. avium cDNA mixture (cDNA mixture 1 [Table 1]). In panel B, the probe was prepared from the macrophage-grown M. avium cDNA that had been subjected to three rounds of subtractive hybridization with a cDNA mixture prepared from M. avium grown to mid-log phase in Middlebrook 7H9 broth-OADC-0.05% Tween 80 to give cDNA mixture 3 (described in Table 1).
We hypothesize that, in the unsubtracted cDNA mixture, in which cDNA corresponding to rRNA comprises more than 90% of the mixture, the amount of isotope incorporated into cDNA molecules representing individual mRNAs is below or just at the level of detection. Subtractive hybridization between cDNA mixtures prepared from broth-grown and macrophage-grown M. avium allowed hybridization between the abundant rRNA cDNA molecules and removal of these hybrids from the cDNA mixture from macrophage-grown bacilli as a consequence of the binding of the biotinylated hybrids with streptavidin-coated magnetic beads. Each round of subtraction removed additional copies of the rRNA cDNA, thereby allowing a larger number of the cDNA molecules derived from mRNA to be labeled to levels at which hybridization signals could be detected.
Comparison of the cDNA subtractive hybridization technique and SCOTS plus enrichment.
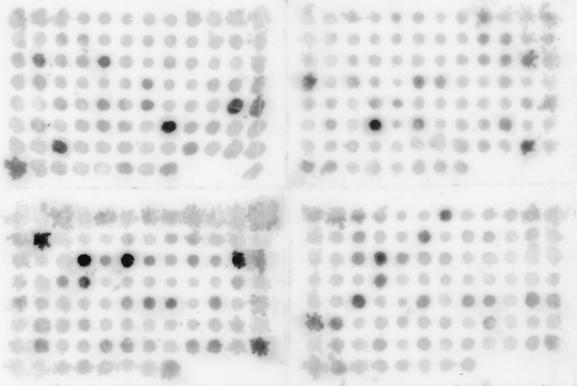
Figure 2 depicts a representative dot blot experiment involving hybridization between the same cosmid clones as in Fig. 1 and a cDNA probe mixture prepared by the SCOTS technique, followed by four rounds of enrichment for cDNA molecules representing genes that were expressed when M. avium had grown for 110 h in human macrophages (cDNA mixture 8, described in Table 1). Comparison of Fig. 2 to Fig. 1B indicates that the SCOTS-prepared, enriched cDNA probe hybridized to 12 of the 13 cosmids to which the subtractive hybridization-prepared cDNA probe hybridized, as well as the three cosmids possessing the rRNA operon. Moreover, the SCOTS-prepared, enriched cDNA probe hybridized to 35 additional cosmid clones (16 with strong intensity, 19 with intermediate intensity).
FIG. 2.
Dot blot hybridization between individual M. avium cosmid clones (the same clones as in Fig. 1) and a cDNA mixture prepared from M. avium grown for 110 h in human macrophages (cDNA mixture 8 [Table 1]). The M. avium cDNA mixture was prepared by three rounds of SCOTS and four rounds of enrichment for cDNA molecules corresponding to M. avium genes expressed during growth in macrophages. Labeling of the cDNA probe and hybridization and wash conditions were as described in Materials and Methods.
Preparation of cDNA libraries from cDNA mixtures obtained by SCOTS.
cDNA mixtures have been prepared from M. avium grown in the following ways: (i) to mid-logarithmic phase in Middlebrook 7H9 broth-OADC-0.05% Tween 80, (ii) for 48 h in human macrophages, and (iii) for 110 h in human macrophages. Following three rounds of SCOTS, the cDNA molecules from broth-grown (mid-log phase) M. avium were cloned into pBluescript SK+ to construct a broth cDNA library.
The cDNA mixtures obtained by SCOTS from macrophage-grown M. avium were enriched for cDNA sequences corresponding to genes that were up-regulated for expression during growth in macrophages by competitive hybridization to M. avium genomic DNA in the presence of excess cDNA from broth-grown M. avium (cDNA mixture 4-1 [Table 1]), as described in Materials and Methods and previously (23). This led to the generation of cDNA mixtures 7 and 8 (Table 1). After three or four rounds of enrichment, the cDNA molecules from cDNA mixtures 7 and 8 were cloned into pBluescript II SK+ to construct cDNA libraries for these two growth conditions (48 and 110 h in macrophages).
A total of 225 individual cDNA clones from the library prepared from cDNA mixture 7 (Table 1) were analyzed. A total of 129 individual cDNA clones from cDNA mixture 8 were analyzed. Only 40 individual cDNA clones from the cDNA library prepared from broth-grown M. avium (cDNA mixture 4-1 [Table 1]) were analyzed, since we were more interested in identifying genes that are expressed by M. avium during growth in macrophages than in identifying those expressed during growth in broth. Individual cDNA clones from the cDNA libraries were grown from stock cultures, plasmid DNA was isolated, and the cDNA inserts were analyzed by Southern hybridizations with cDNA probe mixtures prepared from broth-grown M. avium (cDNA mixture 4-1 [Table 1]), from M. avium grown for 48 h in macrophages (cDNA mixture 5-1 [Table 1]), and from M. avium grown for 110 h in macrophages (cDNA mixture 6-1 [Table 1]). Note that probes prepared from cDNA mixtures from macrophage-grown M. avium (cDNA mixtures 5-1 and 6-1 [Table 1]) were prepared from cDNA mixtures that had been obtained by three rounds of SCOTS but were not enriched for cDNA molecules corresponding to genes that were up-regulated for expression or were expressed by M. avium only during growth in macrophages. These hybridizations were conducted to assess whether individual cDNA clones corresponded to genes that were expressed by M. avium during growth in broth or in macrophages or under both conditions. Although the cDNA clones from macrophage-grown M. avium were from libraries prepared from SCOTS-derived, enriched cDNA mixtures, approximately 10% of the cDNA clones hybridized to the cDNA probe mixture prepared from broth-grown M. avium (data not shown). These results suggest that three rounds of enrichment by competitive hybridization between cDNA mixtures from broth-grown and macrophage-grown M. avium were not sufficient to eliminate all of the cDNA molecules that are expressed by M. avium during growth in both broth and in macrophages. However, an additional round of enrichment did not diminish the presence of constitutively expressed cDNA molecules (data not shown). We have not done a thorough analysis of these “constitutive” cDNA molecules, but several of those that have been sequenced correspond to genes encoding transposases.
When the probe prepared from the SCOTS-derived cDNA mixture from M. avium grown for 48 h in macrophages (cDNA mixture 5-1 [Table 1]) was hybridized to the 40 individual cDNA clones from the cDNA library prepared from broth-grown M. avium, 24 clones were recognized (data not shown). Since this probe was prepared from a cDNA mixture that had not been enriched for cDNA molecules from genes that are up-regulated during growth of M. avium in macrophages, these results may give a clearer indication of the percentage of genes expressed by M. avium under both growth conditions (broth and 48 h in human macrophages). The probe prepared from the SCOTS-derived cDNA mixture from M. avium grown for 110 h in macrophages (cDNA mixture 6-1 [Table 1]) hybridized to only 2 of the 40 cDNA clones from broth-grown M. avium (data not shown). These results suggest that the cDNA mixture from mid-log-phase broth-grown M. avium might not have been the best mixture to use to enrich cDNA mixture 6-1 (Table 1). A number of individual cDNA inserts from each of the cDNA libraries were sequenced and compared to sequences in various databases (see below).
Analysis of individual cDNA clones.
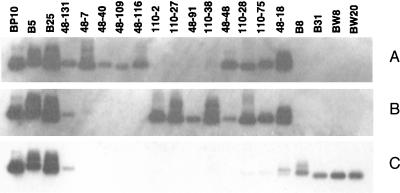
Analysis of individual cDNA clones has enabled us to identify genes that are expressed at different times after infection of macrophages, throughout macrophage infection, only in broth-grown M. avium, and in both macrophage-grown and broth-grown bacteria. Representative examples are illustrated by the Southern hybridization experiments in Fig. 3. The cDNA insert fragments from 20 clones were separated on agarose gels and transferred to nylon filters. The probe for the hybridization depicted in Fig. 3A was derived from the cDNA mixture prepared from M. avium grown for 48 h in macrophages (cDNA mixture 7 [Table 1]). The probe for Fig. 3B was from cDNA prepared from M. avium grown for 110 h in macrophages (cDNA mixture 8 [Table 1]), and the probe for Fig. 3C was cDNA prepared from broth-grown bacteria (cDNA mixture 4-1 [Table 1]). The first four cDNA molecules (BP10, B-5, B25, and 48-131; listed in Table 2 [clone group E], with the prefix Av before the numbers indicated in Fig. 3) represent genes that are expressed by the bacteria under all of these growth conditions. Clones 48-7, 48-40, 48-109, and 48-116 (Table 2, clone group A) represent genes that are expressed at a time during infection of macrophages by which M. avium should be adjusted to the macrophage phagosomal environment and be actively growing (i.e., 48 h after infection). The next four cDNA molecules (110-2, 110-27, 48-91, and 110-38; Table 2, clone group B) represent genes that are expressed late in the macrophage infection model used in these experiments. Clones 48-48, 110-28, 110-75, and 48-18 (Table 2, clone group C) correspond to genes that are expressed throughout the macrophage infection experiments. Three of these genes are also expressed by M. avium during growth in Middlebrook 7H9 broth but appear to be up-regulated for expression during growth in macrophages. This supposition needs to be confirmed by a quantitative analytical method. The last four cDNA clones (B8 [Table 2, clone group D], B31 [not yet sequenced], BW8 [= AvB-108; Table 2, clone group D], and BW20 [not yet sequenced]) represent genes that are only expressed by M. avium during growth in Middlebrook 7H9 broth.
FIG. 3.
Southern hybridizations between individual M. avium cDNA clones and cDNA probes prepared from M. avium grown for 48 (A) or 110 (B) h in human macrophages and from M. avium grown in Middlebrook 7H9 broth (C). The cDNA probes were prepared from cDNA mixtures 7, 8, and 4-1 (all described in Table 1) by incorporation of digoxigenin-11-dUTP with PCR DIG Probe Synthesis kits. Hybridization conditions are described in Materials and Methods.
TABLE 2.
Possible identities of M. avium cDNA clones
| Clonea | Similar protein(s)b | Possible function |
|---|---|---|
| Group A | ||
| Av48-1 | M. tuberculosis LprM (Rv1970) | Part of mce3 operon; similar to several hypothetical M. tuberculosis lipoproteins |
| Av48-5 | M. tuberculosis Rv3896c | Unknown Ala-rich protein; putative p60 homologue |
| Av48-6 (= Av48-76) | M. bovis BCG bramp | Probable manganese transporter |
| M. tuberculosis nramp (Rv0924c) | Probable transmembrane protein | |
| Av48-7 [= Av48(1)-57] | Streptomyces griseus ORF2 | Putative tyrosinase |
| Av48-8 | M. tuberculosis Rv1702c | Similar to other hypothetical proteins inside REP13E12 elements (often in 2 parts) |
| Av48-24 | M. leprae ArgS | Arginine-tRNA synthase |
| M. tuberculosis ArgS (Rv1292) | ||
| Av48-25 | M. tuberculosis Rv1011 | Conserved hypothetical protein |
| Av48-38 (= Av48-120) | M. tuberculosis PPE (Rv0453) | Unknown; M. tuberculosis PPE family |
| Av48-40 | M. tuberculosis Rv0843 | Unknown but similar to various dehydrogenases |
| Av48-41 | M. tuberculosis Pks11 (Rv1665), Pks10 (Rv1660) | Polyketide synthases |
| Av48-44 [Av48-56(1)] | M. tuberculosis MbtF (Rv2379c) | Mycobactin synthesis |
| Av48-45 | M. tuberculosis CobN (Rv2062c) | Cobalt insertion/cobalamin biosynthesis |
| Av48-51 | M. tuberculosis Rv2714 | Unknown (conserved hypothetical protein) |
| Av48-53 | M. tuberculosis MmpL3 (Rv0206c) | Conserved large membrane protein |
| Av48-73 | M. tuberculosis KdpD (Rv1028c) | Probable sensor protein |
| Av48-77 (= Av48-84) | M. tuberculosis Nrp (Rv0101) | Unknown nonribosomal peptide synthase, similar to M. leprae aceA |
| Av48-82 | M. tuberculosis Rv1319c | Some similarity at C terminus to adenylate cyclase |
| Av48-88 | Upstream of M. tuberculosis PE genes (Rv1788, Rv1791) | Unknown |
| Av48-107 | M. tuberculosis CarB (Rv1384) | Carbamoyl-phosphate synthase |
| Av48-109 | M. tuberculosis Rv1802 | M. tuberculosis PPE family |
| Av48-114 | M. tuberculosis Rv0146, M. leprae cosmid B26 MSGB26CS | Unknown |
| Av48-116 | M. tuberculosis Rv0867 | Unknown Ala-Pro-rich protein |
| M. leprae MLCB57.05c | Unknown | |
| Av48-118 | Streptomyces coelicolor putative membrane protein | Unknown |
| Av48-127 | M. tuberculosis UdgA (Rv0322) | UDP-glucose dehydrogenase/GDP-mannose-6-dehydrogenase |
| Av48-135 | M. tuberculosis Rv3496c | Unknown; part of mce4 operon |
| Group B | ||
| Av110-2 | M. tuberculosis Rv0286 | PPE family |
| Av110-10 (= Av110-38) | M. tuberculosis AtpA (Rv1308) | ATP synthase, α chain |
| Av110-19 | M. tuberculosis MbtE (Rv2380c) | Mycobactin synthesis |
| Av110-21 | M. tuberculosis Rv0475 | Unknown |
| Av110-26 | M. leprae RbsF | 30S ribosomal protein S6 |
| Av110-27 | None | Unknown |
| Av110-32 | Agrobacterium tumefaciens ORF3 | GlnP/GlnQ homolog |
| Av110-33 | M. tuberculosis Rv0474 | Probable transcription regulator |
| Av110-50 | M. avium IS1311 gene | Transposase |
| Av110-51 | M. tuberculosis MmsA (Rv0753) | Probable methylmalonate semialdehyde dehydrogenase |
| Av48-91 | E. coli OxcA | Probable oxalyl-CoA decarboxylase |
| AvB-23 | M. tuberculosis Rv3767 or Rv0145 | Unknown |
| Group C | ||
| Av48-18 [= Av48-33(1)] | M. tuberculosis hypothetical protein Rv2094c | Sec-independent protein translocase protein |
| Av48-20c | M. tuberculosis Icd2 (Rv0066c) | Isocitrate dehydrogenase |
| Av48-33(2) | M. tuberculosis Ffh (Rv2916c) | Signal recognition particle protein |
| Av48-48 | M. tuberculosis NirB (Rv0252) | Probable nitrite reductase flavoprotein |
| Av48-128 | M. tuberculosis SucA (Rv1248c) | α-Ketoglutarate dehydrogenase |
| Av48-142 | M. tuberculosis IlvB2 (Rv3470c) | Acetolactate synthase large subunit |
| Av110-6c | M. tuberculosis FbpA (Rv3804) | 32-kDa antigen protein 85-A precursor, mycolytransferase |
| Av110-28c (= Av110-40) | M. tuberculosis (Icl) Rv0467 | Probable isocitrate lyase |
| Av110-75c | M. tuberculosis SecF (Rv2686c) | Protein export membrane protein |
| AvB-29c | M. tuberculosis NarK3 (Rv0261c) | Nitrite extrusion protein |
| AvB-33c | M. tuberculosis Add (Rv3313c) | Probable adenosine deaminase |
| Group D | ||
| AvB-7 | M. tuberculosis Rv0088 | Unknown; putative regulatory protein |
| AvB-8 | M. tuberculosis OtsA (Rv3490) | Probable α,α-trehalose-phosphate synthase |
| AvB-10 | M. tuberculosis PolA | DNA polymerase I |
| AvB-24 | C. jejuni FtsH | Probable membrane-bound zinc metallopeptidase |
| M. tuberculosis FtsH (Rv3610c) | Cell division protein FtsH | |
| AvB-104 (also named AvB-W4) | M. tuberculosis RmlA (Rv0334) | α-d-Glucose-1-phosphate thymidyltransferase |
| AvB-108 (also named AvB-W8) | M. tuberculosis Rv1556 | Putative transcriptional regulator |
| AvB-119 (also named AvB-W19) | M. smegmatis ORF1 | Probable cytochrome P450 |
| Group E | ||
| AvB-5 | M. fortuitum ORF1 (IS219) | Hypothetical protein |
| AvB-25 (= Av110-27) | M. avium strain 2151 IS1601 | Transposase |
| AvB-P10 | M. avium IS1245 | Transposase |
| Av48-131 | M. tuberculosis Rv0287 | Conserved hypothetical protein; probable transcription regulator |
Clone groups: A, genes differentially expressed by M. avium grown for 48 h in human macrophages; B, genes differentially expressed by M. avium grown for 110 h in human macrophages; C, genes expressed by M. avium at both 48 and 110 h after infection of human macrophages; D, clones detected only in broth-grown M. avium cDNA; E, constitutively expressed M. avium cDNA clones.
Proteins were identified if identity scores were ≥40% and positivity scores were ≥50% between deduced amino acid sequences derived from the cDNA inserts and the indicated proteins.
A much lower hybridization signal was detected with the broth-cDNA probe.
We have identified and sequenced a total of 54 cDNA molecules that represent 46 genes that are expressed by M. avium during growth in human macrophages and 7 cDNA molecules that represent genes that are expressed by M. avium during growth in Middlebrook 7H9 broth supplemented with OADC and 0.05% Tween 80. For eight of the genes expressed by intracellular M. avium, we have identified two identical cDNA molecules (apparently sibs). The cDNA clones, proteins to which their gene products are similar, and possible functions of the gene products are summarized in Table 2. In addition, eight genes have been identified that are expressed under all of the growth conditions studied. Four of these genes were identified from cDNA clones (Table 2, clone group E). The genes atpB, clpC, lsr2, and sigA (which were hypothesized to be constitutively expressed) were PCR amplified from M. avium chromosomal DNA to serve as controls in hybridization experiments, although hybridizations with sigA are not shown.
Reproducibility of recovery of cDNA molecules by SCOTS.
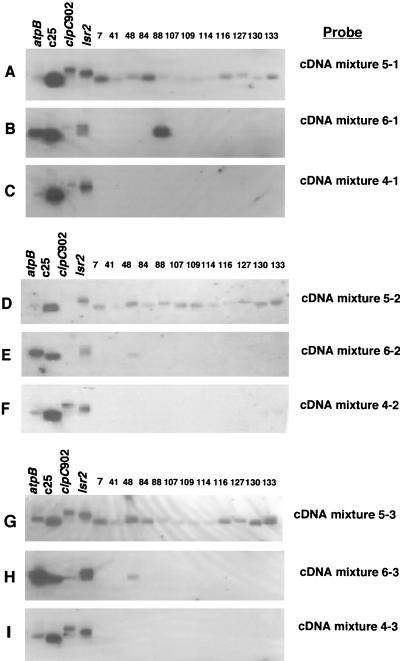
To assess the ability to recover the same cDNA molecules in cDNA mixtures obtained by the SCOTS technique at different times from a single frozen stock of cDNA (reproducibility of recovery of cDNA molecules), Southern hybridization experiments were performed with (i) a subset of 12 individual cDNA clones that were selected from a cDNA library prepared from M. avium that had been grown for 48 h in macrophages and (ii) probes made from several different cDNA mixtures (Fig. 4). The probes were from cDNA mixtures prepared from M. avium grown for 48 h in macrophages (cDNA mixtures 5-1, 5-2, and 5-3 [Table 1]), M. avium grown for 110 h in macrophages (cDNA mixtures 6-1, 6-2, and 6-3 [Table 1]), and broth-grown M. avium (cDNA mixtures 4-1, 4-2, and 4-3 [Table 1]). Samples of cDNA from each of these growth conditions were removed from the original frozen stock cDNA preparations at three different times and were obtained by three rounds of SCOTS each time. For example, samples from the cDNA mixture prepared from M. avium grown for 48 h in macrophages were removed from the frozen stock cDNA preparation at the time the original preparation was made (cDNA mixture 5-1 [Table 1]) and 6 (cDNA mixture 5-2 [Table 1]) and 11 (cDNA mixture 5-3 [Table 1]) months later. Similarly, separate cDNA mixtures were prepared by SCOTS at three different times from the original frozen stock cDNA preparations from M. avium grown for 110 h in macrophages and from broth-grown M. avium.
FIG. 4.
Southern hybridizations between individual M. avium cDNA clones and cDNA probes prepared from M. avium grown for 48 and 110 h in human macrophages and from M. avium grown in Middlebrook 7H9 broth-OADC-0.05% Tween 80. At the time the original stock cDNA mixtures were prepared (cDNA mixtures 4-1, 5-1, and 6-1 [described in Table 1]) and at 6 (cDNA mixtures 4-2, 5-2, and 6-2 [Table 1]) and 11 (cDNA mixtures 4-3, 5-3, and 6-3 [Table 1]) months later, samples were removed from each cDNA mixture and prepared by three rounds of SCOTS. Labeled probes were prepared from each SCOTS-derived mixture by incorporation of digoxigenin-11-dUTP with PCR DIG Probe Synthesis kits (Roche Molecular Biochemicals) and hybridized to 12 individual cDNA clones that had originally been identified from the cDNA mixture prepared from M. avium grown for 48 h in macrophages (cDNA mixture 5-1 [Table 1]). DNA fragments from atpB, clpC902, lsr2, and c25, genes that were expected to be expressed under all of the growth conditions studied, were included in each hybridization assay as positive controls. Hybridization and wash conditions were as described in Materials and Methods.
Samples of each of these cDNA mixtures were labeled (cDNA probe mixtures) and hybridized to 12 individual cDNA clones and to 4 control DNA sequences (sequences from the atpB, clpC, and lsr2 genes and a cDNA insert to which all of the probe mixtures hybridized [c25, which is designated AvB-25; Table 2, clone group E]). The results of these hybridization experiments are illustrated in Fig. 4. It is evident that the probes prepared from cDNA mixtures from M. avium grown for 48 h in macrophages and obtained by SCOTS on three separate occasions (0, 6, and 11 months after the original cDNA mixture was prepared) all hybridized to each of the individual cDNA clones, as well as to two (probe from cDNA mixture 5-2 [Fig. 4D]) or all four (probe from cDNA mixtures 5-1 [Fig. 4A] and 5-3 [Fig. 4G]) of the control sequences. The probes derived from cDNA mixtures from M. avium grown for 110 h in macrophages and prepared by SCOTS on three separate occasions hybridized to three of the four control sequences. Two of these probes also hybridized to one of the individual cDNA clone inserts (clone 48), and the other probe hybridized to a different individual cDNA clone insert. The probes derived from cDNA mixtures from broth-grown M. avium and obtained by SCOTS on three separate occasions each hybridized to all four of the control sequences but did not hybridize to any of the individual cDNA inserts that had been isolated from a cDNA mixture prepared from M. avium grown for 48 h in macrophages. These data demonstrate that more than 87% (14 of 16 to 16 of 16) of the same cDNA molecules were recovered on three separate occasions from a “stock” cDNA preparation by the SCOTS technique.
DISCUSSION
We hypothesize that survival and growth of pathogenic mycobacteria within the phagosomal environment necessitate expression of bacterial genes that are unlikely to be expressed when the bacteria are growing in broth culture in the laboratory. That pathogenic mycobacteria inhibit maturation of phagosomes to phagolysosomes has been well documented (2, 10, 16, 18, 22, 43). However, the mechanism by which mycobacteria accomplish this is not known. The kinds of carbon and energy sources and other nutrients available to mycobacteria growing in the phagosome are not known either. To gain insight into the metabolic activities of intraphagosomally growing mycobacteria, we initially developed a cDNA subtractive hybridization approach (39). These studies led to the identification of an M. avium gene, designated mig, that was apparently expressed by M. avium only when the bacteria were growing in macrophages. Continued analysis by G. Plum and his colleagues has revealed that the mig gene encodes a 58-kDa protein that acts as an acyl coenzyme A (acyl-CoA) synthetase with medium-chain fatty acids and unsaturated long-chain fatty acids as substrates (37). We did not recover cDNA molecules corresponding to mig (CoA) from either cDNA mixture prepared by SCOTS from macrophage-grown M. avium. Two possible explanations for this are (i) differences in donor macrophages and (ii) differences in cultivation of macrophages prior to infection. Different human donors provided macrophages for the studies of Plum and Clark-Curtiss (39) and the studies described here. Plum and Clark-Curtiss incubated PBMC-derived monocytes in RPMI 1640 tissue culture medium for 5 days prior to infection (39), allowing adherence and maturation into macrophages. In the studies described above, the adherent macrophages were incubated for 7 days longer to permit further maturation prior to infection. Additional studies are necessary to resolve these disparate results. We have extended the gene expression studies by the cDNA subtractive hybridization approach and have identified four additional sequences. The DNA sequences of two of these have been determined, but neither corresponds to genes of known function in databases.
A drawback of the cDNA subtractive hybridization technique was that the macrophages were lysed and the mycobacteria were collected by centrifugation prior to isolation of RNA (39). Since the average half-life of mycobacterial mRNA has not been determined but the half-lives of some bacterial mRNAs can be extremely short (31), we devised an alternative approach to overcome this problem, i.e., SCOTS. The SCOTS approach has been successfully used to identify some genes of M. tuberculosis that are expressed by this bacterium during growth in human macrophages (23). In addition, SCOTS has been used to identify Salmonella enterica serovar Typhi genes expressed during growth of the bacteria in the human monocyte-like cell line THP-1 (17). Here we describe studies that used the SCOTS method to identify genes that are expressed by M. avium at different times after infection of human macrophages.
Initially, we compared cDNA mixtures prepared by subtractive hybridization to those prepared by SCOTS plus enrichment for cDNA molecules representing genes up-regulated or uniquely expressed by M. avium during growth in macrophages. These comparisons were done by hybridization of the cDNA mixtures with a set of 368 individual cosmid clones from an M. avium genomic library. Theoretically, this number of clones should represent between three and four M. avium genome equivalents. These experiments indicated that the probe mixtures prepared by SCOTS and enriched for cDNA molecules representing genes differentially expressed during growth of M. avium in macrophages contained cDNA molecules that hybridized to 12 of the 13 cosmids to which the subtractive hybridization cDNA probe mixture hybridized. In addition, the SCOTS-prepared, enriched cDNA probe mixture prepared from M. avium grown for 110 h in macrophages (cDNA mixture 8 [Table 1]) hybridized to 16 other cosmids with strong intensity and to 19 other cosmids with intermediate intensity. It is possible that the differences in cDNA molecules detected by subtractive hybridization and SCOTS are a consequence of the macrophages, which were obtained from two different donors. Further comparisons of the two methods would be necessary to determine whether or not this is the reason. However, we believe that the rapid lysis of infected macrophages and the positive selection for mycobacterial cDNA molecules during the SCOTS procedure result in more efficient recovery of cDNA molecules. We conclude that the SCOTS technique plus enrichment allows recovery of the majority of the cDNA molecules obtained by the cDNA subtractive hybridization method but that SCOTS is superior to cDNA subtractive hybridization in recovering a larger diversity of cDNA molecules.
Studies were also conducted to assess the reproducibility of the SCOTS technique to capture the same set of cDNA molecules. On three separate occasions, cDNA mixtures were prepared by the SCOTS technique from a frozen cDNA stock preparation prepared from M. avium grown for 48 h in human macrophages. Probes generated from each of these SCOTS-purified cDNA preparations hybridized to 87 to 100% of a subset of 16 individual cDNA clones, indicating the high level of reproducible recovery of cDNA molecules by the SCOTS approach.
The majority of our efforts thereafter were directed toward analysis of individual molecules from cDNA libraries prepared from M. avium grown for 48 h in human macrophages, M. avium grown for 110 h in macrophages, and M. avium grown in Middlebrook 7H9 broth supplemented with OADC and Tween 80. Fifty-four molecules representing 46 M. avium genes that are expressed by the bacteria during growth in macrophages have been sequenced. Some correspond to genes that appear to be expressed relatively early (48 h) after infection, whereas others represent genes that are apparently expressed late (110 h) after infection. Still other cDNA molecules have been identified that represent genes that are expressed by M. avium throughout the course of infection in our model system. In addition, cDNA molecules representing genes that are expected to be expressed by M. avium under all growth conditions have also been identified from the cDNA libraries prepared from SCOTS-derived cDNA mixtures. Finally, a small number of cDNAs representing genes that are expressed by M. avium during growth in Middlebrook 7H9 broth, but not during growth in macrophages, have been identified.
It should be noted that we have not identified all of the genes that are expressed by M. avium under any of the growth conditions described in these studies. Among the 54 randomly chosen cDNA molecules from the macrophage-grown cDNA libraries that have been sequenced thus far, only 8 duplicate cDNA molecules have been detected. It is difficult to predict how many M. avium genes will be up-regulated or uniquely expressed by the bacilli during growth in human macrophages because of the paucity of knowledge about mycobacterial physiology during intracellular growth.
Comparison of the cDNA sequences to sequences in the databases has permitted identification of cDNA molecules that represent genes of M. avium that code for proteins similar to those of other organisms (Table 2). Thus, we have identified genes that encode enzymes involved in pyrimidine biosynthesis (carB), intermediary metabolism (udgA), the tricarboxylic acid cycle (icd and sucA), the glyoxalate shunt (icl), nitrogen metabolism (nirB and narK3), salvage of nucleosides and nucleotides (add), polyketide synthesis (pks10 and pks11), mycobactin biosynthesis (mbtE and mbtF), and regulatory proteins. In addition, cDNA molecules corresponding to genes of unknown function have also been identified. In the latter group are genes specifying members of the PPE family of proteins. This is a family with 68 members whose genes, together with those of the PE family, comprise approximately 10% of the coding capacity of the M. tuberculosis H37Rv genome (12). Because these genes are so abundant in the M. tuberculosis genome and because of the highly conserved N-terminal amino acid sequences of the members of each of the families, it is presumed that the PE and PPE families are important M. tuberculosis proteins. It has been hypothesized that these proteins may be a source of antigenic variation among M. tuberculosis isolates (12).
Two other genes encoding proteins whose functions are unknown and for which cDNA molecules were identified from macrophage-grown M. avium were homologues of lprM and a gene of the mce4 operon of M. tuberculosis. These genes are parts of two of the four redundant mce operons found in the M. tuberculosis genome (12). Genes in one of the mce operons have been previously associated with mycobacterial virulence (3, 12), but the precise functions of the proteins specified by the genes in the mce operons remain to be elucidated. In previous experiments in which we used SCOTS to prepare cDNA mixtures from M. tuberculosis grown in macrophages (23), we also identified a cDNA molecule that corresponded to the mceD gene, which is located in the mce1 operon of M. tuberculosis. Do the results of these two studies indicate that expression of different mce operons is dependent upon the species of mycobacteria? Additional studies are necessary to delineate the regulation of expression of the four mce operons.
Analysis of SCOTS-derived cDNA clones has provided interesting insights into the metabolic activities of M. avium during growth within macrophages. For example, cDNA molecules corresponding to genes encoding enzymes of both the citric acid cycle (icd and sucA) and the glyoxalate bypass (icl) were identified. This finding implies that, like E. coli, M. avium utilizes the pathways simultaneously for energy production, amino acid biosynthesis, and replenishment of dicarboxylic acid intermediates (15). Interestingly, identification of a cDNA molecule corresponding to sucA suggests that the complete citric acid cycle is operative and available for oxidative metabolism in macrophage-grown M. avium (4). The enzyme encoded by sucA, α-ketoglutarate dehydrogenase, is unstable and has been difficult to detect in lysates of in vivo-grown mycobacteria; thus, as Barclay and Wheeler pointed out, it has been unclear whether or not M. tuberculosis and M. avium use the complete citric acid cycle during intracellular growth (4). We conclude from our results that they do.
Expression of a homologue of the M. tuberculosis isocitrate lyase gene (icl; Rv0467) was detected in M. avium at both times during growth in macrophages. This result is in agreement with our earlier studies with human macrophage-grown M. tuberculosis (23) and the results of Sturgill-Koszycki et al. (43) and Höner zu Bentrup et al. (25), who demonstrated the presence of this enzyme by two-dimensional gel analysis of proteins produced by M. avium after culture in murine macrophages. Isocitrate lyase is the first enzyme of the glyoxalate bypass. More recent studies have demonstrated the importance of isocitrate lyase for persistence of M. tuberculosis in murine macrophages and in the infected-mouse model (36).
Throughout growth in macrophages, M. avium expressed genes coding for enzymes of biosynthetic pathways such as amino acid (branched-chain) biosynthesis and mycolic acid biosynthesis. These biosynthetic pathways are likely operative when M. avium was grown in Middlebrook 7H9 broth, since this medium is essentially a minimal medium. We hypothesize that, in the macrophage environment, the genes for these pathways are up-regulated for expression. Additional quantitative determinations of specific mRNA levels from the two growth conditions are necessary to test this hypothesis.
Detection of cDNA molecules for genes encoding enzymes involved in the biosynthesis of mycobactins (mbtE and mbtF) in macrophage-grown M. avium demonstrate the importance of this pathway. Mycobactins are siderophores produced by mycobacteria for obtaining iron, a nutrient that is essential for all organisms but is not readily available to intracellular organisms. De Voss et al. have recently presented evidence demonstrating that the ability to produce mycobactins is essential for M. tuberculosis growth in macrophages (19).
In addition to cDNA molecules corresponding to genes whose products are homologous to proteins of other bacteria, numerous cDNA molecules of macrophage-grown M. avium were identified that did not exhibit similarity to any genes or gene products in current databases. It is not surprising that novel genes are expressed by M. avium growing within macrophages, and the products of some of these genes may well be crucial determinants of M. avium virulence. Characterizing these genes and their products and deciphering their roles in intracellular growth should enable us to gain a better understanding of M. avium pathogenesis. Such studies are in progress.
In recent years, investigators have applied molecular genetic approaches to gain a better understanding of the mechanisms of pathogenicity of a number of bacterial pathogens. The underlying rationale has been that bacteria must express specific genes in response to environments encountered in an infected host; the products of such genes would be unnecessary for the organism in in vitro culture (6, 7, 24, 34). Two approaches by which to identify in vivo-expressed bacterial genes that have been developed and successfully employed are in vivo expression technology (6, 34) and signature-tagged mutagenesis (24). Both of these approaches have contributed valuable information relative to the pathogenesis of a number of bacteria (recently reviewed by Chiang et al. [7]). However, drawbacks of these approaches include the necessity of a good animal model for a particular disease and the necessity of well-developed genetic tools with which to manipulate the pathogen. The SCOTS approach, on the other hand, is amenable to any organism (at least any organism in which there is a direct correlation between transcription and translation) since it necessitates only the ability to isolate total RNA and DNA. Another more recently developed method, DNA microarray analysis (46), affords the possibility of identifying a larger number of expressed genes and has the advantage of identifying genes that are down-regulated or repressed in a given environment. However, DNA microarray analysis is an expensive technology and requires relatively large amounts of RNA, thus precluding studies of bacterial gene expression in scarce types of tissues or cells (e.g., PBMC-derived macrophages, alveolar macrophages, or biopsy materials from humans). Thus, the SCOTS approach is an economical, direct approach by which to identify genes expressed by a given organism in response to specific environmental conditions that is widely applicable to virtually any prokaryote and other organisms as well.
Acknowledgments
This work was supported by Public Health Service grants AI38672 and AI46428 from the U.S. National Institutes of Health awarded to J.E.C.-C.
We thank Roy Curtiss III for critical reading and helpful discussions of the manuscript.
Editor: J. T. Barbieri
REFERENCES
- 1.Aberg, J. A., D. Yajko, and M. A. Jacobson. 1998. Eradication of AIDS-related disseminated Mycobacterium avium complex infection after 12 months of antimycobacterial therapy combined with highly active antiretroviral therapy. J. Infect. Dis. 178:1446-1449. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, J. S., and P. D. Hart. 1971. Response of cultured macrophages to Mycobacterium tuberculosis, with observations on fusions of lysosomes with phagosomes. J. Exp. Med. 134:713-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arruda, S., G. Bomfim, R. Knights, T. Huima-Byron, and L. W. Riley. 1993. Cloning of an M. tuberculosis DNA fragment associated with entry and survival inside cells. Science 261:1454-1457. [DOI] [PubMed] [Google Scholar]
- 4.Barclay, R., and P. R. Wheeler. 1989. Metabolism of mycobacteria in tissues, p. 37-106. In C. Ratledge, J. Stanford, and J. M. Grange (ed.), The biology of the mycobacteria, vol. 3. Academic Press, Inc., New York, N.Y.
- 5.Bermudez, L. E., L. S. Young, and H. Enkel. 1991. Interaction of Mycobacterium avium complex with human macrophages: roles of membrane receptors and serum proteins. Infect. Immun. 59:1697-1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camilli, A., D. Beattie, and J. J. Mekalanos. 1994. Use of genetic recombination as a reporter of gene expression. Proc. Natl. Acad. Sci. USA 91:2634-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiang, S. L., J. J. Mekalanos, and D. W. Holden. 1999. In vivo genetic analysis of bacterial virulence. Annu. Rev. Microbiol. 53:129-154. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 9.Clark-Curtiss, J. E., W. R. Jacobs, Jr., M. A. Docherty, L. R. Ritchie, and R. Curtiss III. 1985. Molecular analysis of DNA and construction of genomic libraries of Mycobacterium leprae. J. Bacteriol. 161:1093-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clemens, D. L., and M. A. Horwitz. 1995. Characterization of the Mycobacterium tuberculosis phagosome and evidence that phagosomal maturation is inhibited. J. Exp. Med. 181:257-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coker, R. J., T. J. Helyer, I. N. Brown, and J. N. Weber. 1992. Clinical aspects of mycobacterial infections in HIV infection. Res. Microbiol. 143:369-372. [DOI] [PubMed] [Google Scholar]
- 12.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborn, M. A. Quail, M.-A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 293:537-544. [DOI] [PubMed] [Google Scholar]
- 13.Collins, F. M. 1989. Mycobacterial disease, immunosuppression, and acquired immunodeficiency syndrome. Clin. Microbiol. Rev. 2:360-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper, A. M., R. Appelberg, and I. M. Orme. 1998. Immunopathogenesis of Mycobacterium avium infection. Front. Biosci. 3:141-148. [DOI] [PubMed] [Google Scholar]
- 15.Cronan, J. E., and D. LaPorte. 1996. Tricarboxylic acid cycle and glyoxalate bypass, p. 206-216. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd edition. ASM Press, Washington, D.C.
- 16.Crowle, A. J., R. Dahl, E. Ross, and M. H. May. 1991. Evidence that vesicles containing living, virulent Mycobacterium tuberculosis and Mycobacterium avium in cultured human macrophages are not acidic. Infect. Immun. 59:1823-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daigle, F., J. E. Graham, and R. Curtiss III. 2001. Identification of Salmonella typhi genes expressed within macrophages by selective capture of transcribed sequences (SCOTS). Mol. Microbiol. 41:1211-1222. [DOI] [PubMed] [Google Scholar]
- 18.de Chastellier, C., C. Frehel, C. Offredo, and E. Skamene. 1993. Implication of phagosome-lysosome fusion in restriction of Mycobacterium avium growth in bone marrow macrophages from genetically resistant mice. Infect. Immun. 61:3775-3784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falkinham, J. O., III. 1996. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9:177-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frehel, C., C. de Chastellier, T. Lang, and N. Rastogi. 1986. Evidence for inhibition of fusion of lysosomal and phagosomal compartments with phagosomes in macrophages infected with pathogenic Mycobacterium avium. Infect. Immun. 52:252-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frehel, C., C. de Chastellier, C. Offredo, and P. Berche. 1991. Intramacrophage growth of Mycobacterium avium. Infect. Immun. 59:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 25.Höner zu Bentrup, K., D. L. Swenson, A. Miczak, and D. G. Russell. 1999. Characterization of isocitrate lyase activity and expression in Mycobacterium avium and Mycobacterium tuberculosis. J. Bacteriol. 181:7161-7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horsburgh, C. R. 1991. Mycobacterium avium complex infections in the acquired immunodeficiency syndrome. N. Engl. J. Med. 324:1332-1338. [DOI] [PubMed] [Google Scholar]
- 27.Inderlied, C. B., C. A. Kemper, and L. E. Bermudez. 1993. The Mycobacterium avium complex. Clin. Microbiol. Rev. 6:266-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iseman, M. D. 1996. Pulmonary disease due to Mycobacterium avium complex, p. 45-78. In J.A. Korvik and C. A. Benson (ed.), Mycobacterium avium complex infection: progress in research and treatment. Marcel Dekker, Inc., New York, N.Y.
- 29.Jacobson, M. A., P. C. Hopewell, D. M. Yajko, W. K. Hadley, E. Lazarus, P. K. Mohanty, G. W. Modlin, D. W. Feigl, P. S. Cusick, and M. A. Sande. 1991. Natural history of disseminated Mycobacterium avium complex infection in AIDS. J. Infect. Dis. 164:994-998. [DOI] [PubMed] [Google Scholar]
- 30.Kovacs, J. A., and H. Masur. 2000. Drug therapy: prophylaxis against opportunistic infections in patients with human immunodeficiency virus infection. N. Engl. J. Med. 342:1416-1429. [DOI] [PubMed] [Google Scholar]
- 31.Kushner, S. R. 1996. mRNA decay, p. 849-860. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd edition. ASM Press, Washington, D.C.
- 32.Lennox, E. S. 1955. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190-206. [DOI] [PubMed] [Google Scholar]
- 33.Lodish, H. F. 1970. Secondary structure of bacteriophage f2 ribonucleic acid and the initiation of in vitro protein synthesis. J. Mol. Biol. 50:689-702. [DOI] [PubMed] [Google Scholar]
- 34.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 35.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 36.McKinney, J. D., Höner zu Bentrup, K., E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxalate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 37.Morsczek, C., S. Berger, and G. Plum. 2001. The macrophage-induced gene (mig) of Mycobacterium avium encodes a medium chain acyl-coenzyme A synthetase. Biochim. Biophys. Acta 1521:59-65. [DOI] [PubMed] [Google Scholar]
- 38.Pallela, F. J., K. M. Delancy, A. C. Moorman, M. O. Loveless, J. Fuhrer, G. A. Satten, D. J. Aschman, and S. D. Holmberg. 1998. Declining morbidity and mortality among patients with advanced HIV infection. N. Engl. J. Med. 338:853-860. [DOI] [PubMed] [Google Scholar]
- 39.Plum, G., and J. E. Clark-Curtiss. 1994. Induction of Mycobacterium avium gene expression following phagocytosis by human macrophages. Infect. Immun. 62:476-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rao, S. P., K. Ogata, and A. Catanzaro. 1993. Mycobacterium avium-Mycobacterium intracellulare binds to the integrin receptor αVβ3 on human monocytes and monocyte-derived macrophages. Infect. Immun. 61:663-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schlesinger, L. S., C. G. Bellinger-Kawahara, N. R. Payne, and M. A. Horwitz. 1990. Phagocytosis of Mycobacterium tuberculosis is mediated by human monocyte complement receptors and complement component C3. J. Immunol. 144:2771-2780. [PubMed] [Google Scholar]
- 42.Schlesinger, L. S., and M. A. Horwitz. 1991. Phagocytosis of Mycobacterium leprae by human monocyte-derived macrophages is mediated by complement receptors CR1 (CD35), CR3 (CD11b/CD18) and CR4 (CD11c/CD18) and interferon-gamma inactivation inhibits complement receptor function and phagocytosis of the bacterium. J. Immunol. 147:1983-1994. [PubMed] [Google Scholar]
- 43.Sturgill-Koszycki, S., P. Schlesinger, P. Chakraborty, P. L. Haddix, H. L. Collins, A. Fok, P. Allen, S. Gluck, J. Heuser, and D. L. Russell. 1994. Lack of acidification in Mycobacterium phagosomes produced by exclusion of the vesicular proton ATPase. Science 263:678-681. [DOI] [PubMed] [Google Scholar]
- 44.Swartz, R. D., D. Naai, C. W. Vogel, and H. Yeager, Jr. 1988. Differences in uptake of mycobacteria by human monocytes: a role for complement. Infect. Immun. 56:2223-2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Soolingen, D., P. E. de Haas, P. W. Hermans, and J. D. van Embden. 1994. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 235:196-205. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, M., J. DeRisi, H. H. Kristensen, P. Imboden, S. Rane, P. O. Brown, and G. K. Schoolnik. 1999. Exploring drug-induced alterations in gene expression in Mycobacterium tuberculosis by microarray hybridization. Proc. Natl. Acad. Sci. USA 96:12833-12838. [DOI] [PMC free article] [PubMed] [Google Scholar]