Abstract
Staphylococcus aureus is a major cause of mastitis in bovine and other ruminant species. We here present the results of a comparative genomic analysis between a bovine mastitis-associated clone, RF122, and the recently sequenced human-associated clones, Mu50 and N315, of Staphylococcus aureus. A shotgun sequence survey of ∼10% of the RF122 genome identified numerous unique sequences and those with elevated rates of nonsynonymous substitution. Taken together, these analyses show that there are notable differences in the genomes of bovine mastitis-associated and human clones of S. aureus and provide a framework for the identification of specific factors associated with host specificity in this major human and animal pathogen.
Staphylococcus aureus is a widely distributed animal and human pathogen. Phenotypic analyses of S. aureus strains isolated from human and bovine hosts have revealed important differences between the two populations of isolates, but the underlying genetic basis for the variation among S. aureus strains remains unknown (10, 14, 15). Multilocus enzyme electrophoretic analysis of 2,077 S. aureus isolates from bovine, ovine, and human hosts showed that only 6 of the 33 clusters included isolates from more than one host species (11). Population genetic analysis of 357 S. aureus isolates recovered from cases of bovine mastitis from global sources suggested that only eight clones represented 90% of the isolates, and these clones rarely overlapped with those responsible for human disease (5). To begin identifying the nature and scope of the genetic differences between the bovine and human staphylococcal genomes, we present here a comparative analysis of the genome content of the common bovine S. aureus clone RF122 (4) and the recently sequenced human pathogenic clones Mu50 and N315 (6).
A random shotgun genomic library was prepared as described previously (7), with the exception that RF122 genomic DNA was mechanically sheared with a Hydroshear device (GeneMachines, San Carlos, Calif.). The first 850 random shotgun clones of RF122, encoding 285,077 unique bases and representing approximately 10% of the total genome, were sequenced by using dideoxy terminator chemistry on ABI 3700 DNA sequencers (ABI, Foster City, Calif.) and assembled into 600 contigs consisting of one to six sequences and ranging in size from 82 to 1,828 bases using phredPhrap (P. Green, University of Washington, Seattle) (details of individual genes, gene names, and categories are available at http://www.cbc.umn.edu/ResearchProjects/AGAC/Staph/staphhome.html). The average GC content of the initial sequences was 33.95%, closely approximating the GC content of the completed S. aureus genomes (6). The range of GC contents of individual contigs was 21 to 73%. Fourteen contigs had GC contents greater than 45%. Of these, two had no significant hits (P < e-15) to any GenBank submission based on tBLASTn searches, five had significant homology to S. aureus genomic DNA, and one was homologous to an S. aureus plasmid (1). The remaining five contigs had significant (P < e-15) homologies to sequences of organisms other than staphylococci and may reflect recent horizontal transfer events.
Identification of unique sequences in RF122.
Twelve sequences without homology to any existing sequence in the comprehensive microbial databases were identified in RF122. An additional seven contigs with no homology to S. aureus but with close matches to genes from other organisms were identified, including homologs to a Shigella transposon, a pathogenicity island element from Escherichia coli, a Rhodopseudomonas palustris ferredoxin transporter, and a hypothetical protein from Mesorhizobium loti. Other orthologs matched the gram-positive organisms Streptococcus agalactiae (sodA), Streptococcus pyogenes (putative phage terminase), Enterococcus faecalis (putative invertase), and Bacillus pumilus (Tn 10 transposon). Since the present analysis represents sequences from only ∼10% of the RF122 genome, these data suggest that there are likely to be 100 or more sequences that differentiate the bovine S. aureus clone from its human counterparts.
Recent analysis of RF122 led to the identification of a novel putative pathogenicity island, SaPIbov (4). The 15,981-bp island encodes several virulence factors and toxins (4). As expected, our RF122 contigs included several SaPIbov components. Two of the homologous contigs matched no sequences in either of the finished human S. aureus genomes, indicating possible bovine-specific components of SaPIbov. The genes in these contigs include homologs to sel, whose hypothetical protein product is similar to staphylococcal enterotoxin I, and to a phage terminase homolog.
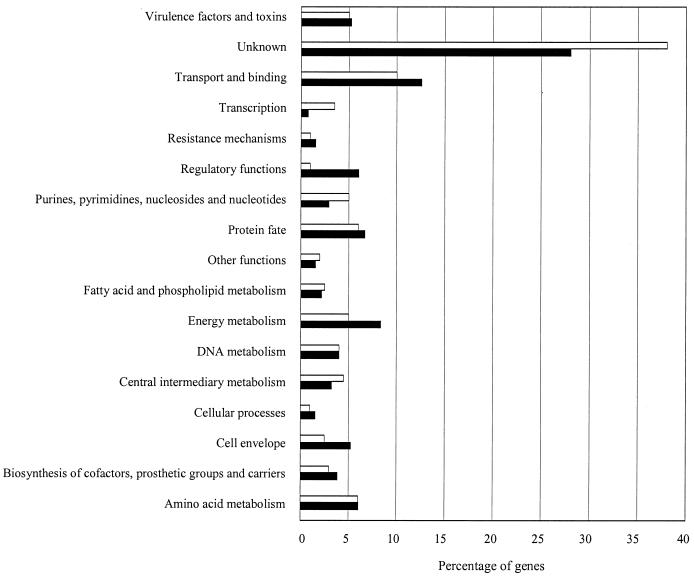
We were able to assign putative functions to many of the gene products based on motifs identified in the homologs from the finished Mu50 and N315 S. aureus genomes and the genome of Bacillus subtilis (Fig. 1). In total, 28% of the identified homologs encoded proteins of unknown function. Other genes of interest include bap, a biofilm-related gene thus far identified only in bovine isolates, drp35, an antibiotic resistance determinant, and rot, a transcriptional repressor regulating toxin expression previously characterized only in human strains (2, 8, 9).
FIG. 1.
Gene functions of RF122 (filled bars) were identified based on homology to existing GenBank submissions in tBLASTx analyses. Functional categories were putatively assigned based on the assignment of gene function in B. subtilis and the completed S. aureus genomes Mu50 and N315. Bars representing Mu50 (open bars) show the approximate percentage of genes in each corresponding category. A breakdown of individual genes, gene names, and categories is available at http://www.cbc.umn.edu/ResearchProjects/AGAC/Staph/staphhome.html.
Patterns of nucleotide substitution.
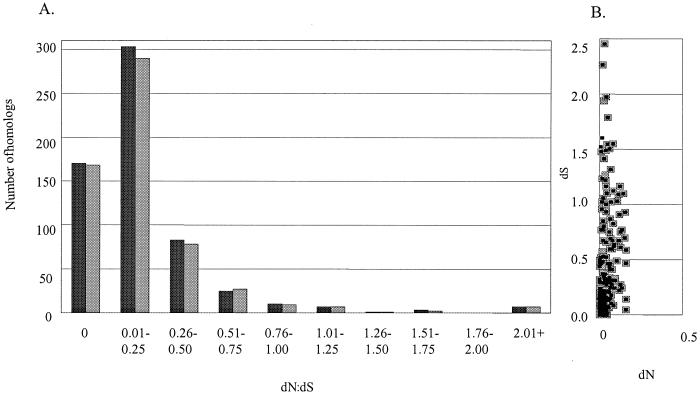
To understand the level and nature of nucleotide variation between RF122 and the recently sequenced human clones of S. aureus, amino acid substitution rates for the aligned sequences were calculated according to the algorithm of Nei and Gojobori (12). For genes with integral roles in bacterial survival and fitness, the rate of nonsynonymous substitution (dN) is often lower than the rate of synonymous substitution (dS) because these genes are under functional constraint and the majority of phenotypic changes will be disadvantageous. The majority of dN:dS ratios of the RF122 S. aureus sequences characterized thus far in comparison with Mu50 and N315 fall in the range of 1:10 to 1:4, corresponding to the most highly conserved sequences (Fig. 2A). The low average ratio of dN:dS for RF122 compared to Mu50 and N315 indicates that the majority of S. aureus genes are under functional constraint (Fig. 2B). Several of the highly conserved genes encoded virulence factors, including clumping factor B, as well as metabolic genes from energy, lipid, nucleic acid, and amino acid pathways (Table 1). The analysis also identified 19 orthologs with dN:dS ratios greater than 1, indicating that mutations that result in a change in the primary structure of the proteins have been conserved in these sequences. While the majority of the protein products from these genes were hypothetical, a urease subunit, an ammonium transporter, the cell division protein FtsH, the surface protein and antibiotic resistance determinant Drp35, and a leukotoxin subunit were also included in this category. The leukotoxin subunit is of particular interest in the context of host specificity, as leukotoxins interact directly with host erythrocytes and phagocytes to cause potentially catastrophic inflammation. It has been suggested that leukotoxins may play a role in causing clinical mastitis, and leukotoxin genes are present in all bovine strains tested, but a causal relationship between leukotoxin production and mastitis remains to be established (3, 13). Should leukotoxins be a significant factor in causing mastitis, positive selection of phenotype-altering mutations in the F subunit of S. aureus leukotoxin may be related to differences in the bovine and human immunological environments.
FIG. 2.
(A) Distribution of dN:dS ratios among genes, calculated according to the algorithm of Nei and Gojobori (12), using the finished S. aureus genomes Mu50 (black bars) and N315 (gray bars) in comparison with the RF122 sequences. Ratios greater than 1.00 represent genes with a higher rate of nonsynonymous than synonymous substitution and constitute possible evidence of positive selection. Ratios of 0.00 result from genes without nonsynonymous substitutions, but the rate of synonymous substitution for these genes varies. (B) Synonymous substitution versus nonsynonymous substitution rates of RF122 compared with Mu50 (black squares) and N315 (gray squares).
TABLE 1.
Substitution rate ratios among RF122 genes and corresponding functional categories
| dN:dS ratio and gene | Ratio | Product | Category | |
|---|---|---|---|---|
| <0.035 (highly conserved) | ||||
| fnb | 0.008 | Fibrinogen-binding protein | Virulence factors | |
| clpL | 0.014 | Clp proteinase chain | Protein fate | |
| sdrC | 0.016 | Conserved hypothetical protein | Putative virulence factors | |
| guaA | 0.017 | GMP synthase | Purines, pyrimidines, nucleosides, nucleotides | |
| sdrD | 0.019 | Conserved hypothetical protein | Putative virulence factors | |
| clfB | 0.019 | Clumping factor B | Virulence factors | |
| gntK | 0.024 | Glucokinase | Energy metabolism | |
| hlgC | 0.025 | Gamma hemolysin component C | Virulence factors | |
| opuCD | 0.025 | Hypothetical protein | Putative transport and binding protein | |
| sdrE | 0.026 | Conserved hypothetical protein | Putative virulence factors | |
| lpl2 | 0.030 | Hypothetical protein | Putative virulence factors | |
| apt | 0.033 | Adenine phosphoribosyl transferase | Purines, pyrimidines, nucleosides, nucleotides | |
| aroC | 0.033 | Chorismate synthase | Amino acid biosynthesis | |
| oppD | 0.034 | Hypothetical protein | Putative transport and binding proteins | |
| >1.00 (positive selection)
|
||||
| lukD | 1.115 | Leukotoxin F subunit | Toxins | |
| ebhB | 1.467 | Hypothetical cell wall enzyme | Cell surface components | |
| tag | 1.560 | DNA-3-methyladenine glycosidase | DNA metabolism | |
| ftsH | 1.587 | Cell division protein | Cellular processes | |
| ureC | 2.198 | Urease alpha subunit | Central intermediary metabolism | |
| SAB0296 | 2.450 | Similar to exotoxin 4 | Putative virulence factors | |
| drp35 | 3.113 | Surface protein | Antibiotic resistance | |
| SAB0711 | 4.417 | Unknown | Unknown | |
| SAB025 | 5.324 | Unknown | Unknown | |
Concluding comments.
In sum, this preliminary analysis of the genome of the common clone of bovine S. aureus reveals the presence of numerous genes and sequences that differentiate this isolate from previously characterized strains of the species. Our investigation also provides strong evidence that the whole genome sequencing approach will build a solid foundation for future investigation of the mechanisms of virulence and host specificity of this important human and animal pathogen.
Acknowledgments
We thank Barbara May, Michael Paustian, and Ling-Ling Li for their assistance with the preparation and analysis of the library and sequences. We also appreciate the support of Megan Lillehei and Todd Markovitz at the Advanced Genetic Analysis Center at the University of Minnesota for assistance with automated DNA sequencing.
This research is funded by a competitive award from the USDA-NRI Sustaining Animal Health and Well-Being Program (to V.K.). L.L.H. is supported by NIH NIGMS Biological Process Technology Institute fellowship GM08347.
Editor: V. J. DiRita
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cucarella, C., C. Solano, J. Valle, B. Amorena, I. Lasa, and J. R. Penades. 2001. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J. Bacteriol. 183:2888-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fitzgerald, J. R., W. J. Meaney, P. J. Hartigan, and C. J. Smyth. 2000. Molecular population and virulence factor analysis of Staphylococcus aureus from bovine intramammary infection. J. Appl. Microbiol. 88:1028-1037. [DOI] [PubMed] [Google Scholar]
- 4.Fitzgerald, J. R., S. R. Monday, T. J. Foster, G. A. Bohach, P. J. Hartigan, W. J. Meaney, and C. J. Smyth. 2001. Characterization of a putative pathogenicity island from bovine Staphylococcus aureus encoding multiple superantigens. J. Bacteriol. 183:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapur, V., W. M. Sischo, R. S. Greer, T. S. Whittam, and J. M. Musser. 1995. Molecular population genetic analysis of Staphylococcus aureus recovered from cows. J. Clin. Microbiol. 33:376-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 7.May, B. J., Q. Zhang, L-L. Li, M. L. Paustian, T. S. Whittam, and V. Kapur. 2000. Complete genomic sequence of Pasteurella multocida, Pm70. Proc. Natl. Acad. Sci. USA 98:3460-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McNamara, P. J., K. C. Milligan-Monroe, S. Khalili, and R. A. Proctor. 2000. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J. Bacteriol. 182:3197-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami, H., H. Matsumaru, M. Kanamori, H. Hayashi, and T. Ohta. 1999. Cell wall affecting antibiotics induce expression of a novel gene, drp35, in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 264:348-351. [DOI] [PubMed] [Google Scholar]
- 10.Musser, J. M., P. M. Schlievert, A. W. Chow, P. Ewan, B. N. Kreiswirth, V. T. Rosdahl, A. S. Naidu, W. Witte, and R. K. Selander. 1990. A single clone of Staphylococcus aureus causes the majority of cases of toxic shock syndrome. Proc. Natl. Acad. Sci. USA 87:225-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Musser, J. M., and R. K. Selander. 1990. Genetic analysis of natural populations of Staphylococcus aureus, p 59-67. In R. Novick and R. A. Skurray (ed.), Molecular biology of the staphylococci. VCH Publishers, New York, N.Y.
- 12.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 13.Schuberth, H., C. Krueger, H. Zerbe, E. Bleckmann, and W. Leibold. 2001. Characterization of leukocytotoxic and superantigen-like factors produced by Staphylococcus aureus isolates from milk of cows with mastitis. Vet. Microbiol. 82:187-199. [DOI] [PubMed] [Google Scholar]
- 14.Sompolinsky, D., Z. Samra, W. W. Karakawa, W. F. Vann, R. Schneerson, and Z. Malik. 1985. Encapsulation and capsular types in isolates of Staphylococcus aureus from different sources and relationship to phage types. J. Clin. Microbiol. 22:828-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sordelli, D. O., F. R. Buzzola, M. I. Gomez, L. Steele-Moore, D. Berg, E. Gentilini, M. Catalano, A. J. Reitz, T. Tollersrud, G. Denamiel, P. Jeric, and J. C. Lee. 2000. Capsule expression by bovine isolates of Staphylococcus aureus from Argentina: genetic and epidemiologic analyses. J. Clin. Microbiol. 38:846-850. [DOI] [PMC free article] [PubMed] [Google Scholar]