Abstract
Expression of type IV pili (Tfp) correlates with the ability of Neisseria gonorrhoeae to colonize the human host, as well as with adherence to human epithelial tissue, twitching motility, competence for natural transformation, and autoagglutination. N. gonorrhoeae PilF (required for Tfp biogenesis) and PilT (required for twitching motility and transformation) share significant identities with members of a family of putative ATPases involved in membrane trafficking of macromolecules. An open reading frame downstream of the pilT locus encoding a 408-amino-acid protein with 33% identity with the gonococcal PilT protein and 45% identity with the PilU protein in Pseudomonas aeruginosa was characterized, and the corresponding gene was designated pilU. Unlike N. gonorrhoeae pilT mutants, pilU mutants express twitching motility and are competent for DNA transformation. However, loss-of-function mutations in pilU increased bacterial adherence to ME-180 human epithelial cells eightfold and disrupted in vitro Tfp-associated autoagglutination. Comparative alignment of N. gonorrhoeae PilU with other members of the TrbB-like family of traffic ATPases revealed a conserved carboxy-terminal domain unique to family members which are not essential for Tfp biogenesis but which specifically modify Tfp-associated phenotypes. Studies of the pilT-pilU locus by using Northern blotting, transcriptional fusions, and reverse transcription-PCR showed that the two genes encoding closely related proteins with dissimilar effects on Tfp phenotypes are transcribed from a single promoter.
Neisseria gonorrhoeae type IV pili (Tfp) act as a primary factor in the colonization of human mucosal epithelium (13, 31). Identification of the precise roles served by these organelles in the host-microbe encounter has been complicated because their expression correlates with the elaboration of diverse phenotypes, each of which may contribute to the interaction, including adherence for human epithelial cells, bacterial autoagglutination, and twitching motility. The correlations have been defined mainly by the absence of the phenotypes in mutants failing to express Tfp, but the molecular nature of the associations remains unclear. It has therefore been of particular interest to isolate mutants which are altered in these properties but retain organelle expression.
Twitching motility is a novel mode of flagellum-independent movement on solid surfaces and is also found in Pseudomonas aeruginosa (4, 37) and Synechocystis species (2), as well as in Myxococcus xanthus, in which it is manifested as social gliding motility (43). In N. gonorrhoeae and P. aeruginosa, twitching motility has been shown to be related mechanistically to Tfp fiber retraction (20, 28). Mutations in the highly conserved pilT genes encoding members of a large family of putative nucleotide binding proteins (TrbB-like proteins) lead to a piliated but nonmotile phenotype in all four organisms (2, 37, 39, 42). Members of this protein family are distinguished by conserved motifs common to proteins with ATP binding and hydrolysis activities (18). It has been well established in systems involving Tfp biogenesis and extracellular protein secretion that mutations in the Walker A consensus sequence (GXXXXGKT/S) and other conserved domains lead to loss of function (3, 22, 34). The PilT proteins share the highest degrees of identity of any of the conserved Tfp biogenesis-function components found in these organisms. The less related BfpF protein of enteropathogenic strains of Escherichia coli appears to be orthologous to PilT since bfpF mutants continue to express Tfp (referred to as bundle-forming pili [Bfp]) but are defective in bacterial aggregate dispersal, an apparently physiological equivalent of twitching motility (3). In addition, pilT mutants of gonococci, M. xanthus, and enteropathogenic strains of E. coli strains each exhibit increased Tfp-dependent cell-cell aggregation (1, 39, 42). Although the roles of PilT proteins in these processes remain unclear, PilT has been demonstrated to function as a conditional antagonist of stable fiber formation (40, 41).
Study of the P. aeruginosa pilT locus revealed the presence of a downstream gene whose product was closely related to PilT but shared less identity with other members of the TrbB-like family of proteins (38). Like P. aeruginosa pilT mutants, pilU mutants were reported to be hyperpiliated and twitching motility defective but remained sensitive to phage infectivity. Both types of mutants are also attenuated in virulence in a mouse model of acute pneumonia (6). In this paper, we describe the identification and characterization of an N. gonorrhoeae gene distal to pilT which encodes a protein exhibiting the highest level of identity to P. aeruginosa PilU. Based on this property and its linkage to pilT, the gonococcal gene was designated pilU. We detail here the influence of PilU on Tfp-dependent human cell adherence and multicellular aggregative behavior. Further characterization of the effects of mutations in the pilT-pilU locus revealed that despite the high degree of conservation in both gene order and protein structure, significant differences appear to exist between P. aeruginosa and gonococci with regard to the biology of the systems. Unique features of the findings for gonococci led us to propose a model in which PilU exerts is effects downstream of those exerted by PilT.
MATERIALS AND METHODS
Bacterial strains, plasmids, and transposon mutagenesis.
Bacterial strains and cloning vectors used in this study are listed in Table 1. Gonococcal strains were propagated on solid Gc medium at 37°C in 5% CO2 or in this medium lacking agar and preincubated overnight in 5% CO2 (15). E. coli strains HB101 (GIBCO BRL) and BL21(DE3) were used for plasmid cloning and T7 expression experiments, respectively. All E. coli strains and their recombinants were maintained in Luria-Bertani medium supplemented with the appropriate antibiotics.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference or source |
|---|---|---|
| N. gonorrhoeae strains | ||
| N400 | Derived from VD300,a contains the IPTG-inducible recA6 alleleb | 11 |
| GF2 (PilF−) | pilF::m-Tn3erm | 11 |
| GT3 | pilT::m-Tn3erm | 40 |
| GT7 | pilT::m-Tn3erm | 40 |
| GT17 | pilT::m-Tn3cm at position 73c | This study |
| GT26 | pilT::m-Tn3erm | 40 |
| GT50 | pilT::m-Tn3erm at position 1105c | This study |
| GT101 | pilTdud1 | 40 |
| GT102 | pilTΔQSL | 40 |
| GT103 | pilT::m-Tn3erm | 40 |
| GT104 | pilTind | 40 |
| GT105 | pilTfs164 | 40 |
| GTU2 | pilT::m-Tn3cm pilU::mTn3erm | This study |
| GTXE1 | pilT::xylE | This study |
| GUXE2 | pilU::xylE | This study |
| GU2 | pilU::m-Tn3erm at position 1591c | This study |
| GU4 | pilU::m-Tn3erm at position 1714c | This study |
| GU5 | pilU::m-Tn3erm at position 2102c | This study |
| GU21 | pilU::m-Tn3erm at position 1314c | This study |
| GU33 | pilU::m-Tn3erm at position 2541c | This study |
| GU127 | pilU::m-Tn3erm at position 1192c | This study |
| E. coli strains | ||
| BL21(DE3) | F−ompT rB− mB− with prophage λ carring the T7 RNA polymerase gene | 32 |
| HB101 | F−mcrB mrr hsdS20(rB− mB−) recA13 supE44 ara14 galK2 lacY1 proA2 rpsL20(Smr) xyl5 λ−leu mtl1 | GIBCO BRL |
| Plasmids | ||
| pBluescript II SK | Cloning vector, Ampr | 32 |
| p6/16/11 | 1.1-kb SalI fragment in pBluescript II SK | This study |
| p11/2/13 | 3.3-kb EagI/XbaI fragment in pBluescript II SK | This study |
| pHSS6 | Vector for shuttle mutagenesis, Kanr | 27 |
| p1-49 | 3.3-kb EagI/XbaI fragment in pHSS6 | This study |
| pT7-5 | T7 RNA polymerase promoter λ10, Ampr | 32 |
| pT7U | pilU in pT7-5 | This work |
| pT7U(ΔSalI) | SalI deletion subclone of pT7U | This work |
Transposon mutagenesis was carried out by using the Tn3 derivative m-Tn3erm, as described previously (27). A 3.3-kb EagI/XbaI fragment was subcloned from λ clone 18/4 (17) into pBluescript II SK to give clone p11/2/13. This 3.3-kb fragment containing most of the pilU open reading frame (ORF) was removed from p11/2/13 by EagI/XbaI digestion. The EagI site was filled in with the Klenow fragment of DNA polymerase I. The fragment was then cloned into ClaI (blunted) and XbaI sites of the mutagenesis vector pHSS6 to create p1-49. The isolation and localization of transposon insertions were determined as described previously (11). The selected insertions were transformed into N400, and the transformants were obtained by selection on agar plates containing 8 μg of erythromycin ml−1.
DNA sequencing and sequence analysis.
Restriction fragments from lambda phage clone 18/4 (17) were gel purified and cloned in the vector pBluescript II SK by using appropriate sites. The plasmid subclones, p11/2/13 and p1/16/11 carrying a 3.3-kb EagI/XbaI fragment and a 1.1-kb SalI DNA fragment, respectively, were used as templates for DNA sequencing. The complete sequences of pilU were determined by the dideoxy chain termination method (25). The sequencing reactions were carried out with a modified form of T7 DNA polymerase (Sequenase 2.0; United States Biochemicals) along with standard vector-based and customized oligonucleotide primers. The nucleotide sequences of the pilE gene of the gonococcal mutants were determined by thermal cycle sequencing of PCR products by using Circumvent (New England Biolabs Inc.) and customized oligonucleotide primers. A partial sequence of pilU spanning nucleotides 765 to 1444 appears in the EMBL/GenBank/DDBJ nucleotide sequence data libraries under accession number L11719. DNA and protein sequence data were compiled and analyzed by computer by using both the MACVECTOR 3.5 (International Biotechnologies Inc.) and University of Wisconsin Genetics Computer Group software packages (8).
Measurement of transformation frequencies.
Competence for natural transformation of gonococcal strains was assessed as described previously by using chromosomal DNA from rifampin- and nalidixic acid-resistant mutants of VD300 (44), as well as plasmid pSY6 DNA carrying a gonococcal gene responsible for low-level nalidixic acid resistance (30). Transformants were selected on agar plates containing 10 μg of nalidixic acid ml−1 or 5 μg of rifampin ml−1 for chromosomal DNA and 1 μg of nalidixic acid ml−1 for plasmid DNA.
ME-180 cell adherence assay.
The adherence assay was performed by using ME-180 cells (human epidermoid carcinoma) obtained from the American Culture Collection. ME-180 cells were maintained in McCoy's 5A medium supplemented with 10% fetal bovine serum. These epithelial cells were seeded into a 24-well tissue culture plate at a density of 5 × 105 cells the day before infection. For assays, 2 × 107 to 5 × 107 bacteria in 1 ml of McCoy's 5A medium supplemented with 1% fetal bovine serum (assay medium) were added to each well containing a monolayer and incubated at 37°C in a 5% CO2 incubator. After 1 h, nonadherent bacteria were removed by washing the wells five times with assay medium. The monolayers and cell-associated bacteria were then recovered by treatment with 0.25% trypsin for 5 min at 37°C. The recovered bacteria were plated on agar after dilution. The number of cell-associated bacteria was determined by determining the number of CFU. Adherence was quantified by determining the ratio of cell-associated CFU to total CFU of the inoculum. The results are presented below as the ratio of the adherence of the strains tested to the adherence of the wild-type controls.
Immunofluorescence microscopy.
Immunofluorescence microscopy was performed to directly visualize the binding of gonococci to human epithelial cells. ME-180 cells were cultivated in 24-well plates on glass coverslips until they were confluent, and they were infected with various gonococcal strains by using the method described above for the adherence assay. The cells were washed five times with phosphate-buffered saline to remove nonadherent bacteria and then fixed with methanol on ice for 5 min. The coverslips were air dried and rinsed again with phosphate-buffered saline. Fixed cells on coverslips were incubated with 30 μl of fluorescein-labeled mouse monoclonal antibodies specific to gonococcus (N. gonorrhoeae culture confirmation test kit; Syva Microtrak) for 15 min at 37°C according to the manufacturer's specifications. The coverslips were rinsed with distilled water and thoroughly air dried before they were mounted on slides. Cells were viewed with a Leitz orthoplan microscope.
Transmission electron microscopy.
Electron microscopy samples were prepared on 3.05-mm Formvar-carbon-coated grids (Tousimis Research Corp.). Prior to use, the grids were discharged in a high-vacuum evaporator (DENTON model DV-502) to facilitate spreading of both bacteria and the stain. Grids were gently touched to individual bacterial colonies grown for 16 h on Gc agar plates. The grids were each air dried for 10 min, floated on a drop of 0.5% ammonium molybdate (pH 7.0) for 30 s, and air dried again. The specimens were examined with a Philips CM-10 transmission electron microscope.
T7 RNA polymerase expression systems.
The PilU proteins were expressed in E. coli by using the T7 expression vector pT7-5 (32). The pilU gene from N400 was cloned after PCR amplification by using the oligonucleotides UL (GCTCTAGAGCCTTCCTGTTGAAACCTGCC), creating an XbaI site (underlined), and UR (CCATCGATACGGCATTGGGTTTTGCGGAT), creating a ClaI site (underlined). Vent DNA polymerase (New England Biolabs Inc.) was used according to the manufacturer's specifications. The PCR products were purified by agarose gel electrophoresis after digestion with ClaI and XbaI and ligated directionally into pT7-5 cleaved with the same enzymes, creating pT7U. In order to generate the deletion in pilU, pT7U was digested with SalI and recircularized. The resultant plasmid, pT7U(ΔSal), contains an in-frame deletion of a SalI fragment (303 bp) in the pilU gene. These constructs, pT7U and pT7U(ΔSalI), were then transformed into E. coli BL21(DE3), which contains a T7 RNA polymerase gene under lacP control. Proteins were expressed under standard conditions, except that Luria-Bertani media instead of defined minimal media were used.
SDS-PAGE and immunoblotting.
For detection of the proteins, whole-cell lysates were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and immunoblotting. Protein samples were separated on 12.5% acrylamide gels and then transferred to Immobilon-PSQ polyvinylidene difluoride membranes (Millipore Corp.). Filters were incubated with rabbit polyclonal pilin-specific antibody GC2-66 (11) or rabbit polyclonal anti-PilT serum (PilT2B), either crude or affinity purified. Antigen detection was performed by using alkaline phosphate-conjugated goat anti-rabbit antibodies (Tago Inc.) and a colorimetric phosphate substrate. Details of the methods used for sample preparation, SDS-PAGE, and antigen detection have been published previously (14). To affinity purify antibodies from rabbit serum, a variation of the method of Weinberger et al. (36) was employed by using truncated PilU (PilUΔSal) expressed in E coli.
Construction of xylE transcriptional reporter fusion.
The pilT::xylE and pilU::xylE chromosomal fusion strains were generated by genomic integration (9) of a plasmid that carried the promoterless xylE gene fused to either 3′ deleted pilT or 3′ deleted pilU. The p11/2/7 plasmid (39), containing the pilT gene and the incomplete pilU gene, was used for fusion of the 1.0-kb xylE cassette, generating pTxylE and pUxylE. The xylE structural gene was derived from plasmid pXYLE10 (29) as an SmaI fragment, and the ends were filled in by using the Klenow fragment of E. coli DNA polymerase. The xylE gene was then inserted into the XmnI site of pilU in p11/2/7, generating pUxylE. Plasmid pUxylE carries a functional pilT gene, the whole intergenic region between pilT and pilU, and 38 bp of the pilU coding region fused to the xylE cassette. Plasmid pTxylE was constructed similarly, by inserting the blunt-ended SmaI xylE cassette into an EcoRI fragment (DNA between the EcoRI site in the pilT gene and the EcoRI site in the polylinker)-deleted vector derived from p11/2/7 (39). The orientation of xylE cassettes in each construct was determined by measuring xylE expression. The fusions carried by pTxylE and pUxylE were introduced with the vector into the genome of Gc strain N400 via transformation as previously described (9) to obtain strains GTXE1 and GUXE2, respectively. Transformants were obtained by selection on agar plates containing 50 μg of kanamycin ml−1.
Plasmid p11/2/7 containing selected insertions in the pilT region was transformed into GUXE2, and the transformants were obtained by selection on agar plates containing 8 μg of erythromycin ml−1. The transposon insertions into either the upstream copy or the downstream copy of pilT were determined by PCR by using primers SS6-EcoRI (5′-GATCCCCACCGGAATTGCG-3′), SS6-Sma (5′-GATCCTCCAGCGCGGGG-3′), and Tn3R (5′-GTCAGAGGCAGAAAACGTTG-3′). Primers SS6-EcoRI and SS6-Sma read out from each end of the vector, and primer Tn3R is derived from the inverted repeat elements at the end of the transposon.
Assay of xylE expression (catechol 2,3-dioxygenase activity).
Functional expression of the xylE gene was detected by either plate assays or enzyme assays. For plate assays, E. coli or gonococcal transformants on antibiotic selection plates were sprayed with an aqueous solution of 0.5 M catechol. Colonies of bacterial cells that expressed xylE became yellow due to the conversion of catechol to 2-hydroxymuconic semialdehyde by catechol 2,3-dioxygenase. For enzymatic assays, gonococcal cultures were grown for 5 h at 37°C in Gc liquid media. Cells were harvested by centrifugation, washed in 50 mM potassium phosphate buffer (KBP) (pH 7.5), and resuspended in a lysis buffer (100 mM KBP [pH 7.5], 20 mM EDTA, 10% [vol/vol] acetone). Cells were lysed by incubation for 15 min at 37°C, 10 μl of 10% Triton X-100 per ml was added, and the extract was vortexed and placed on ice for 15 min. Cell debris was removed by centrifugation for 10 min in a microcentrifuge at 4°C. Catechol dioxygenase specific activities were determined spectrophotomerically as described previously (45). The assay buffer (50 mM KBP [pH 7.5], 1 mM catechol) was preincubated at 37°C, and reactions were initiated by adding cell extract (10 μl) to the assay buffer (290 μl). The optical density at 375 nm was measured over time for saturation (in most cases for 10 min). Enzyme activity was calculated as the rate of change in optical density at 375 nm per minute per microgram of protein. Protein concentrations of cell extracts were determined by the Bradford method by using bovine serum albumin as the protein standard.
Northern analysis.
Total RNA was isolated from various strains grown in liquid Gc media for 4 to 5 h by using the TRIzol reagent (GIBCO BRL) as recommended by the manufacturer. RNA (20 μg) was resolved in 1% (wt/vol) agarose-2.2 M formaldehyde gels and transferred overnight to nylon membranes (positively charged; Boehringer Mannheim). The transferred RNA was immobilized by UV cross-linking by using Stratalinker (Stratagene). The DNA fragments of pilT and pilU generated by PCR with primers complementary to each gene (PT EcoRV [5′-CCGGATATCATTCGGGTTCACGGCGACATGCGG-3′] and PT3′-1 [5′-TGCAGCGTGCCGAAAACCAAGTG-3′] for pilT and UL and UR for pilU) were used as probes. The PCR-generated fragments were gel purified and radiolabeled with [α-32P]dATP (specific activity, 3,000 Ci/mmol; Amersham) by using a random primed DNA labeling kit (Boehringer Mannheim). RNA sizes were determined by using an RNA ladder (United States Biochemicals) as the molecular size standard. Prehybridization of blots was done for 4 h at 42°C in a solution containing 50% formamide, 5× Denhardt's solution, 6× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate, pH 7.0), 0.5% SDS, and 100 μg of denatured salmon sperm DNA ml−1. Hybridization was done overnight at 42°C in the same solution with a probe. Blots were washed in 6× SSC at room temperature twice for 1 to 2 min, in 2× SSC-0.1% SDS at 60°C twice for 15 min, and in 0.1×SSC-0.1% SDS at 68°C twice for 15 min. After they were washed, the blots were rinsed in 6× SSC and exposed to Kodak X-Omat film at −70°C.
Probes were removed from nylon membranes between hybridizations. Each blot was stripped by washing it three times (10 min each) in boiling 0.5% SDS. The membrane was exposed to X-ray film for 2 days to check that the probe had been completely removed.
RT-PCR.
Total RNA was prepared as described above for Northern analysis and was further purified by treatment with RNase-free DNase. Reverse transcription (RT) was performed by using Superscript II reverse transcriptase (GIBCO BRL) with 10 μg of purified RNA per reaction mixture as instructed by the manufacturer. The antisense primer B6U (5′-GTGGAAGTGTGTGCCGATTTC-3′), designed to anneal to the 5′ end of pilU, was used to synthesize cDNA strands. The cDNA products were used as templates for PCR amplification with B6U and sense primer F4T (5′-CGTCATCTCCCAAAACCTGCT G-3′), which anneals to the 3′ end of pilT. The PCR was performed by using Taq polymerase (GIBCO BRL) for 25 cycles with an annealing temperature of 57°C and an extension temperature of 72°C. The resulting products were analyzed in a 0.8% agarose gel. Reactions in which reverse transcriptase was omitted were included as negative controls to eliminate the possibility that genomic DNA served as the template for the PCR.
Primer extension analysis.
Total RNA of Gc medium-grown strains was isolated as described above for Northern analysis. Oligonucleotides complementary to pilT (PET1 [5′-CGAAGG CGAGTAAGTCGGTAA TCTGC-3′] and PET2 [5′-GATATGCCCGAACTCAGGTGAAGGTCG-3′]) and pilU (REU1 [5′-GATGTCGTGCAGGTTATCGGTATTCAT-3′]) were used to map the 5′ termini of pilT and pilU mRNAs in Gc medium. Ten picomoles of each primer was 5′ end labeled with 33P by using 50 μCi of [γ-32P]ATP and phage T4 polynucleotide kinase (GIBCO BRL) according to the manufacturer's specifications. Approximately 0.2 pmol of labeled primer and 10 μg of RNA were mixed, and distilled H2O was added to bring the reaction volume to 12 μl. Reaction mixtures were incubated at 70°C for 10 min and quickly chilled on ice to allow annealing of the primers to the template. After brief centrifugation of each annealed mixture, 4 μl of 5× First Strand buffer, 2 μl of 0.1 M dithiothreitol, and 1 μl of a deoxynucleoside triphosphate mixture (10 mM dATP, 10 mM dGTP, 10 mM dCTP, and 10 mM dTTP) were added, and the preparation was incubated at 44°C for 2 min. One microliter of Superscript II reverse transcriptase (GIBCO BRL) was added, and the preparation was incubated at 44°C for an additional 50 min. The reaction was terminated by adding 5 μl of Sequenase stop buffer (United States Biochemicals). Extension products were analyzed by electrophoresis in a denaturing 5% polyacrylamide gel and were visualized by autoradiography.
Preparation of an anti-PilT polyclonal antiserum.
A protein fusion and purification system was used to produce a polypeptide corresponding to the PilT protein. A polyhistidine-tagged PilT was expressed in E. coli and purified from cell lysates by using an Ni2+-nitrilotriacetic acid-Sepharose column according to the manufacturer's instructions (Qiagen, Chatsworth, Calif.). The purified His-tagged protein was used as an immunogen to produce polyclonal antibodies in rabbits after the protein was analyzed by SDS-PAGE and Coomassie blue staining (16). The protein was emulsified in Freund's adjuvant and injected subcutaneously into rabbits at 2-week intervals for 6 to 8 weeks. After blood was tested for specific antibodies, the animals were sacrificed, and serum was collected.
Nucleotide sequence accession number.
The nucleotide sequences reported herein for N. gonorrhoeae pilU have been appended to EMBL/GenBank/DDB5 accession number S72391.
RESULTS
Identification of the gonococcal pilU gene downstream of pilT.
During a study designed to identify gonococcal genes sharing DNA sequence homology with the P. aeruginosa pilB gene, an ORF whose derived polypeptide was closely related to gonococcal PilT (termed ORF4) was found to overlap and to be divergently oriented relative to pilT (17). Brossay and colleagues (5) later showed that the region 5′ of pilT was rearranged in the plasmid clone from which the DNA sequence was determined and that the rearrangement occurred during subcloning of the pilT gene from a λ phage clone to a high-copy-number plasmid vector. To reexamine the location of these pilT-related sequences, a series of subclones of λ clone 18/4 were constructed by using restriction fragments which hybridized with an ORF4-derived probe (17). Two subclones, one containing a 3.3-kb EagI/XbaI DNA fragment and one containing a 1.1-kb SalI DNA fragment, were sequenced and found to contain overlapping segments of ORF4; the first clone contained the carboxy terminus, and the second contained the true amino terminus for ORF4 and the 3′ end of pilT. Sequencing of PCR products derived by using N400 genomic DNA as the template showed that the DNA sequence for this locus present in the genomic DNA was identical to that found in the lambda phage clone and the new plasmid subclones (data not shown).
By using a putative translational start site for ORF4 preceded by a potential ribosome binding site (5′-AGAAGC-3′) mapping 162 bp downstream of the termination codon for pilT, ORF4 should encode a protein with 408 amino acid residues and a predicted molecular mass of 45,695 Da. The region immediately upstream of ORF4 did not possess any identifiable consensus promoter sequence elements, while a 15-bp inverted repeat sequence containing the gonococcal DNA uptake sequence (12) that could possibly function as a transcriptional terminator was found just 3′ of this region.
In order to assess if ORF4 corresponded to a polypeptide of the size predicted from the nucleotide sequence, PCR-generated DNA fragments encompassing ORF4 and a derivative containing an in-frame deletion that removed 101 residues were cloned into the T7 promoter plasmid pT7-5. As determined by SDS-PAGE analyses of whole-cell lysates from cells containing the vector control, 46- and 34-kDa proteins were expressed from the ORF4 construct and the deletion derivative, respectively, following T7 RNA polymerase derepression. The relative migration positions of these proteins corresponded well with those predicted for ORF4 and the deletion derivative, and both of these species reacted specifically with polyclonal rabbit serum raised against a poly(His)-PilT fusion protein in immunoblots (Fig. 1, lanes 4 and 6). These findings confirmed that ORF4 encodes a polypeptide which shares antigenic determinants with gonococcal PilT, and the corresponding gene was designated pilU.
FIG. 1.
Detection of PilU expressed in E. coli. Plasmid-encoded PilU and truncated PilU were expressed from the T7 promoter and analyzed by immunoblotting with affinity-purified anti-PilT rabbit serum. Whole-cell lysates were prepared from E. coli strain BL21 carrying the pT7-5 vector control, pT7U, or pT7U(ΔSalI) grown in the absence (lanes 1, 3, and 5) or in the presence (lanes 2, 4, and 6) of isopropyl-β-d-thiogalactopyranoside (IPTG). Lanes 1 and 2, pT7-5; lanes 3 and 4, pT7U; lanes 5 and 6, pT7U(ΔSalI).
Construction and characterization of gonococcal PilU mutants.
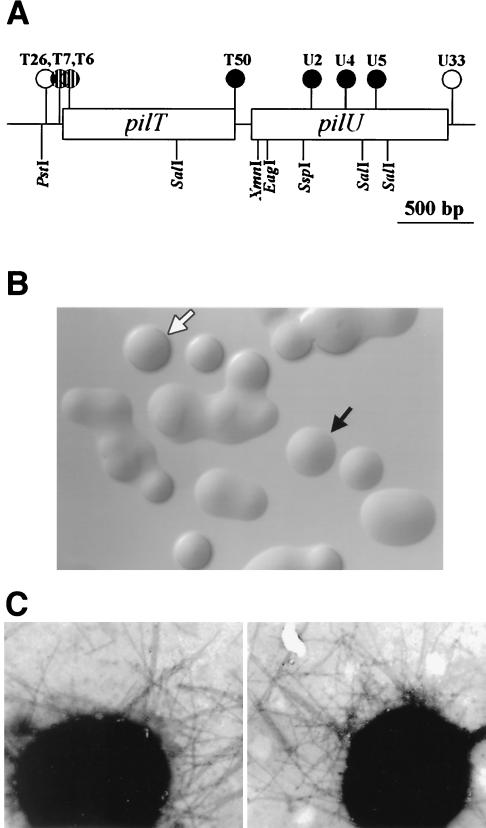
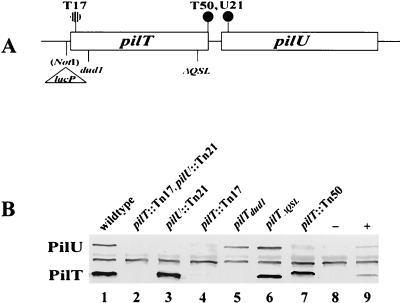
Plasmids containing restriction fragments encompassing pilU were subjected to transposon mutagenesis (26). After the sites of insertion were determined by DNA sequencing, selected mutants (Fig. 2A) were introduced into the chromosome of strain N400 by transformation. All mutants with transposons within the pilU ORF exhibited a flattened colony shape which was characteristic of strains lacking Tfp-associated autoagglutination (Fig. 2B). The nonautoagglutinating phenotype was also evident when the mutants were cultured in liquid media, in which they failed to form clumps of cells and grew as turbid suspensions (data not shown). A mutant carrying an insertion within the second-to-last codon of pilT (strain T50) also displayed the nonautoagglutinating colony phenotype. This was surprising, not only because the insertion mapped over 160 bp in front of the pilU ORF but also because all other transposon insertions in the pilT ORF result in a hyperautoagglutinating phenotype (39). Transformants containing a transposon inserted immediately downstream of the pilU ORF (U33) were indistinguishable from the parent strain in their growth patterns. Therefore, the changes in colony morphology and aggregative behavior observed in the mutants are not the result of effects on distal gene expression.
FIG. 2.
(A) Physical map of the gonococcal pilT-pilU locus. Transposon insertions are indicated by circles. The solid and striped circles indicate insertions resulting in PilU− and PilT− phenotypes, respectively, whereas the open circles indicate transposon insertions that had no effect on phenotypes. Restriction sites are also shown. (B) Colony morphologies of N. gonorrhoeae wild type (domed colony with a defined edge) (open arrow) and PilU− mutant (flat colony lacking a defined edge) (solid arrow). (C) Transmission electron micrographs of N. gonorrhoeae strains N400 (left panel) and GU2 (right panel). Other PilU− strains exhibited phenotypes identical to that exhibited by GU2. Bacteria were negatively stained with 0.5% ammonium molybdate. Magnification, ×39,000.
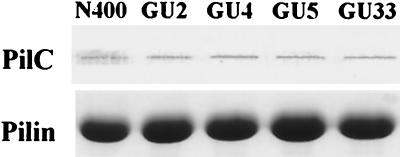
Since changes in Tfp-associated autoagglutination are often found in conjunction with alterations in expression of the pilin subunit, the complete pilE gene in all mutants was sequenced, but no changes were detected. Loss of autoagglutination is also associated with the absence or reduction of piliation. However, all the nonautoagglutinating mutants in this study had levels of piliation as determined by electron microscopy which were indistinguishable from that observed for the parental strain. Moreover, no differences in pilus fiber distribution, morphology, or lateral aggregation could be seen (Fig. 2C). In addition, the yields of purifiable Tfp from these strains were indistinguishable from one another, and no differences in migration of the pilin subunit or in levels of the copurifying PilC protein were seen (Fig. 3). In summary, no quantitative or qualitative differences in Tfp expression could be detected in association with the nonautoagglutinating phenotype of the pilU mutants.
FIG. 3.
Quantitative analysis of Tfp and PilC in pilU mutants. The amounts of samples loaded were equalized based on the total protein concentration of whole cells from which Tfp were purified. (Top) Immunoblot of purified pili obtained by using rabbit antibodies specific for PilC. (Bottom) Coomassie blue-stained SDS-PAGE gel showing the relative amounts of PilE in purified pili.
pilT mutants of the four species noted previously, as well pilU mutants of P. aeruginosa, do not express twitching motility. When colonies of gonococcal pilU mutants were examined microscopically, twitching motility could be detected as groups or flares of cells at the periphery moving in a jerky fashion. Unlike the gonococcal pilT mutants, which are defective in DNA uptake, the gonococcal pilU mutants display levels of transformation which are indistinguishable from those found for the wild-type parent (data not shown).
PilU mutants display increased adherence for human epithelial cells.
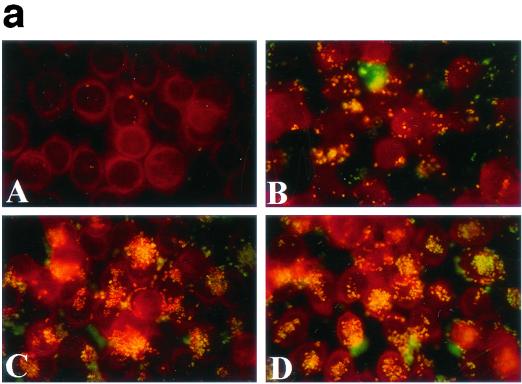
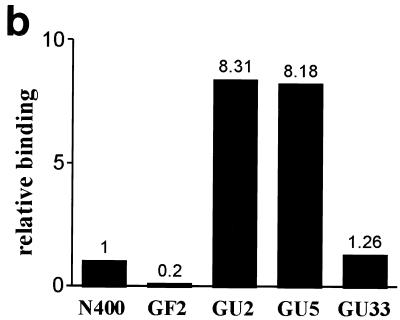
The defect in autoagglutination seen in gonococcal PilU mutants appeared to provide a unique opportunity to assess the contribution of this phenotype to Tfp-mediated interactions with human epithelial cells. An assay performed with human cervical carcinoma cell line ME-180 cells was employed, which measured the number of CFU of adherent bacteria which were recovered 1 h postinfection following washing of the monolayers. To rule out the influence of autoagglutination on the values of recovered CFU, the monolayers and associated bacteria were recovered in the presence of trypsin under conditions under which bacterial aggregates are dissociated. As shown in Fig. 4b, strains carrying insertions in the pilU gene adhered to ME-180 cells at levels more than eightfold greater than the level of adherence of the wild-type isogenic parent or the strain that carried the transposon 3′ of the pilU ORF. To confirm these findings, parallel experiments were performed in which infected monolayers were fixed after washing and stained with fluorescein-conjugated monoclonal antibodies specific for gonococci. As shown in Fig. 4a, this in situ technique visually confirmed that the PilU mutants (panels C and D) adhered to a greater extent than the wild-type parent (panel B). It was also observed that despite the nonaggregating phenotype of PilU mutants propagated on plates and in liquid media, these mutants adhered to epithelial cells in a localized fashion as clumps of bacterial aggregates rather than as isolated diplococci.
FIG. 4.
(a) Patterns of adherence of gonococcal strains to ME-180 cells. ME-180 cells were incubated for 1 h with bacteria, fixed, and stained with fluorescent conjugated monoclonal antibodies. (A) Nonpiliated strain GF2 (PilF−); (B) wild-type strain N400; (C) pilU mutant strain GU2; (D) pilU mutant strain GU5. (b) Increased adherence of PilU mutant strains to ME-180 cells. Adhesion assays were performed with ME-180 cells. The results are means of three experiments and are expressed as the ratios of the values for the mutants to the values for the wild-type control.
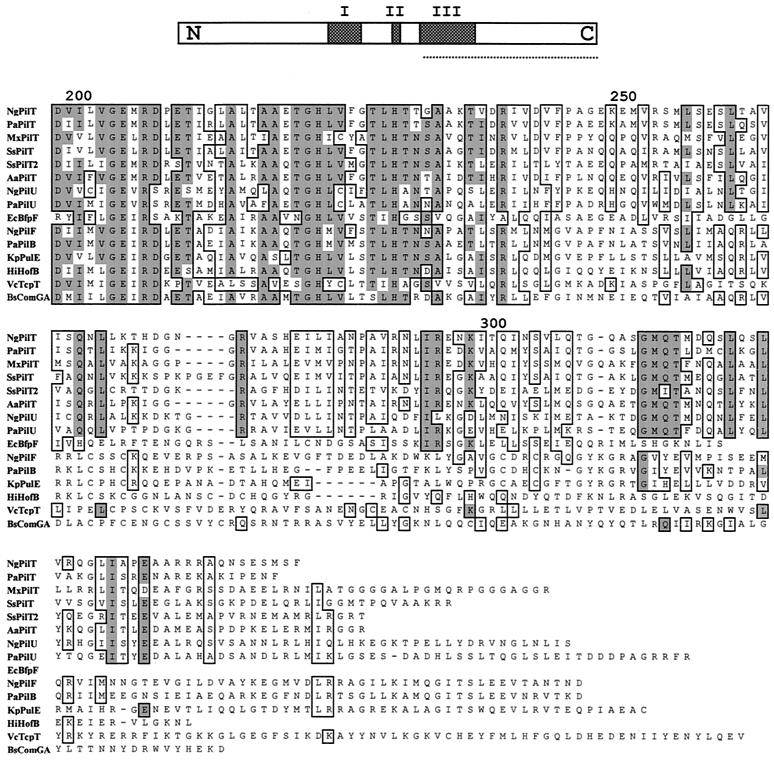
Sequence alignment of PilU with other TrbB-like proteins defines a novel subclass with extended carboxy-terminal residue conservation.
By using the TBLAST algorithm, gonococcal PilU was found to be most related to P. aeruginosa PilU (45% identity) and less related to the PilT proteins (33 to 35% identity), followed by other members of the TrbB-like protein family. TrbB-like proteins share three conserved domains (Fig. 5) related to ATP binding and hydrolysis, and amino acid sequence alignment of the PilU and PilT proteins showed that in addition to the sharing of these domains, a relatively high degree of identity extended into the carboxy-terminal segments. Extended searching and Clustal X alignment revealed that the carboxy-terminal motif is absent in related proteins required for Tfp biogenesis, type II secretion, conjugation, or type IV secretion but present in the PilT1 and PilT2 proteins of Synechocystis sp. strain PCC6803, which are required for phototactic motility associated with Tfp (2). The extended carboxy-terminal motif is also present in other uncharacterized TrbB-like protein ORFs, including a third family member in the genomes of N. gonorrhoeae and Neisseria meningitidis and single family members in Vibrio cholerae, Clostridium perfringens, and many other species (data not shown).
FIG. 5.
Amino acid sequence alignment of the carboxy-terminal segments of the GspE/TrbB family of proteins, determined by using Clustal X. Conserved residues are enclosed in boxes, and identical residues shaded. The alignment begins in the regions corresponding to conserved motif III as defined by Lessl and Lanka (top, underlined) (18), and numbering is based on the N. gonorrhoeae PilU sequence. The identical and similar residues unique to the PilT/PilU family members (dispensable for Tfp biogenesis but essential for associated phenotypes) begin after residue 250. This region contains the sequence motif GMQTXXXXLXXLXXXXXI (residues 313 to 333 of N. gonorrhoeae PilU). The following sequences are shown: NgPilT (N. gonorrhoeae, accession number AAB30824), PaPilT (P. aeruginosa, accession number P24559), MxPilT (M. xanthus, determined by Wu et al. [42]), SsPilT (Synechocystis sp., accession number BAA18564), SsPilT2 (Synechocystis sp., accession number BAA18443), AaPilT (Aquifex aeolicus, accession number AAC06903), NgPilU (N. gonorrhoeae, sequence determined in this study), PaPilU (P. aeruginosa, accession number S54702), EcBfpF (E. coli, accession number S70973), NgPilF (N. gonorrhoeae, accession number P37094), PaPilB (P. aeruginosa, accession number P22608), KpPulE (Klebsiella pneumoniae, accession number C34469), HiHofB (Haemophilus influenzae, accession number P44622), VcTcpT (V. cholerae, accession number P29480), and BsComGA (Bacillus subtilis, accession number B30338).
PilU expression in gonococci is influenced by a subset of pilT alleles.
Despite the high degree of identity shared by N. gonorrhoeae PilT and PilU, the autoagglutination phenotypes of pilT and pilU mutants were opposite, with the former mutants hyperautoagglutinating and the latter mutants lacking autoagglutination. To examine this phenomenon in more detail, a double mutant carrying transposon insertions in both pilT and pilU was constructed, and this mutant was found to be phenotypically identical to the mutants carrying only pilT mutations. It appeared, then, that mutations in pilT were epistatic to those in pilU with regard to all measurable phenotypes. As controls, immunoblotting of the single and double mutants was performed with antibodies raised against the PilT fusion protein which were affinity purified by using the truncated PilU protein expressed in E. coli. Both PilT and PilU were detected in the wild-type strain and were absent in the double mutant (Fig. 6B, lanes 1 and 2), while only PilT was detected in the pilU transposon mutant (lane 3). Surprisingly, the strain carrying the transposon insertions in the pilT ORF failed to express detectable levels of PilU (lanes 4 and 7). In the case of the mutant with the transposon site in the second-to-last codon of the pilT ORF (T50) (lane 7), PilT migration was decreased, apparently because of the extended, alternate reading frame created by the transposon insertion. PilU was detected in strains expressing the dud1 (a frameshift mutation) and the ΔQSL (an in-frame deletion of 9 bp) pilT alleles. In addition, neither PilT nor PilU was detected in a strain in which pilT was expressed from a regulated promoter unless the strain was propagated under conditions under which transcription was derepressed (lanes 8 and 9).
FIG. 6.
PilT and PilU expression in wild-type and mutant strains. (A) Physical map of the strains used. The solid and striped circles indicate transposon insertions resulting in PilU− and PilT− phenotypes, respectively. Names and sites of other mutations, as well as the restriction site created for construction of the pilTind allele (40), are also shown. (B) Immunoblot of whole-cell lysates from the wild type (lane 1), GTU2 (lane 2), GU21 (lane 3), GT2-17 (lane 4), GT101 (lane 5), GT102 (lane 6), GT50 (lane 7), and GT104 (pilTind) (lanes 7 and 8) with (lane +) or without (lane −) induction (200 μM isopropyl-β-d-thiogalactopyranoside). Immunoblots were probed with affinity-purified polyclonal anti-PilT serum.
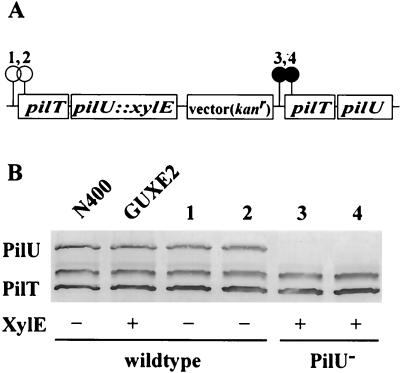
Expression of gonococcal pilU is coupled transcriptionally to pilT.
One possible explanation for the results described above is that the PilT protein is required for stabilization of the PilU polypeptide. For this to be true, however, some truncated versions of PilT must function in that capacity while others do not. A second possible explanation is that the two genes are cotranscribed from a single promoter and that the failure to detect PilU in the transposon mutants is due to the polar effects of the mutations. To distinguish between these two scenarios, strain GUXE2 was constructed, in which a tandem duplication of the pilT-pilU locus was created in conjunction with a transcriptional fusion of an xylE reporter gene to the upstream pilU gene copy (Fig. 7A). This configuration made it possible to assess the effects of two transposon insertions (one in the promoter region and one in the beginning of the pilT ORF) on both PilU expression and its related phenotypes, as well as pilU transcription in an otherwise wild-type background. Transposon mutations in the upstream copy of pilT eliminated XylE activity but had no influence on either PilU expression or the autoagglutination phenotype (Fig. 7B, lanes 1 and 2), while downstream insertions had no effect on XylE activity but eliminated PilU expression and led to the nonautoagglutinating phenotype (Fig. 7B, lanes 3 and 4).
FIG. 7.
Analysis of gene expression in the pilT-pilU locus with an xylE fusion construct. (A) Genomic configuration of strain GUXE2 created by integration of an xylE fusion plasmid (described in the text) into the genome of N400. The positions of transposon insertions in derivatives of strain GUXE2 which carry a duplication of the pilT gene and the flanking region are indicated by circles. The numbers above the circles correspond to lane numbers in panel B. (B) Immunoblot of whole-cell lysates from wild-type and fusion strains obtained by using affinity-purified polyclonal anti-PilT serum. The XylE activity was measured by the spray method as described in Materials and Methods, and the results were scored positive or negative. The phenotypes of each strain analyzed were noted.
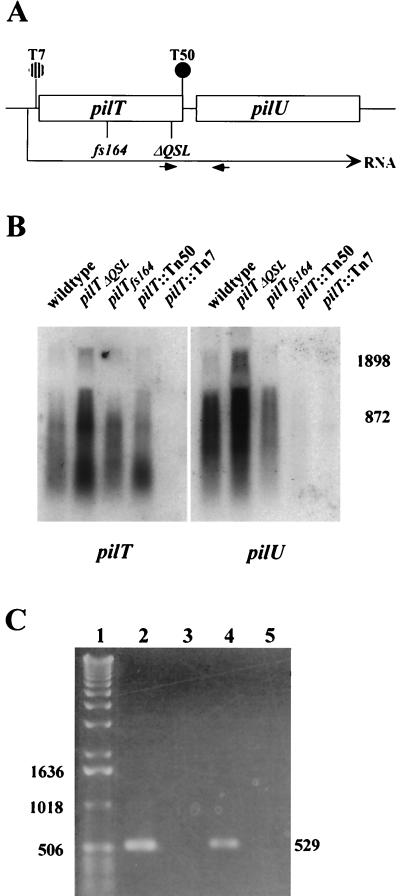
Next, the patterns of transcription within the pilT-pilU locus in wild-type and mutant backgrounds were examined directly. Figure 8 shows Northern blots of total RNAs hybridized with pilT- and pilU-specific probes. When RNA from wild-type cells was used, the pilT probe detected a weakly hybridizing species that was more than 2,300 bases long and reactive species that ranged in size from 1,200 bases down to 300 bases. Similar results were obtained with the RNAs from the pilTΔQSL and and pilTfs164 backgrounds, except that the former RNA sample appeared to contain increased levels of all the hybridizing species. In the RNA from cells with the T50 transposon insertion, there appeared to be a shift in distribution down to the smaller size range. The pilU probe appeared to detect the same weakly hybridizing species at sizes greater than 2,300 bases and a range of reactive species whose sizes were shifted slightly up from the sizes seen with the pilT probe, but only a faint signal was obtained with the RNA from cells with the T50 transposon insertion. As was the case for the pilT probe, the pilTΔQSL RNA contained increased levels of all the pilU hybridizing species. No signals were detected by using either probe and RNA from cells harboring the transposon insertion in the promoter region of pilT. These patterns of hybridization implied that the pilT- and pilU-specific transcripts might be generated by processing of a polycistronic mRNA.
FIG. 8.
Transcription patterns within the pilT-pilU locus in wild-type and mutant strains. (A) Physical map of the locus in the strains analyzed. (B) Northern blot analyses of pilT and pilU expression. Total RNAs (10 μg) from wild-type and mutant strains were electrophoresed on a formaldehyde-agarose gel and blotted onto a nylon membrane. The transferred RNA was hybridized with a pilT probe, yielding the autoradiogram shown on the left. The membrane was then washed and rehybridized with the pilU probe, yielding the autoradiogram shown on the right. The positions of RNA size markers (in base pairs) are shown on the right. (C) Detection of an RNA species spanning pilT and pilU by RT-PCR. Lane 1, 1-kb DNA ladder; lane 2, positive control amplification of intergenic region with wild-type strain chromosomal DNA as the template and with the primers indicated by small arrows in panel A (the expected PCR product is 529 bp long); lane 3, negative control for RT-PCR, in which the conditions were identical to those used to obtain the product in lane 4, except that reverse transcriptase was not added to the reaction mixture; lane 4, RT-PCR with RNA from the wild-type strain; lane 5, RT-PCR with RNA from GT7.
These results, together with the other data, suggested that pilT and pilU are organized in an operon. RT-PCR was used, therefore, to confirm the presence of a single RNA species encompassing the 3′ end of pilT and the 5′ end of pilU. As shown in Fig. 8C, a DNA fragment corresponding to the predicted 529-bp product was obtained after positive control amplification of the intergenic region with genomic DNA as the template (lane 2). The same product was obtained in an RT-PCR when wild-type RNA was used (lane 4) but was not seen in control reactions performed with wild-type RNA in the absence of reverse transcriptase (lane 3) or with RNA from cells harboring the transposon insertion in the promoter region of pilT (lane 5).
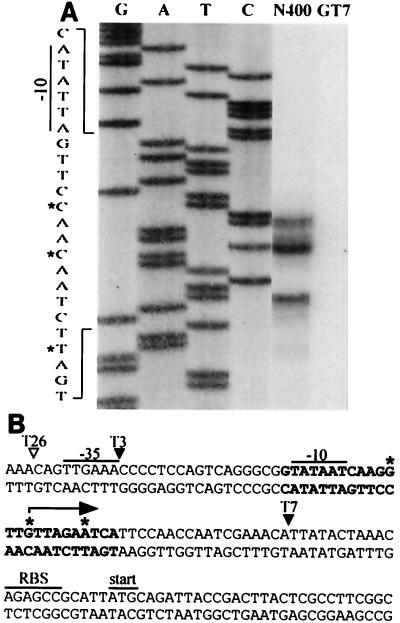
Primer extension analysis of the pilT-pilU region.
In order to map the promoter for the pilT-pilU operon, the location of the transcriptional start site for these genes was determined. RNA isolated from the wild-type strain and cells harboring the transposon insertion T7 in the promoter region of pilT were subjected to primer extension with a radiolabeled primer complementary to sequences early in the pilT ORF. This oligonucleotide primed the synthesis of three DNA products whose endpoints mapped 53, 50, and 44 bases in front of the pilT start codon; the intermediate-size species was most abundant, while no product was seen when RNA from cells harboring the transposon insertion T7 was used (Fig. 9). To confirm these results, the primer extension analysis was repeated with a different primer, and the results were identical to those obtained in the previous analysis (data not shown). The DNA sequence upstream of the sites mapped by primer extension carried a potential −10 promoter element (relative to the most abundant extension product) with a perfect match to the σ70 consensus sequence. A −35 region, with five of six conserved bases relative to the consensus sequence, was identified with proper spacing relative to the −10 region (Fig. 9). These data indicate that this σ70-type promoter is most likely responsible for transcription of the pilT-pilU operon during propagation in vitro. This conclusion is supported by the fact that strains carrying the transposon insertion T3 (mapping immediately 3′ to the −35 sequence) failed to express either pilT or pilU hybridizing RNA, while strains bearing the insertion T26 (mapping 3 bases 5′ to the −35 sequence) were phenotypically wild type and yielded the same primer extension products as the parental strain (Fig. 9). Primer extension experiments also were performed with primers complementary to the N-terminal coding region of the pilU gene, but no distinct DNA products were found (data not shown).
FIG. 9.
(A) Primer extension analysis of 5′ termini of pilT-pilU transcripts. RNA was isolated from strain N400 or GT7 after 5 h of culture in Gc liquid medium. Primer extension reaction mixtures were loaded alongside DNA sequencing reaction mixtures obtained by using wild-type genomic DNA templates with the extension primers. The letters above lanes G, A, T, and C (DNA sequencing reactions) indicate the dideoxynucleotides used to terminate the reactions. The nucleotide sequences surrounding the primer extension products have been expanded. The asterisks indicate nucleotides corresponding to the start points. (B) Nucleotide sequence of the upstream region of the pilT gene. Nucleotide sequences expanded in panel A are indicated by boldface type, and the transcriptional start sites mapped by primer extension analysis are indicated by asterisks. The arrow indicates the major pilT transcriptional start site. The consensus −10 and −35 hexamers, putative ribosome binding site for pilT (RBS), and translation start site are marked. The arrowheads show transposon insertion sites in mutant strains tested or discussed in the text. The solid arrowheads indicate a conferred pilT mutant phenotype, whereas the open arrowhead indicates that the transposon insertion had no effect.
Characterization of the pilT-pilU intergenic sequences.
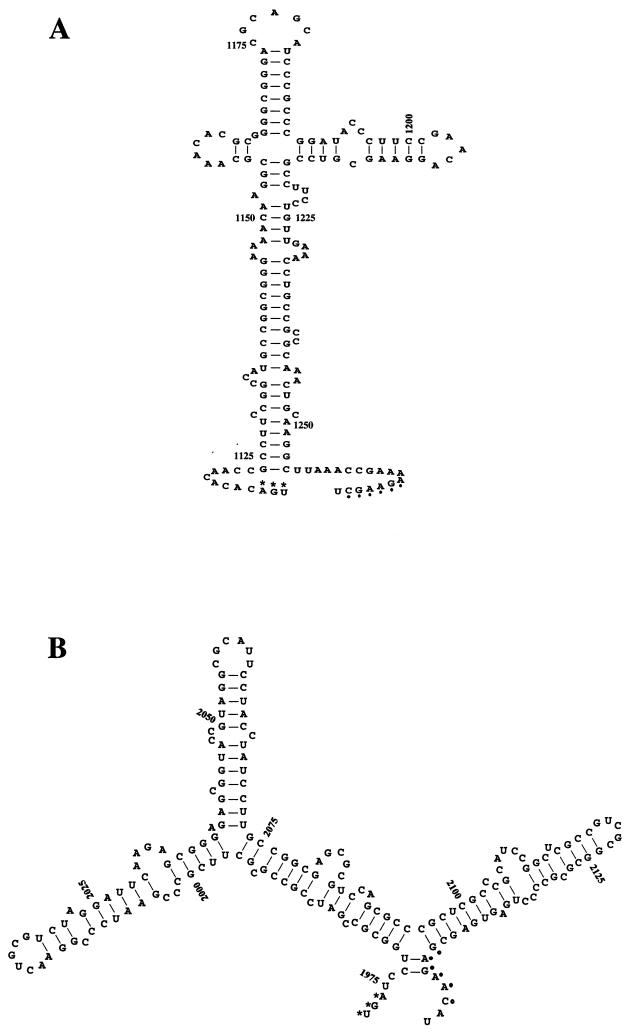
Given the evidence for the organization of the pilT-pilU locus as an operon, it was of interest to examine in more detail the sequences lying between the two ORFs. By using the Genetics Computer Group program FOLD, it was found that an RNA corresponding to the sense strand of the intergenic region was predicted to have an optimal secondary structure with a minimal free energy of −53.2 kcal/mol (Fig. 10A). Thus, these sequences may act as attenuators of transcription through this region or as nucleolytic processing sites for the mRNA. During their original studies of the analogous pilT-pilU locus in P. aeruginosa, Whitchurch and Mattick noted that a similarly sized intergenic region existed, but they concluded based on the data examined that the two genes were transcribed separately. We found, however, in analyzing the P. aeruginosa 171-bp intergenic sequences that although there was no sequence identity with the analogous region in gonococci, RNA corresponding to the sense strand was predicted to have an analogous optimal secondary structure with a minimal free energy of −64.1 kcal/mol (Fig. 10B).
FIG. 10.
Predicted stem-loop structure of RNA derived from the pilT-pilU intergenic sequences generated by the FOLD program of the Wisconsin Genetics Computer Group package. The RNAs corresponding to the sense strand of the intergenic regions have optimal secondary structures with minimal free energies of −53.2 kcal/mol (length, 162 bp) for N. gonorrhoeae (A) and −64.1 kcal/mol (length, 171 bp) for P. aeruginosa (B). The numbering of nucleotides is as in GenBank accession number S72391 for panel A and M55524 for panel B. The asterisks indicate the stop codon of the pilT ORFs, and the dots indicate putative ribosome binding sites for pilU.
To test the possibility that the gonococcal intergenic sequences might function as transcriptional attenuators, strains were constructed which carried fusions of the transcriptional reporter xylE to either pilT or pilU in an otherwise wild-type background, and the levels of reporter activities were determined. The results indicated that there were no significant differences in the levels of activity for the pilT and pilU fusions, suggesting that the intergenic sequences do not mediate attenuation in this reporter system (data not shown).
DISCUSSION
We identified a new gonococcal protein termed PilU as a new member of the TrbB-like family of proteins which are defined by the presence of a highly conserved, centrally localized domain implicated in ATP binding and hydrolysis and appear to function in the trafficking of macromolecules across membranes (18). The primary structure of gonococcal PilU is most closely related to that of the PilU protein of P. aeruginosa, followed by the PilT proteins found in gonococci, P. aeruginosa, and M. xanthus. These proteins are dispensable for Tfp biogenesis but are essential for twitching motility or its functional equivalent (social gliding motility or aggregate dispersal) (37, 39, 42). Gonococcal mutants expressing null alleles of pilU have no discernible qualitative or quantitative defects in Tfp expression and are distinguished only because they lack the autoagglutinating or aggregating properties normally associated with piliated strains propagated in vitro and display increased adherence for the human epithelial cell line ME-180. Thus, a common feature of all of these molecules is that they modify Tfp-associated properties. These similarities are mirrored by the higher degrees of interrelatedness found for these molecules and by the presence of an extended region of identity which extends into the carboxy-terminal segments, as exemplified by the conserved residues GMQTXXXXLXXLXXXXXI. The potential significance of this domain or motif is supported by the PilT− phenotype of gonococcal strains carrying the pilTΔQSL allele, in which the integrity of this motif is perturbed (39). It is of interest that the BfpF protein, which is dispensable for expression of Tfp by enteropathogenic strains of E. coli but modifies the aggregative and twitching motility-like behavior of these strains, has been termed a member of the PilT family (1, 3). However, this molecule lacks the extended carboxy-terminal conservation and signature motif defined here. This situation may reflect the fact that the Bfp proteins belong to a distinct subset of the Tfp family whose subunit and biogenesis components appear to be more divergent from those found in the other species.
The PilT proteins of gonococci and P. aeruginosa share 66% identity in their primary structures, while the PilU proteins of these species are 45% identical. These two pairs of molecules, therefore, represent the most highly conserved orthologues involved in Tfp expression and phenotypes in these species, and the arrangement and spacing of the genes are also highly conserved. In addition, the genes located upstream of the pilT genes in P. aeruginosa, pathogenic Neisseria species, Vibrio vulnificus, and other species are identically arranged, and their products are orthologous (unpublished data). It seems, then, that there has been an incredible degree of conservation within these loci of the species, which presumably reflects both an early origin in eubacterial evolution and strong selective pressures to restrict diversification. It is remarkable in this regard, then, that while we have shown that gonococcal pilT and pilU are organized in an operon, it was concluded that the same genes in P. aeruginosa are separately transcribed (38). A close examination of the results presented by Whitchurch and Mattick reveals, however, that this conclusion was based on indirect data and that the expression of pilU RNA or PilU in the context of classical polar and nonpolar mutations in pilT was never measured (38). Given the high degree of conserved, predicted secondary structure in RNA corresponding to the intergenic sequences in these two species, we propose that the gene organization in P. aeruginosa may in fact be identical to that in gonococci and that this possibility needs to be reexamined. Whatever the case in P. aeruginosa, the organization of the gonococcal pilT-pilU locus is quite curious, particularly given that no evidence was found for transcriptional attenuation in association with the intergenic sequence. In light of the finding that pilT mutations are epistatic to pilU mutations and that the gene products may act on a common pathway, organization as a single transcriptional unit would ensure that the two molecules are expressed in a highly coordinated fashion. Moreover, the processing of the RNA into two species which could have altered stabilities may provide an opportunity for differential expression of gene products in response to conditions which regulate the processing event or influence RNA turnover. This potential form of gene regulation might compensate for the fact that the primary promoter for the locus appears to be recognized by the σ70 transcription system.
It is difficult to understand how gonococcal PilU expression might influence Tfp-associated aggregation since the role of Tfp in this property is unknown and the precise function(s) served by any member of the TrbB-like protein family has yet to be elucidated. However, it has been shown previously that gonococcal PilT influences Tfp expression in a manner which is consistent with its acting as a conditional antagonist of stable fiber formation (40, 41). In addition, pilT mutants hyperautoagglutinate, while pilU mutants are defective in this trait; therefore, PilT appears to inhibit Tfp-associated aggregation, while PilU promotes this property. Furthermore, the influence of PilU is manifest only in the presence of PilT. These lines of evidence lead us to propose that PilU exerts its effect downstream of the processes influenced by PilT (41) and does so by acting as an antagonist of PilT activity. Given the highly related structures of the proteins, PilU might modify PilT activity by competing for a common interactive component or site on the inner membrane (24). Since the related traffic ATPases of the Helicobacter pylori type IV secretion system and conjugative plasmid systems can form hexameric ring structures (35), it is also possible that PilU has an effect by forming nonfunctional multimers with PilT. Further understanding of the functional relationships between PilT and PilU is required in order to test these hypotheses. Two PilT proteins have been reported in Synechocystis sp. strain PCC6803; loss of PilT1 leads to a hyperpiliated, nonmotile phenotype, and loss of PilT2 leads to a piliated, motile but negatively phototactic phenotype (2). Based on our findings, we suggest that PilT2 may act like N. gonorrhoeae PilU and propose that pilT1 mutations are epistatic to mutations in pilT2. These scenarios are consistent with the higher degree of structural relatedness between PilT2 and the PilU proteins of N. gonorrhoeae and P. aeruginosa (data not shown).
The enhanced adherence to human epithelial cell line ME-180 found for the pilU mutants indicates that PilU acts in a manner that inhibits or diminishes this property. This result is in stark contrast to the findings reported for a P. aeruginosa pilU mutant which showed reduced association with both animal- and human-derived epithelial cells (6). Neisserial Tfp-mediated adherence to human epithelial cells has been demonstrated to require expression of the PilC protein and correlates with the ability of this protein to associate with purified fibers. However, there were no detectable changes in the levels of purifiable Tfp or PilC antigen in purified fibers in pilU mutants. It has been suggested that in N. meningitidis high-level Tfp-associated adherence correlates with expression of fibers which form lateral aggregates or bundles and which appear to promote interbacterial interactions (19). Electron microscopic studies of pilU mutants did not reveal any differences between these mutants and the wild type in either the level of piliation or the degree to which Tfp fibers form bundles.
Although pilU mutants display reduced levels of autoagglutination when they are grown in gonococcal or tissue culture media, they adhere to ME-180 cells as aggregates of cells in which many bacteria adhere to one another rather than directly to the epithelial cells. This behavior is very similar to that reported for the interaction of Bartonella henselae with endothelial cells (7), the interaction of Salmonella enterica serovar Typhimurium expressing type 1 fimbriae with enterocytes (33), and the interaction of Klebsiella pneumoniae with human intestinal cells (10). Since microbial clustering in all these cases fails to occur in the absence of host tissue, it seems that eukaryotic cell contact may be responsible for induction or modification of the aggregative phenotype. Two studies of pathogenic Neisseria have shown that PilT can promote the dispersal of bacterial aggregates following adherence (21, 23). If, as proposed in this work, N. gonorrhoeae PilU acts as an antagonist of PilT, the increased adherence seen for the pilU mutants may reflect defectiveness in the dispersal phenomenon associated with pilus retraction triggered by cell contact. This may involve not only modified bacterium-bacterium interactions but also altered recruitment of cortical plaque constituents within the epithelial cells to the site of bacterial contact (21). Future studies will examine if differences in pilus retraction kinetics and force generation or cortical plaque formation are seen with pilU mutants.
Acknowledgments
This work was supported by Public Health Service grant AI 27837 from the National Institutes of Health to M.K. M.W. acknowledges support from National Institutes of Health training grant 2 T32 GM07315.
Editor: A. D. O'Brien
REFERENCES
- 1.Anantha, R. P., K. D. Stone, and M. S. Donnenberg. 1998. Role of BfpF, a member of the PilT family of putative nucleotide-binding proteins, in type IV pilus biogenesis and in interactions between enteropathogenic Escherichia coli and host cells. Infect. Immun. 66:122-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bhaya, D., N. R. Bianco, D. Bryant, and A. Grossman. 2000. Type IV pilus biogenesis and motility in the cyanobacterium Synechocystis sp. PCC6803. Mol. Microbiol. 37:941-951. [DOI] [PubMed] [Google Scholar]
- 3.Bieber, D., S. W. Ramer, C. Y. Wu, W. J. Murray, T. Tobe, R. Fernandez, and G. K. Schoolnik. 1998. Type IV pili, transient bacterial aggregates, and virulence of enteropathogenic Escherichia coli. Science 280:2114-2118. [DOI] [PubMed] [Google Scholar]
- 4.Bradley, D. E. 1980. A function of Pseudomonas aeruginosa PAO polar pili: twitching motility. Can. J. Microbiol. 26:146-154. [DOI] [PubMed] [Google Scholar]
- 5.Brossay, L., G. Paradis, R. Fox, M. Koomey, and J. Hebert. 1994. Identification, localization and distribution of the PilT protein in Neisseria gonorrhoeae. Infect. Immun. 62:2302-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comolli, J. C., A. R. Hauser, L. Waite, C. B. Whitchurch, J. S. Mattick, and J. N. Engel. 1999. Pseudomonas aeruginosa gene products PilT and PilU are required for cytotoxicity in vitro and virulence in a mouse model of acute pneumonia. Infect. Immun. 67:3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehio, C., M. Meyer, J. Berger, H. Schwarz, and C. Lanz. 1997. Interaction of Bartonella henselae with endothelial cells results in bacterial aggregation on the cell surface and the subsequent engulfment and internalisation of the bacterial aggregate by a unique structure, the invasome. J. Cell Sci. 110:2141-2154. [DOI] [PubMed] [Google Scholar]
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Drake, S. L., S. A. Sandstedt, and M. Koomey. 1997. PilP, a lipoprotein essential for type IV pilus biogenesis in Neisseria gonorrhoeae, is required for stable expression of the PilQ multimer. Mol. Microbiol. 23:657-668. [DOI] [PubMed] [Google Scholar]
- 10.Favre-Bonte, S., A. Darfeuille-Michaud, and C. Forestier. 1995. Aggregative adherence of Klebsiella pneumoniae to human intestine-407 cells. Infect. Immun. 63:1318-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freitag, N., H. S. Seifert, and M. Koomey. 1995. Characterization of the pilF-pilD pilus assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16:575-586. [DOI] [PubMed] [Google Scholar]
- 12.Goodman, S. D., and J. J. Scocca. 1988. Identification and arrangement of the DNA sequence recognized in specific transformation of Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 85:6982-6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heckels, J. E. 1989. Structure and function of pili of pathogenic Neisseria species. Clin. Microbiol. Rev. 2:S66-S73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koomey, M., S. Bergstrom, M. Blake, and J. Swanson. 1991. Pilin expression and processing in pilus mutants of Neisseria gonorrhoeae: critical role of Gly−1 in assembly. Mol. Microbiol. 5:279-287. [DOI] [PubMed] [Google Scholar]
- 15.Koomey, M., E. C. Gotschlich, K. Robbins, S. Bergstrom, and J. Swanson. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lauer, P., N. H. Albertson, and M. Koomey. 1993. Conservation of genes encoding components of a type IV pilus assembly/two-step protein export pathway in Neisseria gonorrhoeae. Mol. Microbiol. 8:357-368. [DOI] [PubMed] [Google Scholar]
- 18.Lessl, M., and E. Lanka. 1994. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell 77:321-324. [DOI] [PubMed] [Google Scholar]
- 19.Marceau, M., J. L. Beretti, and X. Nassif. 1995. High adhesiveness of encapsulated Neisseria meningitidis to epithelial cells is associated with the formation of bundles of pili. Mol. Microbiol. 17:855-863. [DOI] [PubMed] [Google Scholar]
- 20.Merz, A., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 21.Merz, A. J., C. A. Enns, and M. So. 1999. Type IV pili of pathogenic neisseriae elicit cortical plaque formation in epithelial cells. Mol. Microbiol. 32:1316-1332. [DOI] [PubMed] [Google Scholar]
- 22.Possot, O., and A. P. Pugsley. 1994. Molecular characterization of PulE, a protein required for pullulanase secretion. Mol. Microbiol. 12:287-299. [DOI] [PubMed] [Google Scholar]
- 23.Pujol, C., E. Eugene, M. Marceau, and X. Nassif. 1999. The meningococcal PilT protein is required for induction of intimate attachment to epithelial cells following pilus-mediated adhesion. Proc. Natl. Acad. Sci. USA 96:4017-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sandkvist, M., M. Bagdasarian, S. P. Howard, and V. J. DiRita. 1995. Interaction between the autokinase EpsE and EpsL in the cytoplasmic membrane is required for extracellular secretion in Vibrio cholerae. EMBO J. 14:1664-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seifert, H. S., R. S. Ajioka, D. Paruchuri, F. Heffron, and M. So. 1990. Shuttle mutagenesis of Neisseria gonorrhoeae: pilin null mutations lower DNA transformation competence. J. Bacteriol. 172:40-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seifert, H. S., M. So, and F. Heffron (ed.). 1986. Shuttle mutagenesis: a method for introducing transposons into transformable organisms. Plenum Publishing Corp., New York, N.Y.
- 28.Skerker, J. M., and H. C. Berg. 2001. Direct observation of extension and retraction of type IV pili. Proc. Natl. Acad. Sci. USA 98:6901-6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stein, D. C. 1992. Plasmids with easily excisable xylE cassettes. Gene 117:157-158. [DOI] [PubMed] [Google Scholar]
- 30.Stein, D. C., R. J. Danaher, and T. M. Cook. 1991. Characterization of a gyrB mutation responsible for low-level nalidixic acid resistance in Neisseria gonorrhoeae. Antimicrob. Agents Chemother. 35:622-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Swanson, J. 1973. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J. Exp. Med. 137:571-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tabor, S., and C. C. Richardson. 1985. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. USA 82:1074-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thankavel, K., A. H. Shah, M. S. Cohen, T. Ikeda, R. G. Lorenz, R. Curtiss III, and S. N. Abraham. 1999. Molecular basis for the enterocyte tropism exhibited by Salmonella typhimurium type 1 fimbriae. J. Biol. Chem. 274:5797-5809. [DOI] [PubMed] [Google Scholar]
- 34.Turner, L. R., J. C. Lara, D. N. Nunn, and S. Lory. 1993. Mutations in the consensus ATP-binding sites of XcpR and PilB eliminate extracellular protein secretion and pilus biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 175:4962-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turner, L. R., J. W. Olson, and S. Lory. 1997. The XcpR protein of Pseudomonas aeruginosa dimerizes via its N-terminus. Mol. Microbiol. 26:877-887. [DOI] [PubMed] [Google Scholar]
- 36.Weinberger, C., S. M. Hollenberg, E. S. Ong, J. M. Harmon, S. T. Brower, J. Cidlowski, E. B. Thompson, M. G. Rosenfeld, and R. M. Evans. 1985. Identification of human glucocorticoid receptor complementary DNA clones by epitope selection. Science 228:740-742. [DOI] [PubMed] [Google Scholar]
- 37.Whitchurch, C. B., M. Hobbs, S. P. Livingston, V. Krishnapillai, and J. S. Mattick. 1991. Characterisation of a Pseudomonas aeruginosa twitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene 101:33-44. [DOI] [PubMed] [Google Scholar]
- 38.Whitchurch, C. B., and J. S. Mattick. 1994. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol. Microbiol. 13:1079-1091. [DOI] [PubMed] [Google Scholar]
- 39.Wolfgang, M., P. Lauer, H. S. Park, L. Brossay, J. Hebert, and M. Koomey. 1998. PilT mutations lead to simultaneous defects in competence for natural transformation and twitching motility in piliated Neisseria gonorrhoeae. Mol. Microbiol. 29:321-330. [DOI] [PubMed] [Google Scholar]
- 40.Wolfgang, M., H. S. Park, S. F. Hayes, J. P. van Putten, and M. Koomey. 1998. Suppression of an absolute defect in type IV pilus biogenesis by loss-of-function mutations in pilT, a twitching motility gene in Neisseria gonorrhoeae. Proc. Natl. Acad. Sci. USA 95:14973-14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wolfgang, M., J. P. van Putten, S. F. Hayes, D. Dorward, and M. Koomey. 2000. Components and dynamics of fiber formation define a ubiquitous biogenesis pathway for bacterial pili. EMBO J. 19:6408-6418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu, S., J. Wu, and D. Kaiser. 1997. The Myxococcus xanthus pilT locus is required for social gliding motility although pili are still produced. Mol. Microbiol. 23:109-121. [DOI] [PubMed] [Google Scholar]
- 43.Wu, S. S., and D. Kaiser. 1995. Genetic and functional evidence that type IV pili are required for social gliding motility in Myxococcus xanthus. Mol. Microbiol. 18:547-558. [DOI] [PubMed] [Google Scholar]
- 44.Zhang, Q. Y., D. DeRyckere, P. Lauer, and M. Koomey. 1992. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc. Natl. Acad. Sci. USA 89:5366-5370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zukowski, M. M., D. F. Gaffney, D. Speck, M. Kauffmann, A. Findeli, A. Wisecup, and J. P. Lecocq. 1983. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc. Natl. Acad. Sci. USA 80:1101-1105. [DOI] [PMC free article] [PubMed] [Google Scholar]