Abstract
Hemorrhagic septicemia (HS) is a fatal systemic disease of cattle and buffaloes. In South Asia HS is caused by infection with Pasteurella multocida serotype B:2. Some control is achieved with alum-precipitated or oil-adjuvanted killed whole-cell vaccines injected subcutaneously, but these vaccines provide only short-term immunity and require annual administration for effective use. Live attenuated vaccines have the advantage of a natural route of entry into the host, but for live strains to be used as vaccines, the mode of attenuation should be well defined. We constructed aroA attenuated derivatives of two P. multocida serotype B:2 strains by allelic exchange of the native aroA sequence with aroA sequences disrupted with a kanamycin resistance cassette or with marker-free aroA sequences containing an internal deletion. These strains were confirmed to be aroA mutants by PCR and Southern blot analysis, enzyme assay, and lack of growth on minimal medium. The aroA derivatives were highly attenuated for virulence in a mouse model of HS. Mouse challenge experiments showed that intraperitoneal or intranasal vaccination of an aroA strain completely protected mice against challenge with a high dose (>1,000 50% lethal doses) of either the parent strain or the other wild-type B:2 strain. The spread of the parent and the aroA derivatives to different organs was compared when the organisms were inoculated by different routes.
Diseases caused by Pasteurella species are economically important worldwide. These bacteria act as primary or secondary pathogens and cause a wide spectrum of diseases, collectively termed pasteurellosis and ranging from septicemia to pneumonia. Pasteurella multocida is recognized as an important veterinary pathogen. Serotype designations are based on capsular antigen-somatic antigen combinations (capsular antigens A to F and somatic antigens 1 to 16). Certain serotypes of P. multocida cause hemorrhagic septicemia (HS), a disease of cattle and buffaloes with high mortality rates and economic significance in Asia and Africa. In South Asia, serotype B:2 predominates (36). HS is a primary pasteurellosis characterized by terminal septicemia and is distinct from other pasteurelloses, in which P. multocida may play only a secondary role. The virulence factors of P. multocida responsible for HS have not been defined, and the antigens responsible for natural immunity are unknown. Some control is achieved with alum-precipitated or oil-adjuvanted killed whole-cell vaccines injected subcutaneously (s.c.), but these vaccines have the disadvantage of providing only short-term immunity (7) and require annual administration for effectiveness (10). The oil-adjuvanted vaccines have the added disadvantage of high viscosity, which makes them unpopular among field users, although improved oil-adjuvanted vaccines with lower viscosities have been described (29, 34, 35). However, all such vaccines suffer from a requirement for high numbers of inactivated cells (1010 to 1011 cells) and consequent problems of reactogenicity.
Naturally acquired immunity is common among survivors of HS outbreaks, and consequently, animals up to 2 years old are most susceptible. The nature of the immune responses to P. multocida is poorly understood, and the relative contributions of cellular immunity and humoral immunity to long-term protection have not been established. It is not known where the organism lodges and multiplies during the early clinical phase of HS, although HS pathogenesis involves rapid translocation of bacteria from the respiratory tract to the blood and lymph. In attempts to mimic natural infection and to elicit long-term humoral and cellular immunity, live vaccines have been developed (22), but these vaccines are ill-defined and of questionable safety.
Live attenuated vaccines in general have the advantage of a natural route of entry into the host, which allows targeting of immunostimulatory factors to the same sites of the immune system that occur in the natural infection. The aroA gene encodes 5-enolpyruvylshikimate-3-phosphate (EPSP) synthase, which is involved in the conversion of shikimic acid to chorismic acid, a common intermediate in the biosynthesis of aromatic amino acids. Mutation in the aroA gene creates a dependence for growth on aromatic compounds that are not available in the host, as this pathway is not operative in mammalian cells. This means that aroA mutants are capable of only limited replication before they are cleared from the host. Attenuated aroA mutants of P. multocida serotype A, which causes fowl cholera, have been described (14, 15) and have been shown to provide protection against challenge in chickens (28). However, serotype A strains do not cross-protect against challenge with serotype B strains (1, 2, 26). The aims of this work were to construct defined mutations in the aroA genes of two serotype B:2 strains, to test these strains in a mouse experimental model to determine their degree of attenuation and their protective properties, and to compare the mutant derivatives with the parent strains for spread and persistence within the host.
MATERIALS AND METHODS
Bacteria, plasmids, and growth conditions.
The bacterial strains and plasmids used are listed in Table 1. P. multocida strains were grown routinely in brain heart infusion (BHI) broth (Oxoid) in flasks shaken at 150 rpm at 37°C or on BHI agar containing 5% (vol/vol) sheep blood (B&E Laboratories, Bonnybridge, United Kingdom) and 1.2% (wt/vol) agar. A chemically defined Pasteurella minimal medium (PMM) (17) was also used; when required, this medium contained an “aromix” supplement consisting of l-tryptophan, l-tyrosine, and l-phenylalanine, each at a final concentration of 40 μg ml−1, and 2,3-dihydrobenzoic acid and ρ-hydroxybenzoic acid, each at a concentration of 10 μg ml−1. Escherichia coli strains were grown in Luria broth in flasks shaken at 150 rpm at 37°C or on Luria agar containing 1% (wt/vol) agar. For E. coli hosts containing low-copy-number plasmids, such as pAKA19 and pEG18.3, Terrific broth (30) supplemented with 1% (wt/vol) yeast nitrogen base (Difco) was used to improve the plasmid yield. Antibiotics (Sigma) were used at the following concentrations: ampicillin, 50 μg ml−1; chloramphenicol, 30 μg ml−1; tetracycline, 15 μg ml−1; kanamycin, 50 μg ml−1; and streptomycin, 100 μg ml−1.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | supE ΔlacU (φ80 lacZΔM15) hsdR recA endA gyrA thi-1 relA | Invitrogen |
| SM10λpir | thi-1 thr leu tonA lacY supE recA::RP4-2Tcr::Mu, Kmr λpir | 21 |
| TOP10 | mcrA Δ(mrr-hsdRMS-mcrBC) (φ80lacZΔM15) ΔlacX74 deoR recA araD Δ(ara-leu) galU galK rspL(Strr) endA nupG | Invitrogen |
| P. multocida strains | ||
| 85020 | Wild type, serotype B:2 | Veterinary Research Institute, Peradeniya, Sri Lanka |
| Quetta | Wild type, serotype B:2 | Vaccine Production Unit, Quetta, Pakistan |
| Plasmids | ||
| PCR2.1-TOPO | Direct cloning vector for PCR amplimers, Apr Kmr | Invitrogen |
| pUC4K | Derivative of pUC4 containing Kmr gene from Tn903 | Pharmacia Biotech |
| pEG18.3 | Suicide vector, Apr Kmr GmrsacBR | P. Cotter, University of California, Santa Barbara, 20 |
| pAKA19 | Suicide vector, Apr, derivative with a ColE1 origin of replication of pAKA16 (α-lac multiple cloning site MCS inserted into pPH843, Apr, from Mannheimia haemolytica) | 4, 5 |
Preparation and manipulation of DNA.
Chromosomal DNA was prepared by the method of Ausubel et al. (3), and plasmid DNA was isolated by using a QIAprep kit (Qiagen) according to the manufacturer's instructions. PCRs were performed with 25-μl reaction mixtures containing 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 3 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, 100 pmol of each primer, 10 ng of template DNA, and 1 U of Taq DNA polymerase (Life Technologies, Paisley, United Kingdom) by using 35 cycles of 94°C for 1 min, 55°C for 1 min, and 72°C for 1 min and a final extension step at 72°C for 6 min. PCR products and restriction endonuclease fragments were fractionated by agarose gel electrophoresis, excised from the gel, and purified by using a GenElute agarose spin column (Supelco) or a QIAEXII gel extraction kit (Qiagen). Restriction endonuclease digestion of DNA and cloning of purified restriction fragments were performed by using standard procedures (3). Purified PCR products were cloned directly into the pCR2.1-TOPO vector (Invitrogen, Paisley, United Kingdom) according to the manufacturer's instructions. Ligated DNAs were introduced into bacteria by electroporation (11, 16) by using a Gene Pulser apparatus (Bio-Rad, Hemel Hempstead, United Kingdom) set at 2 kV, 25 μF, and 200 Ω (field strength, 10 kV cm−1) for E. coli and at 2.5 kV, 25 μF, and 400 Ω (field strength, 25 kV cm−1) for P. multocida. Following electroporation, cells were incubated in broth medium for 1 h in the case of E. coli and for 3 h in the case of P. multocida before aliquots of the cell suspension were spread on the appropriate antibiotic selective agar. Conjugation was performed by plate mating late-exponential-growth-phase cultures as described previously (5). For this purpose, spontaneous streptomycin-resistant derivatives of the parent P. multocida strains were isolated and used as recipients to select against the E. coli strain used as the donor.
Construction of aroA mutants of P. multocida HS strains.
Forward and reverse primers were designed from the aroA sequence of P. multocida serotype A:1 strain PBA100 (14; GenBank accession number Z14100). The aroA genes of P. multocida strains 85020 and Quetta were amplified by PCR as 1.2-kb amplimers by using forward primer AroA1 (TTACTCTCAATCCCATCAGC; nucleotides 315 to 334) and reverse primer AroA2 (ACAATGCGATTAAAGCAAAG; nucleotides 1495 to 1514). The PCR products were purified and cloned into pCR2.1-TOPO. For transfer of cloned genes to P. multocida, the aroA fragments were removed as BamHI/XhoI fragments and cloned into the suicide vector pAKA19 cut with the same enzymes. A cassette encoding kanamycin resistance (Kmr) was removed as a 1.24-kb PstI fragment (nucleotides 421 to 1661) from plasmid pUC4K (Pharmacia) and inserted into the aroA genes at a unique NsiI site (nucleotide 718). Allelic exchange of the Kmr-disrupted aroA genes with the native genes in the P. multocida chromosome was successfully achieved with strain 85020 by electroporation and with the Smr Quetta strain by conjugation. For the latter procedure, the plasmid was first transferred to E. coli SM10λpir by electroporation. Aliquots of bacteria were spread onto BHI blood agar containing only kanamycin for strain 85020 and kanamycin plus streptomycin for Smr strain Quetta in order to select Kmr and Kmr Smr colonies, respectively. For each strain, 50 single colonies were picked and subcultured on BHI blood agar containing kanamycin for 5 days to encourage loss of the pAKA19 plasmid and exchange of the aroA::Kmr insert with the native aroA gene on the recipient chromosome. Chromosomal DNA was then prepared from at least 20 single clones of each strain and checked by PCR by using the AroA1 and AroA2 primers. Clones designated P. multocida JRMT1 for the 85020 strain and JRMT2 for the Quetta Smr strain were chosen for further study. Each of these clones produced ∼2.4-kb amplimers, the pattern of PCR amplimers predicted after allelic exchange with the aroA allele containing the Kmr insert, while ∼1.2-kb amplimers were produced by the parent strains (data not shown).
Marker-free aroA deletion derivatives of the 85020 and Quetta strains were constructed by removing a 142-bp internal AflII/SacII fragment (nucleotides 938 to 1080) from the aroA genes cloned in pCR2.1-TOPO. The deleted genes were exchanged with the wild-type alleles in the chromosome by using the sacB-based allelic exchange procedure (25). The deleted aroA genes were first cloned into pAKA19 as 1.05-kb BamHI/XhoI fragments, and the 3.8-kb Kmr-sacBR cassette from pEG18.3 was then introduced into the constructs at the BamHI site. These plasmids were transferred to P. multocida strains by electroporation, and Kmr colonies were selected after incubation for 48 h on BHI blood agar containing kanamycin. This first step selected for clones in which recombination at the aroA sequence had incorporated the whole plasmid into the recipient chromosome. Resulting colonies were patched onto BHI blood agar containing 5% (wt/vol) sucrose. After incubation for 48 h, sucrose-resistant colonies exhibiting a nonmucoid phenotype were picked and tested for the Kms phenotype, which was indicative of loss of vector sequences. Sucrose-resistant and Kms colonies were then screened by PCR by using the AroA1 and AroA2 primers. Clones designated P. multocida JRMT12 for the 85020 strain and P. multocida JRMT13 for the Quetta strain were chosen for further study. Each of these clones produced amplimers of the predicted size,1.05 kb for the deleted aroA gene, compared with the ∼1.2-kb amplimer for the native gene (data not shown).
The Bacillus subtilis sacBR genes encode a levansucrase whose expression in the presence of sucrose is lethal in most gram-negative bacteria. This feature provides positive selection for directed allelic exchange of unmarked mutations (25). Sucrose metabolism via expression of sacB in P. multocida creates a mucoid and liquefied colony phenotype after 48 h of incubation that has been reported previously (18), but the authors indicated that sucrose metabolism via expression of sacB was not lethal in P. multocida serogroup A1 and therefore not useful as a positive selection procedure for allelic exchange. However, in our hands, lack of expression of sacB and the resulting absence of the mucoid phenotype in the presence of sucrose acted as a good indicator of allelic exchange and loss of the vector sequences containing the sacB gene.
Southern blot hybridization.
Southern blot hybridization was done by standard procedures (3) by using a nylon membrane (Hybond N+; Amersham Pharmacia Biotech, Little Chalfont, United Kingdom) for DNA transfer from agarose gels and DNA probes labeled with a digoxigenin (DIG) random priming kit (Boehringer, Mannheim, Germany) according to the manufacturer's instructions. Hybridization was performed in a rolling hybridization oven (Techne, Cambridge, United Kingdom) under high-stringency conditions, and DIG-labeled hybrid DNA was detected with anti-DIG-alkaline phosphatase conjugate antibody and a DIG luminescence detection kit (Boehringer, Lewes, United Kingdom). Blots were exposed to photosensitive film (X-ray film; Kodak, Hemel Hempstead, United Kingdom) for up to 1 h.
Enzyme assay.
EPSP synthase was assayed by the reverse reaction (19). This reaction couples phosphenolpyruvate release from EPSP to the pyruvate kinase and lactic dehydrogenase reactions and measures NADH oxidation at 340 nm (ɛ = 6,220 M−1 cm−1). One unit of activity oxidized 1 nmol of NADH per min at 25°C. EPSP was kindly provided by J. R. Coggins (Glasgow University), and pyruvate kinase and lactic dehydrogenase were obtained from Boehringer. Cell extracts were prepared from exponential-phase cultures of P. multocida strains grown in BHI broth. Cells were harvested by centrifugation, resuspended (10%, wt/vol) in 100 mM KH2PO4 (pH 7.0), and lysed by sonication (three times, 20 s each) with intermittent cooling. Interfering NADH oxidase activity present in crude extracts was minimized by preparing cytoplasmic S100 extracts by centrifugation at 100,000 × g for 2 h (19). Supernatants were stored at −20°C. Protein concentrations were determined by using a bicinchoninic acid protein assay kit (Pierce Chemical Co., Rockford, Ill.).
Virulence and protection tests.
The mouse provides a good model for HS infection as it manifests a septicemic form of disease similar to HS in the natural hosts (6, 9). Groups of female BALB/c mice (Harlan Olac, Bicester, United Kingdom) that were 5 to 6 weeks old were used for the first set of experiments involving aroA strains JRMT1 and JRMT2, and groups of female BALB/c mice that were 6 to 7 weeks old were used for the second set of experiments involving aroA strain JRMT12. Mice were injected intraperitoneally (i.p.) with 500 μl of 10-fold serial dilutions in phosphate-buffered saline (PBS) of exponential-phase cultures (E540nm, ∼1) grown in BHI broth. For intranasal (i.n.) inoculation, the method of Rushton (27) was used. Mice were anesthetized with halothane, and 50 μl of an appropriate bacterial dilution was applied to the nares and allowed to be inhaled. s.c. vaccination was done by injecting 200 μl of an appropriate bacterial dilution into the nape of the neck of an anesthetized mouse. Mice were weighed daily and checked regularly for up to 10 days, and the mice which became moribund were euthanized in accordance with animal ethics guidelines. For virulence determinations, a 50% lethal dose (LD50) was estimated by direct observation of dose-response data. For collection of blood samples and internal organs, mice were sacrificed and samples were collected aseptically in preweighed bottles. Organs were homogenized in 10-ml portions of PBS, and 10-fold serial dilutions were plated on BHI blood agar and incubated overnight at 37°C. For mouse protection tests, groups of mice vaccinated with attenuated mutant strains were challenged 2 weeks later with different doses of the P. multocida 85020 and Quetta parent strains. Numbers of survivors were recorded at 6 days postchallenge.
RESULTS
Characterization of the aroA mutants.
Three tests were performed to confirm the nature of the aroA mutations in P. multocida strains JRMT1, JRMT2, JRMT12, and JRMT13. After overnight growth in BHI broth, the mutant and parent strains were collected by centrifugation, washed with PBS, diluted into PMM with or without the aromix supplement, and grown for 24 h with shaking at 37°C. All the mutant strains failed to grow in PMM unless it was supplemented with aromix (E540 without aromix, <0.02; E540nm with aromix, ∼0.8). The parent strains had an E540nm of ∼0.9 in PMM alone.
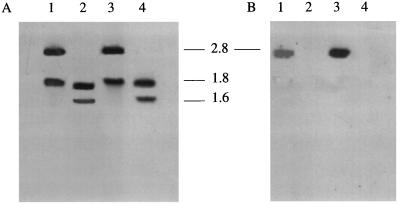
Genomic DNA was prepared from the parent and mutant strains. In the case of JRMT1 and JRMT2 the DNA was digested with EcoRV, and a Southern blot was probed with an aroA probe (the aroA gene of P. multocida 85020 amplified by PCR and DIG labeled) and with the Kmr cassette (the PstI fragment of plasmid pUC4K). The aroA probe hybridized to two bands at ∼1.6 and ∼1.8 kb in the EcoRV-digested DNA from the parent strains, confirming the presence of an EcoRV site in the aroA gene (nucleotide 931) (Fig. 1A), and no hybridization was detected with the Kmr probe (Fig. 1B). For the mutant strains, two bands at ∼1.8 and ∼2.8 kb were visible with the aroA probe (Fig. 1A), and the Kmr probe hybridized only with the larger band (Fig. 1B). This confirmed insertion of the Kmr cassette into the section of aroA represented by the 1.6-kb EcoRV fragment. For Southern blot analysis of JRMT12 and JRMT13, chromosomal DNA was digested with EarI (the EarI site at position 1052 in the native aroA gene was removed by the 142-bp internal AflII/SacII deletion) and, following electrophoresis and blotting onto a membrane, was hybridized with the aroA probe used previously. The probe hybridized to two bands at ∼1.3 and ∼1.6 kb for the parent strains, a pattern predicted from internal digestion of the native aroA gene by EarI, but to only one band at ∼2.75 kb for the aroA deletion mutants (Fig. 2), a pattern predicted after removal of the internal fragment containing the EarI site from the native gene.
FIG. 1.
Southern blot analysis of parent and aroA derivatives JMRT1 and JMRT2. Chromosomal DNA prepared from parent and mutant strains was digested with EcoRV, separated by electrophoresis, and transferred to a nylon membrane. (A) Hybridization with an aroA probe prepared by PCR with the AroA1 and AroA2 primers from chromosomal DNA of the 85020 parent strain. (B) Hybridization with a Kmr cassette prepared from plasmid pUC4K. Lanes 1, aroA derivative JRMT1; lanes 2, parent strain 85020; lanes 3, aroA derivative JRMT2; lanes 4, parent strain Quetta. The positions of size markers (in kilobases) are indicated between the blots.
FIG. 2.
Southern blot analysis of parent and ΔaroA derivatives JMRT12 and JMRT13. Chromosomal DNA prepared from parent and mutant strains was digested with EarI, separated by electrophoresis, and transferred to a nylon membrane. The blot was hybridized with an aroA probe prepared by PCR with the AroA1 and AroA2 primers from chromosomal DNA of the 85020 parent strain. Lane 1, parent strain 85020; lane 2, ΔaroA derivative JRMT12; lane 3, parent strain Quetta; lane 4, ΔaroA derivative JRMT13. The positions of size markers (in kilobases) are indicated on the right.
S100 extracts were prepared for the assay of EPSP synthase. The enzyme specific activities obtained for the P. multocida 85020 and Quetta wild-type strains were 4.7 and 3.6 U mg of protein−1, respectively, whereas no activity was detected in similar extracts prepared from the aroA mutant strains.
Mouse virulence tests with aroA strains JRMT1 and JRMT2.
Initial tests were done with the 85020 parent strain and the aroA derivative JRMT1, and groups of mice were injected i.p. with graded doses of these strains. The parent strain, 85020, was highly virulent by this route (Table 2) and could kill mice within 1 to 2 days with a very small inoculum. The aroA derivative, JRMT1, was greatly attenuated. LD50s of <20 CFU per mouse for the parent strain and >3 × 108 CFU per mouse for JRMT1 were obtained. Similar results were obtained with the Quetta strain and its aroA derivative, JRMT2 (Table 2). The toxicity of very high doses of the attenuated strain was evident from experiment 2 (Table 2), in which all of the mice injected with 3 × 109 CFU died within 48 h. Mice given 3 × 108 CFU showed some weight loss during the first day postchallenge, but by the second day all of these mice had recovered their original weight (data not shown).
TABLE 2.
Mouse virulence testsa
| Challenge strain | CFU per mouse (no. of survivors/no. challenged)
|
||
|---|---|---|---|
| Expt 1 | Expt 2 | Expt 3 | |
| 85020 (wild type) | 2,000 (0/3) | 320 (0/5) | 266 (0/5) |
| 200 (0/3) | 32 (0/5) | 26 (0/5) | |
| 20 (0/3) | 3.2 (2/5) | 2.6 (2/5) | |
| 2 (2/3) | |||
| JRMT1 (aroA strain) | 2 × 105 (3/3) | 3 × 109 (0/5) | 2 × 108 (5/5) |
| 2 × 104 (3/3) | 3 × 108 (5/5) | 2 × 107 (5/5) | |
| 2,000 (3/3) | 3 × 107 (5/5) | 2 × 106 (5/5) | |
| 200 (3/3) | 3 × 106 (5/5) | 2 × 105 (5/5) | |
| 2 × 104 (5/5) | |||
| Quetta (wild type) | 300 (0/5) | ||
| 30 (0/5) | |||
| 3 (0/5) | |||
| JRMT2 (aroA strain) | 2 × 108 (5/5) | ||
| 2 × 107 (5/5) | |||
| 2 × 106 (5/5) | |||
| 2 × 105 (5/5) | |||
| 2 × 104 (5/5) | |||
| None (PBS control) | (3/3) | (5/5) | (5/5) |
Groups of mice were injected i.p. with 0.5-ml portions of dilutions of exponential-phase bacteria grown in BHI broth and resuspended in PBS. The numbers of survivors were recorded 6 days postchallenge.
The virulence of the P. multocida strains delivered by different routes of inoculation was examined next. Groups of mice were inoculated with graded doses of P. multocida strains by the i.p., i.n., and s.c. routes (Table 3). The results indicated that the LD50s varied according to the route of inoculation. The LD50 of P. multocida 85020 delivered by the i.p. and s.c. routes was <20 CFU/mouse, and the LD50 delivered by the i.n. route was ∼103 CFU/mouse. For the aroA mutant derivative JRMT1, the LD50 delivered by the s.c. and i.n. routes was >2 × 109 CFU per mouse, and no obvious toxicity was noted at the highest dose tested (2 × 109 CFU per mouse). Similar results were obtained with the Quetta parent strain and its aroA derivative, JRMT2, except that the Quetta strain appeared to be less virulent than strain 85020 when it was delivered by the i.n. route (the LD50 was ∼105 CFU per mouse for the Quetta strain, compared to ∼103 CFU per mouse for strain 85020) (data not shown).
TABLE 3.
Mouse virulence tests in which different routes of inoculation were useda
| Challenge strain | CFU per mouse (no. of survivors/no. challenged)
|
|||
|---|---|---|---|---|
| Expt 1
|
Expt 2 (i.n. route) | |||
| i.p. route | s.c. route | i.n. route | ||
| 85020 (wild type) | 2,000 (0/3) | 2,000 (0/3) | 2,000 (1/3) | 104 (1/5) |
| 200 (0/3) | 200 (0/3) | 200 (2/3) | 1,000 (3/5) | |
| 20 (0/3) | 20 (0/3) | 20 (3/3) | 100 (5/5) | |
| JRMT1 (aroA strain) | 2 × 109 (0/3) | 2 × 109 (3/3) | 2 × 109 (3/3) | |
| 2 × 108 (3/3) | 2 × 108 (3/3) | 2 × 108 (3/3) | ||
| 2 × 107 (3/3) | 2 × 107 (3/3) | 2 × 107 (3/3) | ||
Groups of mice were inoculated with graded doses of bacteria by the i.p., i.n., and s.c. routes. The numbers of survivors were recorded on day 4 postchallenge in experiment 1 and on day 10 postchallenge in experiment 2.
Mouse protection tests with aroA strains JRMT1 and JRMT2.
Due to some adverse effects on mouse weight gain resulting from a dose of 3 × 108 CFU per mouse, immunization with a smaller dose was considered preferable. A vaccination dose of ∼107 CFU per mouse was used as it offered good protection and resulted in no apparent toxicity. The protective properties of the aroA strains after inoculation via different routes and with one-and two-dose vaccination regimens were compared. Mice given two doses of 2 × 107 CFU of JRMT1 i.p. or i.n. were completely protected against i.p. challenge after an additional 2 weeks with high doses (1,000 or 10,000 LD50s) of the parent 85020 strain (Table 4). In fact, one i.p. inoculation of JRMT1 was sufficient to protect all the mice against challenge. However, this was not the case for i.n. inoculation of JRMT1 followed by i.p. challenge, where two doses were required for full protection. With the s.c. route of inoculation, mice given one dose of JRMT1 showed no protection and mice given two doses showed some protection, but this was the least efficient route examined. Similar results were obtained after vaccination with JRMT2 and challenge with the parent Quetta strain (data not shown). Good cross-protection by each aroA mutant against challenge with the heterologous parent strain was also evident (Table 4 and data not shown).
TABLE 4.
Protective properties of aroA mutant JRMT1 inoculated by different routesa
| Expt | First vaccination (CFU per mouse) | Second vaccination (CFU per mouse) | Route | Challenge dose
|
No. of survivors/no. challenged | |
|---|---|---|---|---|---|---|
| LD50 | Parental strain | |||||
| 1 | 2.3 × 107 | 1.1 × 107 | i.p. | 1,000 | 85020 | 6/6 |
| 2.3 × 107 | 1.1 × 107 | i.p. | 1,000 | Quetta | 6/6 | |
| 1.1 × 107 | i.p. | 1,000 | 85020 | 6/6 | ||
| 2.3 × 107 | 1.1 × 107 | i.p. | 10,000 | 85020 | 6/6 | |
| 1.1 × 107 | i.p. | 10,000 | 85020 | 6/6 | ||
| i.p. | 100 | 85020 | 0/6 | |||
| i.p. | 100 | Quetta | 0/6 | |||
| 2 | 2.4 × 107 | 2.2 × 107 | i.n. | 1,000 | 85020 | 4/4 |
| 2.4 × 107 | 2.2 × 107 | i.n. | 10,000 | 85020 | 4/4 | |
| 2.2 × 107 | i.n. | 1,000 | 85020 | 1/4 | ||
| 2.2 × 107 | i.n. | 10,000 | 85020 | 0/4 | ||
| 2.4 × 107 | 2.2 × 107 | i.n. | 1,000 | Quetta | 4/4 | |
| 2.4 × 107 | 2.2 × 107 | s.c. | 1,000 | 85020 | 2/4 | |
| 2.4 × 107 | 2.2 × 107 | s.c. | 10,000 | 85020 | 1/4 | |
| 2.2 × 107 | s.c. | 1,000 | 85020 | 0/4 | ||
| 2.2 × 107 | s.c. | 10,000 | 85020 | 0/4 | ||
| 2.4 × 107 | 2.2 × 107 | s.c. | 1,000 | Quetta | 1/4 | |
Groups of mice were vaccinated once and challenged i.p. 2 weeks later or vaccinated with two doses separated by a 2-week interval and challenged 2 weeks later. The numbers of survivors were recorded on day 6 postchallenge.
The effects of different challenge routes were compared after i.p., i.n., and s.c. vaccination with a single dose of JRMT1 and challenge 2 weeks later with 100 LD50s of the parent strain by the various routes (Table 5). The data showed that mice vaccinated i.p. were completely protected against challenge by all routes. Mice vaccinated i.n. showed complete protection against i.n. challenge but little or no protection against i.p. or s.c. challenge. Mice vaccinated s.c. did not show protection against challenge by any route. In a separate experiment, groups of mice were vaccinated with one dose of JRMT1 by the i.p., i.n., or s.c. route; then one-half of the groups were challenged i.p. 2 weeks later and one-half of the groups were challenged i.p. 4 weeks later with 100 LD50s of strain 85020. Results similar to those shown in Table 4 were obtained (data not shown); mice given the vaccine i.p. were fully protected, but mice vaccinated i.n. or s.c. were not protected, and there was no obvious benefit in terms of the survival rate derived from allowing an additional 2 weeks after vaccination before challenge.
TABLE 5.
Effects of different vaccination routes and different challenge routesa
| Vaccination route | Challenge route | No. of survivors/no. challenged |
|---|---|---|
| i.p. | i.p. | 3/3 |
| i.n. | 3/3 | |
| s.c. | 3/3 | |
| i.n. | i.p. | 1/3 |
| i.n. | 3/3 | |
| s.c. | 0/3 | |
| s.c. | i.p. | 0/3 |
| i.n. | 0/3 | |
| s.c. | 0/3 |
Groups of mice were vaccinated with a single dose of 2.6 × 107 CFU of aroA mutant strain JRMT1 per mouse. After 2 weeks, each group was challenged with 100 LD50s of the parent 85020 strain. The numbers of survivors were recorded 6 days postchallenge.
Mouse virulence and protection tests with aroA strain JRMT12.
A vaccine strain intended for use in the natural target species should not carry any antibiotic resistance genes. Thus, having established the attenuated and protective properties of the aroA strains JRMT1 and JRMT2, we constructed strains with aroA gene deletions, namely, JRMT12 from parent strain 85020 and JRMT13 from parent strain Quetta. These derivatives exhibited attenuation and protective properties in mice that were very similar to those of JRMT1 and JRMT2. Strain JRMT12 had LD50s of >2.6 × 107 and >3.8 × 108 CFU per mouse when it was inoculated i.p. and i.n., respectively (Table 6). Mice that survived i.p. and i.n. inoculation with the graded doses of JRMT12 were challenged by the same routes with 1,000 and 100 LD50s, respectively, of parent strain 85020 (Table 6). All the survivors from the initial i.p. inoculation survived challenge with the parent strain, giving results similar to those obtained with JRMT1, for which a single i.p. vaccination was fully protective against subsequent challenge (Tables 4 and 5). One i.n. vaccination was partially protective only at the highest dose, and again the results were similar to those obtained with JRMT1 (Tables 4 and 5).
TABLE 6.
Virulence and protective properties of aroA mutant JRMT12a
| Strain | Route of inoculation | Dose (CFU per mouse) | (No. of survivors/ no. inoculated) | No. of survivors/no. challenged with parent strain 85020 |
|---|---|---|---|---|
| 85020 (wild type) | i.p. | 10 | 0/3 | NDb |
| 1 | 2/3 | ND | ||
| <1 | 4/4 | ND | ||
| JRMT12 (aroA strain) | i.p. | 2.6 × 108 | 0/5 | |
| 2.6 × 107 | 5/5 | 5/5 | ||
| 2.6 × 106 | 5/5 | 5/5 | ||
| 2.6 × 105 | 5/5 | 5/5 | ||
| 85020 (wild type) | i.n. | 3.1 × 107 | 0/4 | |
| 3.1 × 106 | 1/4 | ND | ||
| 3.1 × 105 | 4/4 | ND | ||
| 3.1 × 104 | 4/4 | ND | ||
| JRMT12 (aroA strain) | i.n. | 3.8 × 108 | 4/4 | 2/4 |
| 3.8 × 107 | 4/4 | 0/4 | ||
| 3.8 × 106 | 4/4 | 0/4 | ||
| 3.8 × 105 | 4/4 | 0/4 |
Groups of mice were inoculated with graded doses of the wild-type or aroA strain. After 2 weeks, mice that had survived the initial i.p. inoculation with the aroA strain were challenged i.p. with 1,000 LD50s of the parent strain, while survivors from the i.n. inoculation were challenged with 100 LD50s of the parent strain. The numbers of survivors were recorded 6 and 10 days postchallenge for the mice challenged i.p. and i.n., respectively.
ND, not done.
Spread and persistence of attenuated strains in vivo.
The spread to different organs of the parent 85020 strain and its aroA derivative, JRMT1, was examined when the organisms were inoculated by different routes (Table 7). For the parent strain, the results showed that i.p. inoculation allowed proliferation and spread to all of the tissues examined at 24 h, and the mice died by 36 h. i.n. inoculation also led to spread of the parent strain to all tissues, but at a lower level. By 48 h the bacterial loads were appreciably lower than those at 24 h in the liver, spleen, and blood, although not in the lungs, perhaps indicating that the bacteria were beginning to be cleared from the former sites. However, the lung counts increased by 72 h and persisted at 96 h without death of the mice. Similar results were obtained with the Quetta parent strain, except that following i.n. inoculation the bacteria were cleared from all sites examined by 96 h (data not shown). For the aroA derivative of strain 85020, i.p. inoculation allowed distribution of the bacteria to all tissues except the lungs by 24 h, but at much reduced levels compared with the levels of the parent strain even though the mutant was administered at a 104-fold-higher dose (Table 7). The bacteria were cleared from all tissues by 48 h, and the mice survived. i.n. inoculation allowed persistence of the bacteria in the lungs for up to 72 h, and some bacteria were detected in the blood at 24 h but not at later times. Similar results were obtained with the Quetta aroA derivative, JRMT2, except that bacteria were still found in low numbers in the lungs at 96 h and some bacteria were detected in the liver as well as in the blood at 24 h; however, like JRMT1, no bacteria were detected in any tissues except the lungs after 24 h.
TABLE 7.
Viable counts of P. multocida strains in different organs following inoculation by two different routesa
| Inocu- lation route | Organ | No. of bacteria (CFU per g of tissue or per ml of blood)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Parent strain 85020
|
Mutant strain JRMT1
|
||||||||
| 24 h | 48 h | 72 h | 96 h | 24 h | 48 h | 72 h | 96 h | ||
| i.p. | Liver | 5.1 × 1010 (3.4 × 1010) | —b | — | — | 100 (2.0) | <100 | <100 | <100 |
| Lungs | 1.1 × 1011 (0.4 × 1011) | — | — | — | <100 | <100 | <100 | <100 | |
| Spleen | 2.7 × 1012 (2.0 × 1012) | — | — | — | 1.0 × 104 (0.3 × 104) | <100 | <100 | <100 | |
| Blood | 1.2 × 109 (0.2 × 109) | — | — | — | 8,000 (7.5) | <50 | <50 | <50 | |
| i.n. | Liver | 1.4 × 104 (1.0 × 104) | <100 | 1.4 × 109 (0.7 × 109) | 4.8 × 109 (4.2 × 109) | <100 | <100 | <100 | <100 |
| Lungs | 4.2 × 104 (3.2 × 104) | 3.1 × 108 (2.1 × 108) | 1.3 × 1011 (1.0 × 1011) | 3.3 × 109 (2.3 × 109) | 5.8 × 104 (2.2 × 104) | 1.8 × 104 (0.8 × 104) | 4.7 × 104 (0.3 × 104) | <100 | |
| Spleen | 2.8 × 105 (2.2 × 105) | 500 (50) | 3.2 × 1010 (1.2 × 1010) | 2.5 × 109 (1.5 × 109) | <100 | <100 | <100 | <100 | |
| Blood | 1,500 (100) | 50 (5) | 1.1 × 107 (1.1 × 107) | 3.0 × 104 (1.8 × 104) | 50 (10) | <50 | <50 | <50 | |
Groups of eight mice were inoculated with the 85020 parent strain at a challenge dose of 723 CFU per mouse i.p. or 7.23 × 104 CFU per mouse i.n. and with the aroA derivative JRMT1 at a challenge dose of 4.1 × 107 CFU per mouse i.p. or 4.1 × 109 CFU per mouse i.n. At 24-h intervals, two mice from each group were sacrificed, the relevant organs were removed aseptically, and the numbers of CFU of bacteria were determined as described in Materials and Methods. Heart blood (100 μl) was also collected aseptically and diluted into 5 ml of PBS, and 10-fold serial dilutions were plated on BHI blood agar. The values are the means (standard deviations) of duplicate platings from two mice.
—, mice died before this time.
DISCUSSION
The control of HS remains a problem because current vaccines are not sufficiently efficacious and require repeated administration. The vaccines are administered parenterally and may not effectively induce mucosal immunity. A live attenuated vaccine, which would mimic the early stages of the natural infection, might be expected to confer more solid and long-term protective immunity, particularly if it is amenable to oral or i.n. administration that would favor induction of mucosal immunity in a way similar to the natural infection. For live strains to be used as vaccines, the mode of attenuation should be well defined and constructed in such a way that the possibility of reversion to virulence is minimized. Shuttle vectors developed in this laboratory (5) enabled us to construct aroA attenuated derivatives of two P. multocida serotype B:2 strains, one from a case of HS in Sri Lanka (strain 85020) and another from Pakistan (Quetta strain). Primers were designed based on the published aroA sequence of P. multocida serotype A, and the amplified aroA sequences were disrupted by insertion of a Kmr cassette, transferred to the B:2 strains on a shuttle suicide vector, pAKA19, and incorporated into the chromosome via allelic exchange. The P. multocida aroA Sri Lankan B:2 strain (JRMT1) and the aroA Quetta B:2 strain (JRMT2) were confirmed to be aroA mutants by PCR and Southern blot analysis, enzyme assay, and a lack of growth on minimal medium.
For the constructs JRMT1 and JRMT2, repeated subculturing in the presence of kanamycin but in the absence of ampicillin was sufficient to promote loss of the vector plasmid. When the attenuated and protective properties of these strains were established, marker-free aroA mutants containing a deletion of a central portion of the aroA gene were constructed. Selection for allelic exchange with the native aroA sequence on the P. multocida chromosome was done by using sucrose sensitivity as a marker for elimination of vector sequences carrying the sacB gene. These strains are more suitable as vaccine candidates than the strains containing the Kmr insert in the aroA gene as they possess no antibiotic-encoding genes and deletion within the aroA gene eliminates the possibility of reversion to the wild type.
HS working parties set up by the United Nations Food and Agricultural Organisation have recommended the use of the mouse for testing new HS vaccines (6). Survival of mice and the LD50s demonstrated that the P. multocida aroA mutants are highly attenuated for virulence and for colonization and persistence in internal organs compared to the wild-type parent strains. When inoculated i.p. or i.n. into mice, the aroA mutant strains showed an obvious loss of virulence, and no illness was observed following administration of 107 and 109 CFU per mouse by the i.p. and i.n. routes, respectively. This compares with LD50s of less than 20 and 103 CFU per mouse for the parent strains inoculated by the i.p. and i.n. routes, respectively. The inability to isolate the aroA mutants from peripheral blood of mice 48 h after i.p. injection indicated that these strains had greatly reduced abilities to spread and survive in vivo.
Immunization with two i.p. or i.n. doses of P. multocida JRMT1 (aroA mutant of strain 85020) completely protected mice against homologous and heterologous i.p. challenge with 1,000 LD50s of the wild-type strains (Table 4). Similar results were obtained after vaccination with JRMT2 (aroA mutant of strain Quetta) followed by challenge with either parent strain. The route of natural infection by P. multocida is probably via the respiratory tract. However, unlike i.p. inoculation, one i.n. dose of live aroA vaccine of either strain induced relatively poor levels of protection against i.p. challenge with the homologous wild-type P. multocida strains. This protection was not affected by increasing the time between vaccination and challenge. However, mice vaccinated i.n. with a single dose were better protected against challenge via the same route (Tables 5 and 6). After a second dose of live cells inoculated i.n., there was a sharp increase in the number of survivors from a homologous or heterologous i.p. challenge (Table 4). Mice given two doses s.c. showed only partial protection against challenge, but a single s.c. dose provided no protection against challenge by any route examined. Thus, the route of inoculation has a significant effect on the protective efficacy of attenuated aroA strains in the mouse model.
The BALB/c mice were highly susceptible to infection by wild-type P. multocida B:2 strains whether they were introduced i.p. or i.n.. The growth of the challenge organism in the liver, lung, spleen, and blood was quantified daily for up to 4 days. P. multocida parent strains were able to multiply very rapidly in vivo, so that introduction of a small number of viable bacteria into the peritoneal cavities of nonvaccinated mice quickly resulted in an in vivo population of >109 viable organisms per g of liver, lung, spleen, or blood, which resulted in death of the mice within 36 h. As suggested by Collins (8), it is probable that unrestricted extracellular growth of the unopsonized organisms occurs within the peritoneal cavity. This is due to the virtual absence of phagocytosis and inactivation of the challenge inoculum by the host macrophages, which allows the organism to grow in the tissue at rates normally achievable only in vitro. When mice were inoculated i.n. with the parent strains at an initial challenge dose of 104 CFU, bacteria spread into the liver, spleen, and blood by 24 h, but the numbers were much lower than those after i.p. inoculation. The bacterial load persisted in all tissues up to 96 h but not at a level that resulted in death of the mice. A similar situation was reported by Collins (8), who used a wild-type P. multocida serotype 5:A turkey isolate which, when inoculated i.n. into mice, spread into the internal organs by 24 h.
With the attenuated JRMT1 and JRMT2 aroA mutant strains, after a single high i.n. challenge dose (>109 CFU per mouse), large numbers of bacteria were detected in the lungs by 24 h, but only small numbers were present in the blood and no bacteria were present in the liver and spleen (Table 5). By contrast, bacteria were clearly present in the blood, liver, and spleen after a single high i.p. dose (>107 CFU per mouse), but the numbers of viable bacteria were greatly reduced compared to the numbers of cells of the parent strains. In the lungs, however, no colonies were detected. Following administration of a single dose by either route, attenuated bacteria were cleared from the blood, liver, and spleen by 48 h, but after i.n. inoculation the attenuated bacteria persisted in the lungs for up to 96 h, although the numbers did not appear to increase significantly like the numbers of the parent strain. The protection studies (Table 4) showed that after one dose, mice were fully protected after i.p. vaccination but were only partially protected after i.n. vaccination, when they were challenged by the i.p. route. Yet, one i.n. dose fully protected against i.n. challenge (Table 5). These differences in efficacy between the i.p. and i.n. routes of vaccination may be a reflection of the smaller numbers of bacteria able to pass from the lungs into the blood and other tissues after a single i.n. inoculation in order to stimulate a fully protective immune response.
HS pathogenesis in cattle and buffaloes involves the rapid translocation of bacteria from the respiratory tract to the blood, liver, and spleen (10, 36), suggesting that the bacteria are able to invade via the mucosal epithelial layers. A similar disease progression occurs in fowl cholera caused by serotype A isolates of P. multocida, which can manifest as an acute septicemia. An avian strain of P. multocida serotype A:3 has been shown to invade polarized epithelial cells in an actin-dependent manner (24), and several studies with avian isolates have shown uptake by and survival in turkey and chicken macrophages and neutrophils (12, 13, 23). Thus, dissemination to the deeper tissues may well occur by vascular migration facilitated by association with phagocytic cells (32, 33). Truscott and Hirsh (31) reported that a P. multocida strain of avian origin produced a substance(s) that interfered with the function of phagocytic cells. The potential for cell invasion and intracellular survival of P. multocida B:2 strains in macrophages and other cell types deserves further study, because significant persistence in an intracellular environment, where humoral immune responses should be ineffective, might indicate that cellular immune mechanisms play a vital role in clearing the infection.
The aroA derivatives of the P. multocida B:2 strains are thus candidate organisms for a live attenuated vaccine against HS as the safety and efficacy of these strains have been demonstrated in a mouse model of infection. Vaccine trials with either cattle or buffaloes, however, are needed in order to establish that the safety and protective properties demonstrated in the mouse are reflected in similar properties in the target species.
Acknowledgments
This work was supported by The Wellcome Trust, Biotechnology and Biological Sciences Research Council, United Kingdom, and by the Iranian government in the form of a scholarship to M.T.
Editor: R. N. Moore
REFERENCES
- 1.Adler, B., R. Chancellor, P. Homchampa, M. Hunt, C. Roffolo, R. Strugnell, and D. Wapling. 1996. Immunity and vaccine development in Pasteurella multocida infection. J. Biotechnol. 44:139-144. [DOI] [PubMed] [Google Scholar]
- 2.Adler, B. R., D. Bulach, J. Chung, S. Doughty, M. Hunt, K. Rajakumar, M. Serrano, A. V. Zanden, Y. Zhang, and C. Ruffolo. 1999. Candidate vaccine antigens and genes in Pasteurella multocida. J. Biotechnol. 73:83-90. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. A., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, and K. Struhl. 1990. Current protocols in molecular biology. Green Publishing Associates and Wiley Interscience, New York, N.Y.
- 4.Azad, A. K., J. G. Coote, and R. Parton. 1992. Distinct plasmid profiles of P. haemolytica serotypes and the characterization and amplification in E. coli of ampicillin-resistance plasmids encoding ROB-1 β-lactamase. J. Gen. Microbiol. 138:1185-1196. [DOI] [PubMed] [Google Scholar]
- 5.Azad, A. K., J. G. Coote, and R. Parton. 1994. Construction of conjugative shuttle and suicide vectors for Pasteurella haemolytica and P. multocida. Gene 145:81-85. [DOI] [PubMed] [Google Scholar]
- 6.Bain, R. V. S., M. C. L. De Alwis, G. R. Carter, and B. Gupta. 1982. Haemorrhagic septicaemia. FAO Animal Production and Health paper 33. Food and Agricultural Organisation, Rome, Italy.
- 7.Chandrasekaran, S., L. Kennett, P. C. Yeap, N. Muniandy, B. Rani, and T. K. S. Mukkur. 1994. Characterisation of immune response and duration of protection in buffaloes immunised with haemorrhagic septicaemia vaccines. Vet. Microbiol. 41:213-219. [DOI] [PubMed] [Google Scholar]
- 8.Collins, F. M. 1973. Growth of Pasteurella multocida in vaccinated and normal mice. Infect. Immun. 8:868-875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dawkins, R. H., R. B. Johnson, T. L. Spencer, and B. Adler. 1990. Pasteurella multocida infections in mice with reference to haemorrhagic septicaemia in cattle and buffalo. Immunol. Cell Biol. 68:57-61. [DOI] [PubMed] [Google Scholar]
- 10.De Alwis, M. C. L. 1992. Haemorrhagic septicaemia—a general review. Br. Vet. J. 148:99-112. [DOI] [PubMed] [Google Scholar]
- 11.Dower, W. J., J. F. Miller, and C. W. Regsdale. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127-6145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harmon, B. G., J. R. Glisson, K. S. Latimer, W. L. Stevens, and J. C. Nunnally. 1991. Resistance of Pasteurella multocida A:3,4 to phagocytosis by turkey macrophages and heterophils. Am. J. Vet. Res. 52:1507-1511. [PubMed] [Google Scholar]
- 13.Harmon, B. G., J. R. Glisson, K. S., and J. C. Nunnally. 1992. Turkey macrophage and heterophil bactericidal activity against Pasteurella multocida. Avian Dis. 36:989-991. [PubMed] [Google Scholar]
- 14.Homchampa, P., R. A. Strugnell, and B. Adler. 1992. Molecular analysis of the aroA gene of Pasteurella multocida and vaccine potential of a constructed aroA mutant. Mol. Microbiol. 6:3585-3593. [DOI] [PubMed] [Google Scholar]
- 15.Homchampa, P., R. A. Strugnell, and B. Adler. 1997. Cross protective immunity conferred by a marker-free aroA mutant of Pasteurella multocida. Vaccine 15:203-208. [DOI] [PubMed] [Google Scholar]
- 16.Jablonski, L., N. Sriranganathan, S. M. Boyle, and G. R. Carter. 1992. Conditions for transformation of Pasteurella multocida by electroporation. Microb. Pathog. 12:63-68. [DOI] [PubMed] [Google Scholar]
- 17.Jablonski, P. E., M. Jaworski, and C. J. Hovde. 1992. A minimal medium for growth of Pasteurella multocida. FEMS Microbiol. Lett. 140:165-169. [DOI] [PubMed] [Google Scholar]
- 18.Jost, B. H., P. Homchampa, R. A. Strugnell, and B. Adler. 1997. The sacB gene cannot be used as a counter-selectable marker in Pasteurella multocida. Mol. Biotechnol. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 19.Lewendon, A., and J. R. Coggins. 1987. 3-Phosphoshikimate 1-carboxypyruvyltransferase from Escherichia coli. Methods Enzymol. 142:342-348. [DOI] [PubMed] [Google Scholar]
- 20.Martinez de Tejada, G. M., J. F. Miller, and P. A. Cotter. 1996. Comparative analysis of the virulence control systems of Bordetella pertussis and Bordetella bronchiseptica. Mol. Microbiol. 22:895-908. [DOI] [PubMed] [Google Scholar]
- 21.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Myint, A., and G. R. Carter. 1989. Prevention of haemorrhagic septicaemia in buffaloes and cattle with a live vaccine. Vet. Rec. 124:508-509. [DOI] [PubMed] [Google Scholar]
- 23.Poermadjaja, B., and A. Frost. 2000. Phagocytic uptake and killing of virulent and avirulent strains of Pasteurella multocida of capsular serotype A by chicken macrophages. Vet. Microbiol. 72:163-171. [DOI] [PubMed] [Google Scholar]
- 24.Rabier, M. J., N. K. Tyler, N. J. Walker, L. M. Hansen, D. C. Hirsh, and F. Tablin. 1997. Pasteurella multocida enters polarized epithelial cells by interacting with host F-actin. Vet. Microbiol. 54:343-355. [DOI] [PubMed] [Google Scholar]
- 25.Reid, J. L., and A. Collmer. 1987. A nptl-sacB-sacR cartridge for constructing directed unmarked mutations in Gram-negative bacteria by marker exchange-eviction mutagenesis. Gene 57:239-246. [DOI] [PubMed] [Google Scholar]
- 26.Rimler, R. B. 1996. Passive immune cross-protection in mice produced by rabbit antisera against different serotypes of Pasteurella multocida. J. Comp. Pathol. 114:347-360. [DOI] [PubMed] [Google Scholar]
- 27.Rushton, B. 1978. Induction of pneumonia in mice with Pasteurella haemolytica. J. Comp. Pathol. 88:477-480. [DOI] [PubMed] [Google Scholar]
- 28.Scott, P. C., J. F. Markham, and K. G. Whitear. 1999. Safety and efficacy of two live Pasteurella multocida aroA mutant vaccines in chickens. Avian Dis. 43:83-88. [PubMed] [Google Scholar]
- 29.Shah, N. H., N. H. Shah, and F. K. De Graaf. 1997. Protection against haemorrhagic septicaemia induced by vaccination of buffalo calves with an improved oil adjuvant vaccine. FEMS Microbiol. Lett. 155:203-207. [DOI] [PubMed] [Google Scholar]
- 30.Tartof, K. D., and C. A. Hobbs. 1987. Improved media for growing plasmid and cosmid clones. Bethesda Res. Lab. Focus 9:12. [Google Scholar]
- 31.Truscott, W. M., and D. C. Hirsh. 1988. Demonstration of an outer membrane protein with antiphagocytic activity from Pasteurella multocida of avian origin. Infect. Immun. 56:1538-1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsuji, M., and T. Matsumoto. 1989. Pathogenesis of fowl cholera: influence of encapsulation on the fate of Pasteurella multocida after intravenous inoculation into turkeys. Avian Dis. 33:238-247. [PubMed] [Google Scholar]
- 33.Tsuji, M., and T. Matsumoto. 1990. Immune defense mechanism against blood-borne Pasteurella multocida in turkeys. Res. Vet. Sci. 48:344-349. [PubMed] [Google Scholar]
- 34.Verma, R., and T. N. Jaiswal. 1997. Protection, humoral and cell-mediated immune responses in calves immunised with multiple emulsion haemorrhagic septicaemia vaccine. Vaccine 15:1254-1260. [DOI] [PubMed] [Google Scholar]
- 35.Verma, R., and T. N. Jaiswal. 1998. Haemorrhagic septicaemia vaccines. Vaccine 16:1184-1192. [DOI] [PubMed] [Google Scholar]
- 36.Wijewardana, T. G. 1992. Haemorrhagic septicaemia. Rev. Med. Microbiol. 3:59-63. [Google Scholar]