Abstract
The liver is an important site of host-microbe interaction. Although hepatocytes have been reported to be responsive to lipopolysaccharide (LPS), the global gene expression changes by LPS and mechanism(s) by which LPS stimulates cultured hepatocytes remain uncertain. Cultures of primary mouse hepatocytes were incubated with LPS to assess its effects on the global gene expression, hepatic transcription factors, and mitogen-activated protein (MAP) kinase activation. DNA microarray analysis indicated that LPS modulates the selective expression of more than 80 genes and expressed sequence tags. We have shown previously that hepatocytes express CD14, which is required both for uptake and responsiveness to LPS. In other cells, responsiveness to microbial products requires expression of Toll-like receptors (TLR) and their associated accessory molecules. Hepatocytes expressed TLR1 through TLR9 as well as MyD88 and MD-2 transcripts, as shown by reverse transcriptase PCR analysis, indicating that hepatocytes express all known microbe recognition molecules. The MAP kinase extracellular signal-regulated kinase 1/2 was phosphorylated in response to LPS in mouse hepatocytes, and the levels of phosphorylation were lower in hepatocytes from TLR4-null mice. NF-κB activation was reduced in TLR4-mutant or -null hepatocytes compared to control hepatocytes, and this defect was partially restored by adenoviral transduction of mouse TLR4. Thus, hepatocytes respond to nanogram concentrations of LPS through a TLR4 response pathway.
Lipopolysaccharide (LPS), a glycolipid constituent of the outer membrane of gram-negative bacteria, initiates signaling cascades in cells such as macrophages and endothelial cells, leading to the release of cytokines and other inflammatory mediators during sepsis. Excessive production of these mediators can cause septic shock and multiple organ failure (55).
A decade ago, CD14, a 55-kDa glycoprotein and monocyte differentiation antigen, was identified as an important LPS recognition molecule (60). CD14 alone, however, is unable to transduce the intracellular LPS signal, since CD14 is only tethered to the cytoplasmic membrane by a glycosyl phosphatidylinositol anchor and lacks a membrane-spanning domain (17). Members of a family of proteins, the mammalian homologues of the Drosophila Toll protein, were found to act as transmembrane coreceptors to CD14 in the cellular response to LPS (34). These Toll-like receptors (TLR) contain ectodomains with leucine-rich repeats, and their intracellular motifs are highly homologous to intracellular signaling domains of interleukin-1 receptor type I (IL-1RI) and IL-1RI accessory protein (reviewed in reference 5). Following dimerization of the TLR, these domains attract the adapter protein MyD88, which in turn recruits the IL-1R-associated kinase. Following this association, IL-1R-associated kinase phosphorylates tumor necrosis factor receptor-associated factor 6, which in turn attracts two more protein tyrosine kinases, transforming growth factor beta-activated kinase 1 (TAK-1) and TAK-1-binding protein 1 (TAB-1) (25). These events eventually lead to the activation of the transcription factors NF-κB or AP-1 (5, 25). Another molecule, MD-2, is associated with TLR4 and is absolutely required for the TLR4 signaling (9).
So far, at least 10 TLR (TLR1 to -10) have been discovered, but only TLR2 and TLR4 have been implicated in the cellular signaling response to LPS. Recent studies suggest that a defective murine TLR4 is responsible for the LPS-hyporesponsiveness in two mouse strains (C3H/HeJ and C57BL10/ScCr) (41). Furthermore, studies in TLR4-deficient mice indicate that TLR4 is essential for LPS-mediated signaling (53). In contrast, accumulating evidence indicates that TLR2 is not sufficient to confer LPS responsiveness (18). Instead, recent data indicate that gram-positive bacterial products such as Borrelia burgdorferi lipoproteins and lipopeptides (20), lipoteichoic acid, peptidoglycan (49), Mycobacterium avium, and heat-killed gram-positive bacteria (Staphylococcus aureus) (30) are TLR2 ligands. We have shown recently that TLR2 is induced in vivo following challenge with LPS and that hepatocytes upregulate the expression of TLR2 in response to tumor necrosis factor alpha (TNF-α) and IL-1 in vitro (32).
The liver is a major target organ of LPS and other microbial products. Early observations demonstrated that the liver is a primary site of uptake and clearance of microbial products, including LPS (reviewed in reference 54). Kupffer cells were shown to be as responsive as other macrophage populations to LPS, producing TNF-α and IL-1 that activate hepatocytes to express proinflammatory products such as the inducible nitric oxide synthase (28). Hepatocytes were initially viewed as passive from an immune standpoint (reviewed in reference 57). Recent data, however, suggest that hepatocytes in fact do play an important role in inflammation during sepsis (56). Previous studies have indicated that LPS leads to induction of the following by monocultures of primary hepatocytes: serum amyloid A, an acute-phase protein (22, 50); cytochrome P450 (37); iron uptake (43); binding of the transcription factor NF-κB (13); superoxide dismutase activity (42); release of the chemoattractant Kc/gro (51); alpha 2 macroglobulin, TNF-α, and IL-6 (47); LPS binding protein (LBP) (58); adhesion molecule expression (48); the production of l-arginine (24); and metallothionein (1). However, most of these studies demonstrated these effects at relatively high doses of LPS (e.g., 1 to 10 μg/ml). We (31) and others (12) have reported that hepatocytes express CD14 in a regulated manner. Hepatocyte CD14 expression is markedly upregulated during endotoxemia (31). We also repeated similar observation in hepatocyte TLR2 (32). These observations suggest that direct response to microbial products by hepatocytes may be a component of the innate immune response.
In the present study, we hypothesized that hepatocytes would respond in a robust fashion to moderate (1- to 1,000-ng/ml) concentrations of LPS by modulating a defined profile of genes. Furthermore, we hypothesized that hepatocytes would respond to LPS using the same molecular mechanisms used by monocytes/macrophages. Accordingly, we sought to evaluate the response of hepatocytes to LPS in the absence of cytokines and nonparenchymal cells, with focus on the role of TLR4 as a critical LPS recognition molecule. Hepatocytes from TLR4-mutant or -null mice (C3H/HeJ and C57BL/10ScN, respectively) and their respective control mice (C3H/HeN and C57BL/10SnJ) were compared with respect to their LPS-mediated effects on activation of NF-κB and mitogen-activated protein (MAP) kinase phosphorylation. Our results indicate that LPS alters the expression of numerous genes, some of them known to be modulated by either AP-1 or NF-κB or both. Finally, our studies demonstrate a defect in NF-κB activation in hepatocytes from TLR4-deficient or TLR4-null mice. Taken together with our previous studies, these results clearly show that hepatocytes possess all the molecular machinery to respond to LPS and other microbial products in a manner at least qualitatively similar to that of monocytes/macrophages.
MATERIALS AND METHODS
Reagents.
LPS (Escherichia coli 0111:B4) was purchased from List Biological Laboratories, Inc. (Vandell Way, Calif.). This LPS dose not contain a significant amount of contaminating proteins that could stimulate TLR2 nonspecifically (20). Williams medium E was purchased from Gibco (Grand Island, N.Y.); fetal calf serum was purchased from HyClone Laboratories (Logan, Utah). All tissue culture plates and flasks were purchased from Corning (Corning, N.Y.). NF-κB and AP-1 consensus oligonucleotides were ordered from Santa Cruz Biotechnology Inc. (Santa Cruz, Calif.). The HepG2 cell line was ordered from ATCC.
Animals.
TLR4-mutant or -null mice (C3H/HeJ and C57BL/10ScN, respectively), which were pathogen free and weighed approximately 20 g, were purchased from Jackson Laboratories (Bar Harbor, Maine) and Harland Laboratories (Indianapolis, Ind.), respectively. The control mice, C57BL/10SnJ and C3H/HeN, were ordered from Jackson Laboratories and Charles River Laboratories (Wilmington, Mass.), respectively. The mice were exposed each day to 12 h of light and darkness. Rodent chow and water were provided ad libitum. Experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Hepatocyte isolation.
Hepatocytes were isolated from mice by an in situ collagenase (type VI; Sigma, St. Louis, Mo.) perfusion technique, modified as described previously (59). Hepatocytes were separated from the nonparenchymal cells by two cycles of differential centrifugation (50 × g for 2 min) and further purified over a 30% Percoll gradient. The purity of these hepatocyte cultures exceeded 98% by light microscopy, and viability was typically more than 95% by trypan blue exclusion assay.
Cell culture and treatment.
Hepatocytes (4 × 105) were plated onto six-well gelatin-coated plastic tissue plates. Culture medium was Williams medium E containing 10% calf serum, 15 mM HEPES, 10−6 M insulin, 2 mM l-glutamine, and penicillin and streptomycin (both at 100 U/ml). Hepatocytes were allowed to attach to plates overnight prior to treatment. Both hepatocytes from control and TLR4-deficient or -null mice were harvested at the same time and plated in equivalent numbers.
cDNA microarray hybridization, scanning, and data analysis.
Total RNA extracted from cultured hepatocytes of normal or LPS-treated C3H/HeN mice was submitted to Incyte Pharmaceuticals (Fremont, Calif.) for further processing. Total RNA was extracted as described previously (31). cDNA microarray setup, probe labeling, hybridization, and signal scanning were performed by Incyte Pharmaceuticals. Hepatocyte RNAs from normal and LPS-treated mice were designated for labeling probe 1 (cyanine-3 dUTP labeled; green) and probe 2 (cyanine-5 dUTP labeled; red), respectively. Differential expression is the ratio of an element's probe 1 signals to probe 2 signals. Data were analyzed using GEMTool software (Incyte Pharmaceuticals).
Preparation of cell lysates and Western blotting analysis.
Hepatocytes were cultured on six-well plates, were washed once with ice-cold phosphate-buffered saline (PBS), and were lysed in 1× lysis buffer (New England Biolabs, Inc., Beverly, Mass.) containing 20 mM Tris-HCl (pH 7.4), 150 mM NaC1, 1 mM Na2-EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and leupeptin (1 μg/ml), phenylmethylsulfonyl fluoride (PMSF) (1 μg/ml), pepstatin (1 μg/ml), and aprotinin (1 μg/ml) on ice for 20 min. Cell lysates were scraped and transferred into microcentrifuge tubes, and lysates were clarified by centrifugation at 15,000 × g for 15 min. The supernatant was collected, and protein content was determined by a BCA protein assay kit (Pierce, Rockford, Ill.). For Western blotting analysis, equal protein amounts (50 μg per well) were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis and transferred onto a nitrocellulose membrane. The membrane was immunostained with the anti-phospho-p42/44, anti-phospho-p38 or anti-phospho-c-Jun N-terminal protein kinase antibody (Santa Cruz or New England Biolabs) and detected with horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin G in a standard enhanced chemiluminescence reaction according to the instructions of the manufacturer (Pierce). The films were scanned by a scanner (HP ScanJet 6100C), and signal intensity was analyzed by the imaging analysis software UN-SCAN-IT gel (Silk Scientific, Orem, Utah). Multiple exposures were performed to make certain the films used for scanning were within linear-exposure range.
Total RNA isolation and Northern blot analysis.
Total RNA was extracted as described previously (31). For Northern blot analysis, total RNA (20 μg per well) was resolved by electrophoresis in a 1% agarose gel containing 2.2 M formaldehyde prior to being transferred to a Genescreen membrane (Dupont, NEN Research Products, Boston, Mass.). The RNA was cross-linked to the membrane with UV Stratalinker (Stratagene, San Diego, Calif.) and then hybridized with a 32P-labeled rat TLR4 probe (provided by Ralph A. Kelly, Cardiovascular Division, Brigham and Women's Hospital, Boston, Mass.). The probe was labeled using a random primed labeling kit (Roche Molecular Biochemicals, Indianapolis, Ind.). Hybridization was carried out at 43°C for 16 to 18 h in a buffer containing 50% deionized formamide, 0.25 M sodium phosphate (pH 7.2), 0.25 M NaCl, 1 mM EDTA, 7% SDS, and denatured salmon sperm DNA (100 μg/ml). Blots were washed for 5 min four times in 2× SSC (1× SSC is 0.015 M NaCl plus 0.015 M sodium citrate)-0.5% SDS at room temperature and three times in 0.1× SSC-0.5% SDS for 10 min at 53°C prior to exposure to X-ray film for autoradiography
Transient-transfection assay.
Cells (4 × 105 cells per well) were placed on six-well plates the day before the transfection. Plasmid DNA prepared by a Qiagen Maxi Kit (Qiagen, Santa Clara, Calif.) was transiently transfected into cells by Lipofectin according to the instructions of the manufacturer (Gibco-BRL). Plasmid pIEPlacZ served as an internal-control vector to normalize the transfection efficiency. Cells were transfected with pELAM (0.1 μg/well) (8), an NF-κB luciferase reporter vector generated by cloning a fragment (bp −241 to −54) of the NF-κB-responsive E-selectin promoter into the pGL3 reporter plasmid (Promega, Inc.), (provided by Douglas T. Golenbock, Boston University Medical School, Boston, Mass.), or AP-1-luc, an AP-1 luciferase reporter vector containing four copies of TRE binding site upstream of luciferase gene in pBR vector (provided by Richard A. Flavell, Section of Immunobiology, Yale University School of Medicine, New Haven, Conn.) plus pIEPlacZ (0.5 μg/well). Cells then recovered overnight in complete growth medium. At 24 h after the start of transfection, cells were treated with LPS for 6 h and then were washed twice with PBS and dissolved in 250 μl of 1× reporter lysis buffer (Promega). Twenty microliters of lysate was used for assay by an Autolumat LB953 luminometer (EG&G Flow Technology Inc., Nashua, N.H.), and luciferase activity of pELAM or AP-1-luc was normalized to β-galactosidase activity.
Preparation of nuclear extracts and electrophoretic mobility shift assay (EMSA).
Cells were washed with cold PBS and then scraped, centrifuged, resuspended in at least 5 pellet volumes of buffer A (10 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 0.5 mM dithiothreitol [DTT], 0.2 mM PMSF, and 0.5% NP-40), and incubated on ice for 15 min before being vigorously vortexed for 10 s at a maximum speed. The nuclei were washed once with buffer A. Nuclear proteins were extracted by gently resuspending the nuclei with an appropriate volume of buffer C (20 mM HEPES [pH 7.9], 10 mM KCl, 1.5 mM MgCl2, 10% glycerol, 0.2 mM EDTA, 0.5 mM DTT, 0.2 mM PMSF) along with buffer D (same as buffer C but has 400 mM KCl). The ratio of buffer C to buffer D was 3 to 1. Buffer D was added in a dropwise fashion. After incubating the nuclei in buffer C plus D for 1 h at 4°C, supernatants were collected by centrifugation at 13,800 × g for 15 min. Double-stranded NF-κB-specific oligonucleotide was end labeled with [γ-32P]ATP using T4 polynucleotide kinase (U.S. Biochemicals, Cleveland, Ohio) and purified on a G-50 Sephadex spin column. Nuclear proteins (5 μg per well) were incubated with 50,000 cpm of 32P-labeled oligonucleotide for 30 min at room temperature in a reaction mixture containing 1 μg of poly(dI-dC), 12.5 mM Tris-HCl (pH 7.5), 50% glycerol, 0.25 mM EDTA, 1.25% NP-40, and 0.25 mM DTT (final volume 20 μl). The DNA-protein complexes were resolved on a 4% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA (TBE) buffer. The gels were dried and then subjected to autoradiography.
Reverse transcriptase PCR (RT-PCR).
Total RNA was extracted as described previously (31). Expression of mRNAs for human TLR1 to TLR9 as well as MyD88 and MD-2 was assessed by first-strand cDNA synthesis from 5 μg of total RNA by extension of oligo(dT) 15 primers (Roche Molecular Biochemicals) with 200 U of Moloney murine leukemia virus RT (Gibco-BRL). PCR of the cDNA was performed in a 50-μl final volume containing 1.5 mM MgC12, a 200 μM concentration of deoxynucleoside triphosphate mix, a 0.6 μM concentration of each gene-specific primer, and 0.4 μl of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). The primer design was assisted by using the primer design software Amplify. All primer sequences are shown in Table 1. All PCR products were size fractionated by 2% agarose gel electrophoresis, and DNA bands were visualized by staining the gel with ethidium bromide.
TABLE 1.
TLR RT-PCR primer sequences
| Gene name | Size (nt) | Primer direction | RT-PCR sequence |
|---|---|---|---|
| Human TLR1 | 439 | Sense | 5′-AGTTGTCAGCGATGTGTTCGG-3′ |
| Antisense | 5′-GATCAAGTACCTTGATCCTGGG-3′ | ||
| Human TLR2 | 294 | Sense | 5′-GGCTTCTCTGTCTTGTGACC-3′ |
| Antisense | 5′-GGGCTTGAACCAGGAAGACG-3′ | ||
| Human TLR3 | 270 | Sense | 5′-AGCCACCTGAAGTTGACTCAGG-3′ |
| Antisense | 5′-CAGTCAAATTCGTGCAGAAGGC-3′ | ||
| Human TLR4 | 438 | Sense | 5′-TTGTATTCAAGGTCTGGCTGG-3′ |
| Antisense | 5′-GCAACCTTTGAAACTCAAGCC-3′ | ||
| Human TLR5 | 338 | Sense | 5′-AGCCATCTGACTGCATTAAGG-3′ |
| Antisense | 5′-GACTTCCTCTTCATCACAACC-3′ | ||
| Human TLR6 | 592 | Sense | 5′-CCTGGGAGGTAAACATCTGA-3′ |
| Antisense | 5′-CCCTCAACCACATAGAAACGA-3′ | ||
| Human TLR7 | 321 | Sense | 5′-GATAACAATGTCACAGCCGTCC-3′ |
| Antisense | 5′-GTTCCTGGAGTTTGTTGATGTTC-3′ | ||
| Human TLR8 | 309 | Sense | 5′-GTGTCACCCAAACTGCCAAGCTCC-3′ |
| Antisense | 5′-GATCCAGCACCTTCAGATGAGGC-3′ | ||
| Human TLR9 | 233 | Sense | 5′-TACCAACATCCTGATGCTAGACTC-3′ |
| Antisense | 5′-TAGGACAACAGCAGATACTCCAGG-3′ | ||
| Human MyD88 | 358 | Sense | 5′-CCGCGCTGGCGGAGGAGATGGAC-3′ |
| Antisense | 5′-GCAGATGAAGGCATCGAAACGCTC-3′ | ||
| Mouse TLR2 | 329 | Sense | 5′-GGCATTAAGTCTCCGGAATTATC-3′ |
| Antisense | 5′-CCATTGAGGGTACAGTCGTCG-3′ | ||
| Mouse TLR4 | 439 | Sense | 5′-GGAAGGACTATGTGATGTGACC-3′ |
| Antisense | 5′-GCTCTTCTAGACCCATGAAATTGG-3′ | ||
| Mouse MD-2 | 293 | Sense | 5′-CTGAATCTGAGAAGCAACAGTGG-3′ |
| Antisense | 5′-CAGTCTCTCCTTTCAGAGCTCTGC-3′ |
Recombinant adenovirus.
Adenovirus vectors with E1 and E3 deleted were constructed through Cre-lox recombination with reagents generously provided by S. Hardy (Somatix, Alameda, Calif.). An EagI/SmaI fragment containing the mouse C3H/HeN TLR4 cDNA (provided by Bruce Beutler, Department of Immunology, IMM-31, The Scripps Research Institute, La Jolla, Calif.) was blunted and inserted into the SmaI site of the shuttle vector pAdlox. An EagI/KpnI fragment containing the TLR4dl cDNA was inserted into the XbaI/KpnI site of the shuttle vector pAdlox. Recombinant adenoviruses were generated by cotransfection of SfiI-digested pAdlox-TLR4wt or pAdlox-TLR4dn and Ψ5 helper virus DNA into the adenovirus packaging cell line CRE8, which expresses Cre recombinase. Recombinant adenoviruses were propagated on CRE8 or 293 cells, purified by cesium chloride density gradient centrifugation and subsequent dialysis according to standard protocols, and stored at −70°C.
Statistical analysis.
Data are presented as means ± standard errors (SE). Experimental results are analyzed for their significance by analysis of variance (Fisher's protected least-significant difference test) by using the statistics software Statview (Abacus Concept Inc., Berkeley, Calif.). Significance was established at the 95% confidence level (P < 0.05).
RESULTS
Effects of LPS on hepatocytes in vitro: a DNA microarray analysis.
The effects of LPS on hepatic metabolism and function of the host are well known (44). In contrast, the direct responses of hepatocytes to moderate doses of LPS are not well characterized. We reasoned that one measure of the responsiveness of hepatocytes to microbial products is the extent of changes in gene expression in response to LPS in isolated cell culture. We utilized the technique of DNA microarray analysis (45) to address this issue. Hepatocytes isolated from C3H/HeN LPS-responsive mice were either untreated or stimulated in vitro for 8 h with LPS (100 ng/ml). Subsequently, mRNA was isolated and hybridized to the mouse GEM array (Incyte Pharmaceuticals), which contains 9,622 known genes and expressed sequence tags (ESTs). Of these, 85 transcripts (0.88%) showed differential expression using 1.5-fold (balanced differential expression) as a cutoff (Tables 2 and 3). The range of differential expression varied from +2.9 (ESTs similar to phosphatidylinositol 3-kinase [PI 3-kinase] catalytic subunit beta; GenBank accession number AA779957) to −2.2 (EST unknown; GenBank accession number AI430081). Of the total 85 transcripts, 68 gene (80%) expression levels were increased (≥1.5-fold), and only 17 (20%) gene expression levels were decreased by in vitro LPS treatment in hepatocytes. In all of those differentially expressed genes, according to our inclusion criteria 21 encoded ESTs with no definitive homology to known genes, while 64 encoded known genes.
TABLE 2.
DNA microarray analysis of gene expression changes in mouse hepatocytes stimulated with LPS: increased gene expressiona
| Function | Fold change | Gene product(s) and description |
|---|---|---|
| Cell death or apoptosis | 2.9 | ESTs, moderately similar to PI-3 catalytic subunit, beta isoform (Homo sapiens) |
| 1.5 | Calpastatin | |
| Liver metabolizing enzyme | 1.6 | Aldehyde dehydrogenase family 1, subfamily A4 |
| 1.6 | Selenophosphate synthetase 2 | |
| 1.6 | Cytochrome P450, 2a5 | |
| 1.6 | Alcohol dehydrogenase 5 | |
| 1.6 | Spermine synthase | |
| 1.6 | Triosephosphate isomerase | |
| 1.6 | Uridine phosphorylase | |
| 1.5 | Phosphoglycerate kinase 1 | |
| 1.5 | Mitochondrial acyl coenzyme A thioesterase | |
| 1.5 | Cytochrome P450, 2c29 | |
| 1.5 | Cytochrome P450, 2a4 | |
| 1.5 | Cytochrome P450, 2c37 | |
| 1.5 | Paraoxonase 1 | |
| Signal transduction | 2.4 | Inhibitor of DNA binding 2 |
| 1.7 | RFG | |
| 1.6 | Mus musculus calcium/calmodulin-dependent protein kinase II delta mRNA, partial cds | |
| 1.5 | Protein tyrosine phosphatase, nonreceptor type 13 interacting protein | |
| 1.5 | Signal recognition particle, 9 kDa | |
| 1.5 | ESTs, highly similar to putative adenosine kinase (Saccharomyces cerevisiae) | |
| 1.5 | Protein kinase, interferon-inducible double-stranded RNA-dependent inhibitor | |
| 1.5 | Disabled homolog 1 (Drosophila) | |
| 1.5 | T-cell-specific GTPase | |
| 1.5 | Aplysia ras-related homolog N (RhoN) | |
| 1.5 | Bright and dead ringer gene product homologous protein Bdp | |
| Stress response or antioxidant | 1.9 | Glutathione-S-transferase, alpha 3 |
| 1.8 | Glutathione-S-transferase, alpha 2 (Yc2) | |
| 1.6 | Glutathione-S-transferase, alpha 4 | |
| 1.6 | Glutathione-S-transferase, mu 6 | |
| 1.5 | Glutathione transferase, zeta 1 (maleylacetoacetate isomerase) | |
| 1.6 | Selenoprotein P, plasma, 1 | |
| Liver protein or acute-phase protein | 1.7 | Serum albumin variant |
| 1.6 | Selenium binding protein 1 | |
| 1.6 | Group-specific component | |
| 1.6 | Complement component 4 binding protein | |
| 1.6 | ESTs, highly similar to fibrinogen gamma-a and -b chain precursors (Rattus norvegicus) | |
| 1.6 | ESTs, highly similar to insulin-induced growth response protein cl-6 (Rattus norvegicus) | |
| 1.5 | Complement component factor I | |
| 1.5 | Apolipoprotein H | |
| 1.5 | Sterol carrier protein 2, liver | |
| 1.5 | Orosomucoid 1 (AGP) | |
| DNA binding protein | 1.5 | Histone H2A.Z |
| Glucose response | 1.5 | Pleiotropic regulator 1, PRL1 homolog (Arabidopsis) |
| Transcription factor | 1.5 | ESTs, highly similar to hypothetical 18.5-kDa protein c12g12.05c (Schizosaccharomyces pombe) |
| 1.5 | LIM only 2 | |
| Reverse transcriptase | 1.5 | ESTs, weakly similar to open reading frame (M. musculus) |
| Cell adhesion | 1.6 | Desmoglein 2 |
| 1.5 | P40-8, functional | |
| Cysteine proteinase | 1.6 | Cathepsin L |
| Membrane protein | 1.5 | Flotillin 1 |
| 1.5 | Lymphocyte antigen 6 complex | |
| Cell locomotion | 1.5 | Hyaluronan-mediated motility receptor (RHAMM) |
| Tight junction protein | 1.5 | Claudin 1 |
| Protease inhibitor | 1.5 | Calpastatin |
Total RNA from cultured hepatocytes from C3H/HeN mice was subjected to DNA microarray analysis. The mRNA from normal and LPS (100-ng/ml)-treated (8 h) mice was labeled with cyanine-3-dUTP (green) and cyanine-5-dUTP (red), respectively. Positive fold change means gene expression was increased following treatment with LPS. The genes are sorted by function according to PubMed search. Only genes whose expression has been increased by at least 50% (1.5-fold) are shown here.
TABLE 3.
DNA microarray analysis of gene expression changes in mouse hepatocytes stimulated with LPS: decreased gene expressiona
| Function | Fold change | Gene product and description |
|---|---|---|
| Signal transduction | −1.5 | Interferon-induced protein with tetratricopeptide repeats 1 |
| ATP synthase | −1.5 | Mus musculus domesticus mitochondrial DNA, complete genome |
| Matricellular protein | −1.6 | Thrombospondin 1 |
| Chemokine or secreted cytokine | −1.7 | Small inducible cytokine A7 |
| −1.6 | Small inducible cytokine A2 | |
| Unknown | −1.6 | F box only protein 14 |
DNA microarray analysis was carried out as described in footnote a to Table 2. A negative ratio indicates genes whose expression was decreased following treatment with LPS. Expression changes in known genes are shown. Additional changes were observed in ESTs.
Categorizing genes by related functions allowed assessment of the pathophysiological effect of LPS on hepatocytes. The 68 positively modulated genes were sorted by functions and are summarized in Table 2. Table 3 contains six genes downregulated by at least 50%. In general, LPS increased the expression of several stress response or antioxidant genes (e.g., glutathione-S-transferase, selenophosphate synthetase 2); genes involved in energy synthesis (e.g., aldehyde dehydrogenase, triosephosphate isomerase, phosphoglycerate kinase 1) and signal transduction; and several ESTs, including the one with the greatest fold change that is homologous to the PI 3-kinase catalytic subunit, beta isoform. Numerous other ESTs with unknown homology or function were also modulated in LPS-treated hepatocytes (data not shown).
Liver cells express all known microbial recognition and signaling molecules.
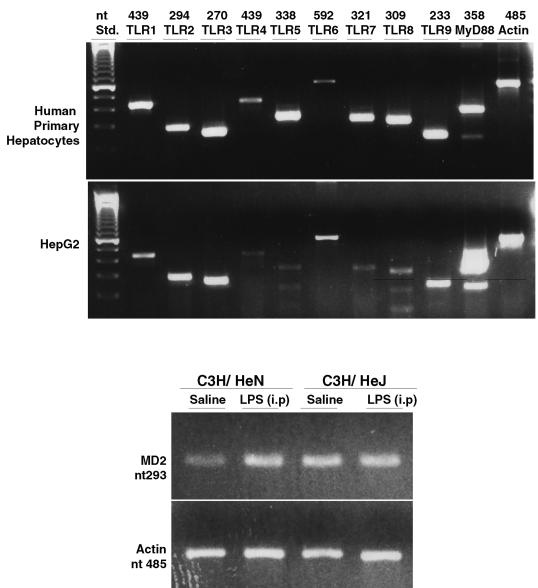
The above data support the concept that hepatocytes possess the necessary machinery to recognize and respond to LPS. We have previously shown that hepatocytes express both CD14 and TLR2 (31, 32). Experiments were undertaken to more fully characterize the expression of TLR and other associated molecules. Accordingly, we designed PCR primers and carried out RT-PCR analysis to determine the expression of transcripts for TLR1 to -9 by human hepatocytes as well as the human hepatoma cell line, HepG2 cells. As indicated in Fig. 1 (top panel), gene expression for all nine TLR tested was identified in both the primary cells and cell line. TLR signaling is known to involve the accessory molecule MyD88 (26), and we also confirmed the presence of MyD88 in these cell types. TLR4 is known to require the adapter molecule MD-2 for function, and RT-PCR also identified MD-2 mRNA in both C3/HeN and C3/HeJ hepatocytes (Fig. 1, bottom panel). Therefore, human hepatocytes constitutively express transcripts for all known TLR and MyD88, and mouse hepatocytes express MD-2. MD-2 expression by human hepatocytes was not examined by RT-PCR in this study because the GenBank sequence was not available.
FIG. 1.
Expression of TLR, MyD88, and MD-2 in human hepatocytes by RT-PCR. (Top panel) Total RNA from both cultured human primary hepatocytes and HepG2 cells were reverse transcribed with an oligo(dT) primer, followed by PCR with primers specific to TLR1 to TLR9, MyD88, or β-actin (as a loading control). (Bottom panel) Total RNA from freshly isolated hepatocytes from saline- or LPS-injected (2.5 mg/kg of body weight) (intraperitoneally [i.p.] administered at 24 h) C3H/HeN or C3H/HeJ mice were subjected to RT-PCR and analysis of MD-2 and β-actin.
LPS modulates ERK1/2 MAP kinase activity.
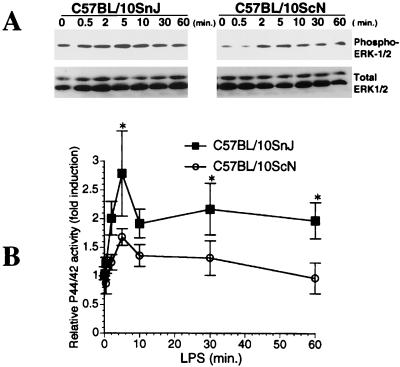
We next sought evidence for activation of signaling events following LPS stimulation by examining activation of MAP kinases. The extracellular signal-regulated kinase (ERK) signaling pathway is activated in response to LPS in both macrophage and nonmyeloid cell types (reviewed in reference 46). Accordingly, we wished to investigate whether LPS treatment leads to ERK activation and to establish if MAP kinase activation is dependent on TLR4. Total hepatocyte cell lysates from TLR4-null (C57BL/10ScN) or control (C57BL/10SnJ) mice obtained at various time points following LPS treatment were subjected to Western blotting analysis for phosphorylated ERK1/2. As indicated in Fig. 2A, LPS (500 ng/ml)-stimulated mouse hepatocytes exhibited increased ERK1/2 (p42/p44) phosphorylation by 2 to 5 min. Quantitative analysis indicates that ERK1/2 activation was significantly lower in C57BL/10ScN hepatocytes compared to that in C57BL/10SnJ hepatocytes (Fig. 2B). Unlike monocytes/macrophages, in which p38 and JNK/stress-activated protein kinase phosphorylation following stimulation with LPS can be easily detected by Western blotting (7), we failed to detect either phosphorylated p38 or JNK in mouse hepatocytes by Western blotting (data not shown).
FIG. 2.
LPS activates ERK1/2 in mouse hepatocytes. (A) Total cell lysates from LPS (500 ng/ml)-stimulated hepatocytes, treated for the indicated time periods, were subjected to Western blotting analysis with anti-phospho-ERK1/2 antibody. The blot was then stripped and immunostained with anti-total ERK1/2 antibody. (B) The films were scanned (HP ScanJet 6100C scanner), and band intensity was analyzed by using the digitized imaging analysis software UN-SCAN-IT gel (Silk Scientific). Multiple exposures were performed to ascertain that the films used for scanning were within linear-exposure range. Data (means ± SE [error bars]) are combined results from nine independent experiments (∗, P < 0.05 [for C57BL/10SnJ versus C57BL/10ScN at 5, 30, and 60 min]).
LPS modulates the activity of NF-κB, but not AP-1.
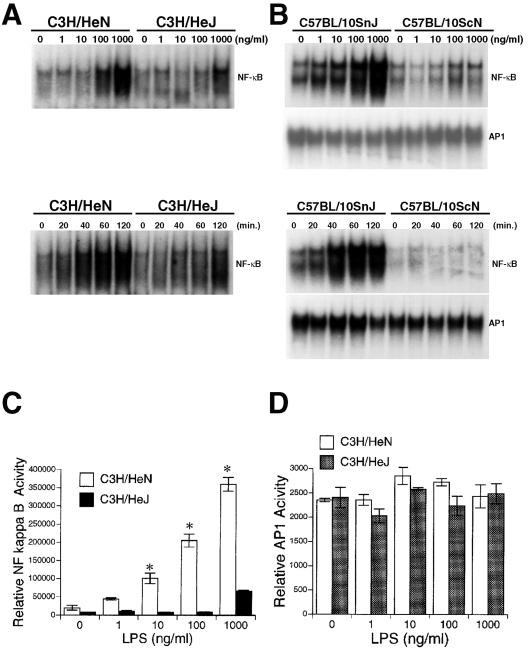
Several of the genes modulated by LPS in hepatocytes in our DNA microarray analysis are known to be regulated by the transcription factors NF-κB and AP-1 (27, 35, 39). The ability of LPS to activate NF-κB or AP-1 in hepatocytes was examined by DNA binding and NF-κB- or AP-1-dependent transcriptional assay. As indicated in Fig. 3A (lower panel), LPS induced NF-κB translocation in control hepatocytes after 20 min of stimulation; maximal induction was observed between 60 and 120 min with LPS at a dose of 500 ng/ml. Dose dependency experiments demonstrated NF-κB translocation at doses of LPS as low as 100 ng/ml, with greater NF-κB binding at 1,000 ng/ml (the maximum dose used) (Fig. 3A, upper panel). Hepatocytes from TLR4-mutant (C3H/HeJ) and TLR4-null (C57BL/10ScN) mice both exhibited decreased LPS-induced NF-κB activation compared to their control mice (C3H/HeN and C57BL/10SnJ, respectively). Hepatocyte AP-1 DNA binding activity did not significantly change in TLR4-null or control mice by EMSA (Fig. 3B). The specificity of hepatocyte NF-κB and AP-1 binding was confirmed by cold and mutant oligonucleotide competition assays (data not shown).
FIG. 3.
Effect of LPS on NF-κB and AP-1 binding and transcriptional activation in hepatocytes. (A) Hepatocytes from C3H/HeN, C3H/HeJ mice were treated with the indicated concentrations of LPS for 60 min or for the indicated time periods with LPS at 500 ng/ml. Nuclear extracts were prepared and the binding of nuclear proteins to a 32P-labeled NF-κB consensus oligonucleotide was analyzed by EMSA. Data are representative of at least three independent experiments. (B) Hepatocytes from C57BL/10SnJ, C57BL/10ScN mice were treated with the indicated concentrations of LPS for 60 min or for the indicated time periods with LPS at 500 ng/ml. Binding of nuclear proteins to a 32P-labeled NF-κB or AP-1 consensus oligonucleotide was analyzed by EMSA. Data are representative of at least three independent experiments. (C and D) Hepatocytes from C3H/HeN or C3H/HeJ mice on six-well plates were transiently transfected with an NF-κB (pELAM, 0.1 μg/well) or AP-1 (AP-1-luc, 0.2 μg/well) luciferase reporter vector and cotransfected with a β-galactosidase reporter vector (pIEPlacZ, 0.5 μg/well). The data (means ± SE [error bars]) shown here are a representative of six independent experiments with similar results (∗, P < 0.05 [for C3H/HeN versus C3H/HeJ]).
We also transfected hepatocytes with an AP-1 (AP-1-luc) or NF-κB luciferase reporter construct (pELAM) to confirm EMSA results with a functional assay. LPS increased NF-κB activity at concentrations as low as 10 ng/ml in hepatocytes from control mice, and this activity was notably lower in hepatocytes from TLR4-deficient mice (Fig. 3C). Similar to the results of the EMSA, AP-1 activation did not show any difference between cells from TLR4-deficient and control mice (Fig. 3D).
Adenovirus gene transfer of TLR4 to hepatocytes slightly increases NF-κB activity.
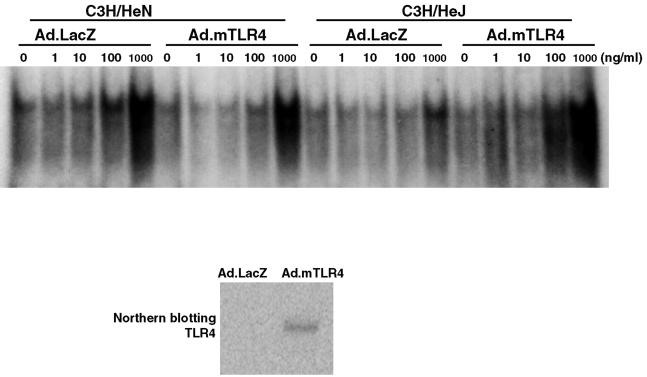
We next carried out experiments in which TLR4 was overexpressed in hepatocytes in order to determine (i) if the defective NF-κB activation observed in C3H/HeJ mice could be reversed and (ii) if TLR4 levels limit the response to LPS in wild-type hepatocytes. The mouse TLR4 cDNA was placed in an adenovirus vector with E1 and E3 deleted (Ad.mTLR4) and used to transduce hepatocytes. The expression of TLR4 mRNA in mouse hepatocytes after Ad.mTLR4 infection was confirmed by Northern blotting (Fig. 4, bottom panel). Following transduction with either Ad.mTLR4 or an adenovirus carrying β-galactosidase (Ad.LacZ) (as a control), hepatocytes were treated with LPS over a 1,000-fold range of concentrations and NF-κB activation was assessed by EMSA. As indicated in Fig. 4 (right half of the top panel), Ad.mTLR4-transduced C3H/HeJ hepatocytes displayed increased NF-κB activation, whereas Ad.LacZ-transduced hepatocytes did not. TLR4 overexpression did not increase NF-κB activation over the Ad.LacZ-transduced C3H/HeN hepatocytes (left half of the top panel). These data provide additional proof that hepatocytes utilize TLR4 to respond to LPS and also suggest that the basal TLR4 levels in C3H/HeN hepatocytes provide near-maximal LPS responsiveness.
FIG. 4.
NF-κB activation by LPS in mouse hepatocytes infected with Ad.mTLR4. (Top panel) Hepatocytes from C3H/HeN or C3H/HeJ mice were infected with Ad.mTLR4 (multiplicity of infection = 10) or Ad.LacZ for 3 h and recovered overnight. At 24 h after the start of infection, cells were treated with the indicated concentrations of LPS for 60 min. Nuclear extracts were prepared, and the binding of nuclear proteins to NF-κB consensus oligonucleotide was analyzed by EMSA. Data are representative of three independent experiments. (Bottom panel) Total RNA (10 μg per sample) was subjected to Northern blot analysis and hybridized with a 32P-labeled rat TLR4 cDNA.
DISCUSSION
The liver is an immunocompetent organ that plays a key role in the innate immune response to pathogens (57). The liver produces both inflammatory mediators and acute-phase reactants and functions to remove pathogens and microbial products from the blood (54). The prevailing opinion is that the response of hepatocytes to LPS is complex, requiring cell-cell interaction between hepatocytes and Kupffer cells, sinusoidal endothelial cells, and stellate cells (21). Our previous studies, however, raised the possibility that hepatocytes can respond directly to LPS (56). Accordingly, we undertook the present study to assess the expression of the LPS response pathway in hepatocytes and the capacity of hepatocytes to utilize TLR4 to respond to LPS. We demonstrated that hepatocytes express transcripts for TLR1 to -9, MyD88, and MD-2 (Fig. 1). Furthermore, hepatocytes respond to submicrogram concentrations of LPS with the activation of ERK1/2 and NF-κB. DNA microarray analysis shows that LPS exposure leads to the up- and downregulation of a number of genes in cultured hepatocytes (Tables 2 and 3).
Studies over the past few years have suggested that hepatocytes can respond directly to microbial products (33, 40, 56). The effect of LPS in vivo on hepatic function is also well known. LPS increases liver mass and hepatocyte volume (44), as well as modulating the synthesis of acute-phase proteins through the release of cytokines (TNF-α and IL-6) (38, 47).
We demonstrate that LPS modulates a distinct profile of genes in mouse hepatocytes (Tables 2 and 3). These genes and their products are segregated into distinct groups, namely, cell death or apoptosis genes, liver metabolism genes, stress response genes, acute-phase proteins, signal transduction proteins, and other liver proteins. While there is intrinsic interest in the profile of genes that hepatocytes express at this time point in response to LPS, this experiment serves to highlight the fact that the hepatocytes can respond to LPS by themselves, without the presence of cytokine-secreting Kupffer cells. Furthermore, it is notable that more genes were upregulated than were downregulated (compare Tables 2 and 3). Most of the positively regulated genes code for metabolic enzymes of the liver, signal transduction proteins, and stress response proteins. The relatively small fold change for most genes can be explained by the time point chosen (8 h). Data from many pervious studies indicate that LPS can stimulate hepatocytes directly, in the absence of cytokines, to modulate various products of hepatocytes (1, 13, 22, 24, 37, 42, 43, 47, 48, 50, 51, 58).
As a first step to characterize the hepatocyte's response to LPS, we examined transcripts of all known receptors for microbial products by RT-PCR. Our result indicates that both human primary hepatocytes and the human hepatoma cell line HepG2 expressed all known microbial recognition molecules (TLR1 to -9), as well as the accessory protein MyD88, and mouse hepatocytes expressed MD-2. Therefore, hepatocytes can express the components needed to respond to diverse stimuli, such as LPS (TLR4) (23), gram-positive bacterial products (TLR2), CpG DNA (TLR9) (19), and flagellin (TLR5) (16). The ligands for the other TLR are not known at present.
One of the tests for the role of a given cell in the response to LPS is the triggering of well-characterized cascades of signal transduction that lead to changes in gene expression. The Ras/Raf-1/MEK/MAP kinase pathway is responsible for transduction of LPS signals (46). The MAP kinases are a group of related serine/threonine protein kinases that participate in transmitting extracellular signals to the cell nucleus. Activation of each of the three major MAP kinase subfamilies (ERK1/2, JNK1/2, and p38) involves dual phosphorylation of tyrosine and threonine residues within the catalytic domain of these enzymes (6). LPS-stimulated MAP kinase activation has predominantly been studied in monocytes and macrophages, with no current information regarding their role, the specific pathways, and downstream targets in LPS-stimulated hepatocytes. In macrophages derived from C3H/HeJ mice, ERK2 activity is found to be refractory to induction by LPS (10, 14). Our data indicate that hepatocytes from TLR4-null mice (C57BL/10ScN) had significantly lower ERK1/2 phosphorylation compared to control mice in response to LPS (Fig. 2), suggesting that TLR4 may play a major role in MAP kinase activation by LPS in hepatocytes.
The transcription factor NF-κB is required for the production of cytokines such as IL-1, IL-6, and TNF-α, as well as for the expression of some acute-phase proteins (2, 4). The activation of NF-κB also protects hepatocytes from apoptosis during liver regeneration and embryonal development (3). Recent studies also indicate that NF-κB-dependent inducible nitric oxide synthase expression protects hepatocytes from TNF-α- and Fas-mediated apoptosis during sepsis (15). Furthermore, the recent generation of hepatocyte-specific inducible IκB transgenic mice indicates that NF-κB is critical for hepatocyte proliferation and bacterial clearance (29). Although the NF-κB responsive genes that protect hepatocytes are unknown, several LPS-inducible genes in hepatocytes from our DNA microarray analysis (Tables 2 and 3), such as PI 3-kinase (52) and selenoprotein P (36), could be potential hepatocyte-protective genes. Some genes demonstrated to be upregulated by LPS in our DNA microarray analysis are clearly either involved with or regulated by NF-κB activation. It has been reported that the PI 3-kinase/Akt pathway mediates hepatocyte survival (52). Selenoprotein P, which functions as a phospholipid hydroperoxide glutathione peroxidase, could protect TNF-α-induced hepatocyte apoptosis (36).
Essani et al. (11) have observed that hepatocytes are the only cell type in the liver to exhibit LPS-inducible and prolonged NF-κB activation during endotoxemia, compared to Kupffer cells and liver endothelial cells. In the data presented herein as well in a study performed by Essani et al. (11), it was observed that LPS-inducible NF-κB activation is lower in hepatocytes from TLR4-mutant or -null mice (C3H/HeJ and C57BL/10ScN, respectively [Fig. 3 and 4]) than in controls, highlighting the functional importance of both TLR4 and NF-κB in the hepatocyte response to LPS.
In summary, our results demonstrate that hepatocytes express all known microbial recognition molecules and respond to LPS by triggering defined signal transduction pathways that culminate in the modulation of a distinct gene profile. Moreover, TLR4 plays a critical role in this signaling. In separate studies not presented here, we have shown that rodent hepatocytes respond to gram-positive bacterial products, as well as undergoing LPS-mediated desensitization in a manner similar to that of monocytes/macrophages (S. Liu, Y. Vodovotz, and T. R. Billiar, unpublished observations). Taken together, all of these elements demonstrate clearly that hepatocytes are capable of responding directly to microbial products in a manner qualitatively similar to that of monocytes/macrophages. The importance of a direct response to LPS by hepatocytes remains uncertain, especially in view of the juxtaposition of these cells to cells that are highly responsive to microbial products (e.g., Kupffer cells). We propose that hepatocytes have retained a capacity to respond to LPS to assume a rapid response in the face of acute and excessive systemic microbial infection, thus bypassing the time needed for Kupffer cells to respond and synthesize cytokines. This may, in part, explain the need for higher concentrations of LPS to initiate signaling events in hepatocytes.
Acknowledgments
This work was supported in part by NIH grant R01-GM-05441.
We thank Stephen C. Strom (Department of Pathology, University of Pittsburgh School of Medicine) for providing the primary human hepatocytes for our TLR RT-PCR analysis.
Editor: R. N. Moore
REFERENCES
- 1.Arizono, K., S. Kagawa, H. Hamada, and T. Ariyoshi. 1995. Nitric oxide mediated metallothionein induction by lipopolysaccharide. Res. Commun. Mol. Pathol. Pharmacol. 90:49-58. [PubMed] [Google Scholar]
- 2.Beg, A. A., and D. Baltimore. 1996. An essential role for NF-κB in preventing TNF-alpha-induced cell death. Science 274:782-784. [DOI] [PubMed] [Google Scholar]
- 3.Beg, A. A., W. C. Sha, R. T. Bronson, S. Ghosh, and D. Baltimore. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-kappa B. Nature 376:167-170. [DOI] [PubMed] [Google Scholar]
- 4.Begbie, M., C. Notley, S. Tinlin, L. Sawyer, and D. Lillicrap. 2000. The factor VIII acute phase response requires the participation of NFκB and C/EBP. Thromb. Haemost. 84:216-222. [PubMed] [Google Scholar]
- 5.Beutler, B. 2000. Tlr4: central component of the sole mammalian LPS sensor. Curr. Opin. Immunol. 12:20-26. [DOI] [PubMed] [Google Scholar]
- 6.Cano, E., and L. C. Mahadevan. 1995. Parallel signal processing among mammalian MAPKs. Trends Biochem. Sci. 20:117-122. [DOI] [PubMed] [Google Scholar]
- 7.Chakravortty, D., Y. Kato, T. Sugiyama, N. Koide, M. M. Mu, T. Yoshida, and T. Yokochi. 2001. Inhibition of caspase 3 abrogates lipopolysaccharide-induced nitric oxide production by preventing activation of NF-κB and c-Jun NH2-terminal kinase/stress-activated protein kinase in RAW 264.7 murine macrophage cells. Infect. Immun. 69:1315-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 9.da Silva Correia, J., K. Soldau, U. Christen, P. S. Tobias, and R. J. Ulevitch. 2001. Lipopolysaccharide is in close proximity to each of the proteins in its membrane receptor complex: transfer from CD14 to TLR4 and MD-2. J. Biol. Chem. 276:21129-21135. [DOI] [PubMed] [Google Scholar]
- 10.Ding, A., E. Sanchez, and C. F. Nathan. 1993. Taxol shares the ability of bacterial lipopolysaccharide to induce tyrosine phosphorylation of microtubule-associated protein kinase. J. Immunol. 151:5596-5602. [PubMed] [Google Scholar]
- 11.Essani, N. A., G. M. McGuire, A. M. Manning, and H. Jaeschke. 1996. Endotoxin-induced activation of the nuclear transcription factor kappa B and expression of E-selectin messenger RNA in hepatocytes, Kupffer cells, and endothelial cells in vivo. J. Immunol. 156:2956-2963. [PubMed] [Google Scholar]
- 12.Fearns, C., V. V. Kravchenko, R. J. Ulevitch, and D. J. Loskutoff. 1995. Murine CD14 gene expression in vivo: extramyeloid synthesis and regulation by lipopolysaccharide. J. Exp. Med. 181:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freedman, A. R., R. J. Sharma, G. J. Nabel, S. G. Emerson, and G. E. Griffin. 1992. Cellular distribution of nuclear factor kappa B binding activity in rat liver. Biochem. J. 287:645-649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geppert, T. D., C. E. Whitehurst, P. Thompson, and B. Beutler. 1994. Lipopolysaccharide signals activation of tumor necrosis factor biosynthesis through the ras/raf-1/MEK/MAPK pathway. Mol. Med. 1:93-103. [PMC free article] [PubMed] [Google Scholar]
- 15.Hatano, E., B. L. Bennett, A. M. Manning, T. Qian, J. J. Lemasters, and D. A. Brenner. 2001. NF-kappaB stimulates inducible nitric oxide synthase to protect mouse hepatocytes from TNF-alpha- and Fas-mediated apoptosis. Gastroenterology. 120:1251-1262. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi, F., K. D. Smith, A. Ozinsky, T. R. Hawn, E. C. Yi, D. R. Goodlett, J. K. Eng, S. Akira, D. M. Underhill, and A. Aderem. 2001. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature 410:1099-1103. [DOI] [PubMed] [Google Scholar]
- 17.Haziot, A., S. Chen, E. Ferrero, M. G. Low, R. Silber, and S. M. Goyert. 1988. The monocyte differentiation antigen, CD14, is anchored to the cell membrane by a phosphatidylinositol linkage. J. Immunol. 141:547-552. [PubMed] [Google Scholar]
- 18.Heine, H., C. J. Kirschning, E. Lien, B. G. Monks, M. Rothe, and D. T. Golenbock. 1999. Cutting edge: cells that carry A null allele for toll-like receptor 2 are capable of responding to endotoxin. J. Immunol. 162:6971-6975. [PubMed] [Google Scholar]
- 19.Hemmi, H., O. Takeuchi, T. Kawai, T. Kaisho, S. Sato, H. Sanjo, M. Matsumoto, K. Hoshino, H. Wagner, K. Takeda, and S. Akira. 2000. A Toll-like receptor recognizes bacterial DNA. Nature 408:740-745. [DOI] [PubMed] [Google Scholar]
- 20.Hirschfeld, M., C. J. Kirschning, R. Schwandner, H. Wesche, J. H. Weis, R. M. Wooten, and J. J. Weis. 1999. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J. Immunol. 163:2382-2386. [PubMed] [Google Scholar]
- 21.Hoebe, K. H., R. F. Witkamp, J. Fink-Gremmels, A. S. Van Miert, and M. Monshouwer. 2001. Direct cell-to-cell contact between Kupffer cells and hepatocytes augments endotoxin-induced hepatic injury. Am. J. Physiol. Gastrointest. Liver Physiol. 280:G720-G728. [DOI] [PubMed] [Google Scholar]
- 22.Hoffman, J. S., and E. P. Benditt. 1982. Secretion of serum amyloid protein and assembly of serum amyloid protein-rich high density lipoprotein in primary mouse hepatocyte culture. J. Biol. Chem. 257:10518-10522. [PubMed] [Google Scholar]
- 23.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 24.Inoue, Y., B. P. Bode, D. J. Beck, A. P. Li, K. I. Bland, and W. W. Souba. 1993. Arginine transport in human liver. Characterization and effects of nitric oxide synthase inhibitors. Ann. Surg. 218:350-363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Irie, T., T. Muta, and K. Takeshige. 2000. TAK1 mediates an activation signal from toll-like receptor(s) to nuclear factor-κB in lipopolysaccharide-stimulated macrophages. FEBS Lett. 467:160-164. [DOI] [PubMed] [Google Scholar]
- 26.Kawai, T., O. Adachi, T. Ogawa, K. Takeda, and S. Akira. 1999. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity 11:115-122. [DOI] [PubMed] [Google Scholar]
- 27.Kim, S. A., S. J. Um, J. H. Kang, and K. J. Hong. 2001. Expression of thrombospondin-1 in human hepatocarcinoma cell lines and its regulation by transcription factor Jun/AP-1. Mol. Cell. Biochem. 216:21-29. [DOI] [PubMed] [Google Scholar]
- 28.Kurose, I., S. Miura, H. Higuchi, N. Watanabe, Y. Kamegaya, M. Takaishi, K. Tomita, D. Fukumura, S. Kato, and H. Ishii. 1996. Increased nitric oxide synthase activity as a cause of mitochondrial dysfunction in rat hepatocytes: roles for tumor necrosis factor alpha. Hepatology 24:1185-1192. [DOI] [PubMed] [Google Scholar]
- 29.Lavon, I., I. Goldberg, S. Amit, L. Landsman, S. Jung, B. Z. Tsuberi, I. Barshack, J. Kopolovic, E. Galun, H. Bujard, and Y. Ben-Neriah. 2000. High susceptibility to bacterial infection, but no liver dysfunction, in mice compromised for hepatocyte NF-κB activation. Nat. Med. 6:573-577. [DOI] [PubMed] [Google Scholar]
- 30.Lien, E., T. J. Sellati, A. Yoshimura, T. H. Flo, G. Rawadi, R. W. Finberg, J. D. Carroll, T. Espevik, R. R. Ingalls, J. D. Radolf, and D. T. Golenbock. 1999. Toll-like receptor 2 functions as a pattern recognition receptor for diverse bacterial products. J. Biol. Chem. 274:33419-33425. [DOI] [PubMed] [Google Scholar]
- 31.Liu, S., L. S. Khemlani, R. A. Shapiro, M. L. Johnson, K. Liu, D. A. Geller, S. C. Watkins, S. M. Goyert, and T. R. Billiar. 1998. Expression of CD14 by hepatocytes: upregulation by cytokines during endotoxemia. Infect. Immun. 66:5089-5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, S., A. N. Salyapongse, D. A. Geller, Y. Vodovotz, and T. R. Billiar. 2000. Hepatocyte toll-like receptor 2 expression in vivo and in vitro: role of cytokines in induction of rat TLR2 gene expression by lipopolysaccharide. Shock 14:361-365. [DOI] [PubMed] [Google Scholar]
- 33.Mazuski, J. E., K. Tolman, and M. J. Shapiro. 1997. Effects of cytokine antagonists on the hepatic acute-phase response. J. Surg. Res. 68:161-169. [DOI] [PubMed] [Google Scholar]
- 34.Medzhitov, R., P. Preston-Hurlburt, and C. A. Janeway, Jr. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 35.Mettouchi, A., F. Cabon, N. Montreau, P. Vernier, G. Mercier, D. Blangy, H. Tricoire, P. Vigier, and B. Binetruy. 1994. SPARC and thrombospondin genes are repressed by the c-jun oncogene in rat embryo fibroblasts. EMBO J. 13:5668-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osawa, Y., M. Nagaki, Y. Banno, Y. Yamada, M. Imose, Y. Nozawa, H. Moriwaki, and S. Nakashima. 2001. Possible involvement of reactive oxygen species in D-galactosamine-induced sensitization against tumor necrosis factor-alpha-induced hepatocyte apoptosis. J. Cell. Physiol. 187:374-385. [DOI] [PubMed] [Google Scholar]
- 37.Pagani, R., M. T. Portoles, M. A. Bosch, I. Diaz-Laviada, and A. M. Municio. 1987. Direct and mediated Escherichia coli lipopolysaccharide action in primary hepatocyte cultures. Eur. J. Cell Biol. 43:243-246. [PubMed] [Google Scholar]
- 38.Panesar, N., K. Tolman, and J. E. Mazuski. 1999. Endotoxin stimulates hepatocyte interleukin-6 production. J. Surg. Res. 85:251-258. [DOI] [PubMed] [Google Scholar]
- 39.Peron, P., M. Rahmani, Y. Zagar, A. M. Durand-Schneider, B. Lardeux, and D. Bernuau. 2001. Potentiation of Smad transactivation by Jun proteins during a combined treatment with epidermal growth factor and transforming growth factor-beta in rat hepatocytes. role of phosphatidylinositol 3-kinase-induced AP-1 activation. J. Biol. Chem. 276:10524-10531. [DOI] [PubMed] [Google Scholar]
- 40.Pittner, R. A., and J. A. Spitzer. 1992. Endotoxin and TNF alpha directly stimulate nitric oxide formation in cultured rat hepatocytes from chronically endotoxemic rats. Biochem. Biophys. Res. Commun. 185:430-435. [DOI] [PubMed] [Google Scholar]
- 41.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 42.Portoles, M. T., M. J. Ainaga, and R. Pagani. 1993. The induction of lipid peroxidation by E. coli lipopolysaccharide on rat hepatocytes as an important factor in the etiology of endotoxic liver damage. Biochim. Biophys. Acta 1158:287-292. [DOI] [PubMed] [Google Scholar]
- 43.Potter, B. J., B. Blades, T. A. McHugh, R. M. Nunes, O. Beloqui, P. A. Slott, and J. H. Rand. 1989. Effects of endotoxin on iron uptake by the hepatocyte. Am. J. Physiol. 257:G524-G531. [DOI] [PubMed] [Google Scholar]
- 44.Qian, D., and J. T. Brosnan. 1996. Administration of Escherichia coli endotoxin to rat increases liver mass and hepatocyte volume in vivo. Biochem J. 313:479-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramsay, G. 1998. DNA chips: state-of-the art of interstitial collagens in the space of Disse. Nat. Biotechnol. 16:40-44. [DOI] [PubMed] [Google Scholar]
- 46.Rao, K. M. 2001. MAP kinase activation in macrophages. J. Leukoc. Biol. 69:3-10. [PubMed] [Google Scholar]
- 47.Saad, B., K. Frei, F. A. Scholl, A. Fontana, and P. Maier. 1995. Hepatocyte-derived interleukin-6 and tumor-necrosis factor alpha mediate the lipopolysaccharide-induced acute-phase response and nitric oxide release by cultured rat hepatocytes. Eur. J. Biochem. 229:349-355. [DOI] [PubMed] [Google Scholar]
- 48.Satoh, S., A. K. Nussler, Z. Z. Liu, and A. W. Thomson. 1994. Proinflammatory cytokines and endotoxin stimulate ICAM-1 gene expression and secretion by normal human hepatocytes. Immunology 82:571-576. [PMC free article] [PubMed] [Google Scholar]
- 49.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 50.Selinger, M. J., K. P. McAdam, M. M. Kaplan, J. D. Sipe, S. N. Vogel, and D. L. Rosenstreich. 1980. Monokine-induced synthesis of serum amyloid A protein by hepatocytes. Nature 285:498-500. [DOI] [PubMed] [Google Scholar]
- 51.Shiratori, Y., Y. Hikiba, E. Mawet, Y. Niwa, M. Matsumura, N. Kato, S. Shiina, M. Tada, Y. Komatsu, T. Kawabe, et al. 1994. Modulation of KC/gro protein (interleukin-8 related protein in rodents) release from hepatocytes by biologically active mediators. Biochem. Biophys. Res. Commun. 203:1398-1403. [DOI] [PubMed] [Google Scholar]
- 52.Suzuki, A., M. Hayashida, H. Kawano, K. Sugimoto, T. Nakano, and K. Shiraki. 2000. Hepatocyte growth factor promotes cell survival from fas-mediated cell death in hepatocellular carcinoma cells via Akt activation and Fas-death-inducing signaling complex suppression. Hepatology 32:796-802. [DOI] [PubMed] [Google Scholar]
- 53.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 54.Tanikawa, K., Y. Mimura, S. Sakisaka, and K. Noguchi. 1998. Role of hepatocytes in the clearance of lipopolysaccharide and its clinical significance. Prog. Clin. Biol. Res. 397:191-198. [PubMed] [Google Scholar]
- 55.Ulevitch, R. J. 1993. Recognition of bacterial endotoxins by receptor-dependent mechanisms. Adv. Immunol. 53:267-289. [DOI] [PubMed] [Google Scholar]
- 56.Vodovotz, Y., S. Liu, C. McCloskey, R. Shapiro, A. Green, and T. R. Billiar. 2001. The hepatocyte as a microbial product-responsive cell. J. Endotoxin Res. 7:365-7357. [PubMed] [Google Scholar]
- 57.Volpes, R., J. J. van den Oord, and V. J. Desmet. 1992. Can hepatocytes serve as "activated' immunomodulating cells in the immune response? J. Hepatol. 16:228-240. [DOI] [PubMed] [Google Scholar]
- 58.Wan, Y., P. D. Freeswick, L. S. Khemlani, P. H. Kispert, S. C. Wang, G. L. Su, and T. R. Billiar. 1995. Role of lipopolysaccharide (LPS), interleukin-1, interleukin-6, tumor necrosis factor, and dexamethasone in regulation of LPS-binding protein expression in normal hepatocytes and hepatocytes from LPS-treated rats. Infect. Immun. 63:2435-2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.West, M. A., T. R. Billiar, R. D. Curran, B. J. Hyland, and R. L. Simmons. 1989. Evidence that rat Kupffer cells stimulate and inhibit hepatocyte protein synthesis in vitro by different mechanisms. Gastroenterology 96:1572-1582. [DOI] [PubMed] [Google Scholar]
- 60.Wright, S. D., R. A. Ramos, P. S. Tobias, R. J. Ulevitch, and J. C. Mathison. 1990. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249:1431-1433. [DOI] [PubMed] [Google Scholar]