Abstract
We report the isolation of a Coccidioides immitis gene (SOWgp) which encodes an immunodominant, spherule outer wall glycoprotein that is presented as a component of a parasitic phase-specific, membranous layer at the cell surface. The open reading frame of the gene from C. immitis isolate C735 translates a 422-amino-acid (aa) polypeptide that contains 6 copies of a 41- to 47-residue tandem repeat enriched in proline (20.4 mol%) and aspartate (19.7%). Two additional isolates of C. immitis produce SOWgps of different molecular sizes (328 and 375 aa) and show a corresponding difference in the number of tandem repeats (four and five, respectively). The accurate molecular sizes of these proline-rich antigens, as determined by surface-enhanced laser desorption/ionization mass spectrometry, are comparable to the predicted sizes from the translated protein sequences rather than the estimated sizes based on gel-electrophoretic separation. The results of Northern hybridization confirmed that SOWgp expression is parasitic phase specific, and immunoblot studies showed that elevated levels of production of this antigen occurred during early spherule development. The recombinant polypeptide (rSOWp) was shown to bind to mammalian extracellular matrix (ECM) proteins in an in vitro assay (laminin > fibronectin > collagen type IV), suggesting that the parasitic cell surface antigen may function as an adhesin. Deletion of the SOWgp gene by using a targeted gene replacement strategy resulted in partial loss of the ability of intact spherules to bind to ECM proteins and a significant reduction in virulence of the mutant strain. The wild-type gene was restored in the mutant by homologous recombination, and the revertant strain was shown to be as virulent as the parental isolate in our murine model of coccidioidomycosis. The parasitic cell surface glycoprotein encoded by the SOWgp gene appears to function as an adhesin and contributes to the virulence of C. immitis.
Coccidioides immitis is a desert soil-inhabiting fungal pathogen that is found in the southwestern United States and in certain arid regions of Mexico, Central America, and South America (15). Air-dispersed spores (arthroconidia) produced by the saprobic phase of the fungus may be inhaled by humans who reside in or travel through these desert areas. The tiny, barrel-shaped arthroconidia (ca. 2 by 6 μm) convert into multinucleate round cells (spherules) within the lungs of the host and grow isotropically to produce large parasitic cells (60 to >100 μm in diameter). The latter undergo an elaborate process of endogenous wall growth and cytoplasmic compartmentalization, which culminates in production and the release of a multitude of endospores (each ca. 4 to 10 μm in diameter [6]). Endospores grow isotropically and differentiate into a second generation of spherules. Although young parasitic cells that are newly released from maternal spherules may be engulfed by host phagocytes, in vitro studies of host-pathogen interactions suggest that many of these fungal cells survive initial attack by the innate immune system (10). Mature spherules most likely escape phagocytosis simply because they are too large to be taken up by neutrophils and macrophages. We know from a variety of studies of coccidioidal infections in mice that the incubation period from arthroconidium inoculation to development of symptomatic disease is typically 10 to 15 days (47). It is reasonable to assume that survival of the fungus in vivo during this early period of infection and its ability to establish disease are dependent on the success of the pathogen to colonize respiratory tissue. However, no information on the mechanisms of host colonization by C. immitis have so far appeared in the literature.
Several lines of evidence have suggested that cell surface and secreted proteins with proline-rich regions (PRRs) play important roles in protein-protein interactions and protein folding (4, 23). These PRRs frequently occur as multiple tandem repeats and are widely distributed among prokaryotes and eukaryotes (44). Cell-surface expressed fungal and bacterial proteins which contain proline-rich tandem repeats have been suggested to function in adherence to host tissue (33, 39). An example is a glycoprotein of Colletotrichum lindemuthianum (CIH1) produced at the surface of intracellular hyphae which infects the tissues of bean plants. The N-terminal domain of the mature glycoprotein is rich in proline, contains several short repetitive motifs, and forms cross-links with the plant cell wall. The PRRs of CIH1 appear to be essential for establishment and maintenance of this fungus-host relationship (33). Candida albicans produces a glycoprotein (HWP1) at its hyphal surface that is characterized by tandemly repeated, proline- and glutamine-rich amino acid motifs (39). Like CIH1, it has been shown that this antigen is expressed in vivo and is developmentally regulated. The hyphal phase-specific HWP1 attaches to buccal epithelial cells by an enzyme-mediated process of adhesion (40). The proline-rich motifs of HWP1 are suggested to dictate a conformation of the polypeptide in which the glutamine residues are presented to and recognized by the host transglutaminase as a substrate. Digestion of the surface antigen apparently results in formation of cross-links between the hyphal and epithelial cell surfaces. A cell surface adhesin of Streptococcus mutans (P1) contains a conserved proline-rich repeat domain which is suggested to be a necessary component of conformational epitopes within the central region of the antigen (4). Deletion of this domain resulted in complete absence of the surface-localized P1 antigen. We have previously reported a major parasitic cell surface-expressed antigen produced by C. immitis that elicits both humoral and cellular immune responses in patients with coccidioidal infection (20). In the present study we demonstrate that this spherule outer wall glycoprotein (SOWgp) contains tandemly repeated proline- and aspartic acid-rich motifs, functions as an adhesin, and contributes to the pathogenicity of C. immitis in a murine model of coccidioidomycosis.
MATERIALS AND METHODS
Fungal media and growth conditions.
The C. immitis isolates C735, C634, and Silveira used in this study were originally obtained from patients with disseminated coccidioidomycosis. The saprobic and parasitic phases of the fungus were grown in vitro under conditions previously described (20). Parasitic-phase cultures were harvested at various times after inoculation with arthroconidia (36 to 144 h) as reported elsewhere (21).
SOWgp gene isolation and sequence analysis.
The previously reported N-terminal and internal amino acid sequences of the purified 82-kDa SOWgp (SOWgp82 [20]) of C. immitis C735 were used to design degenerate sense and antisense primers. These oligonucleotide primers were used in a PCR with template genomic DNA of C. immitis C735 to amplify a fragment of the putative SOWgp82 gene. The nucleotide sequence of the sense primer deduced from the previously reported N-terminal amino acid sequence (GATSHKEH) was 5′-GGIGCNACNWSNCAYAARGARCA-3′ (2,048-fold degeneracy; I, inosine; R is A or G; S is C or G; W is A or T; and Y is C or T). The antisense primer was designed on the basis of an amino acid sequence derived from a cyanogen bromide digestion product (KPEPPKK) (20) of the native SOWgp82 glycoprotein. The sequence of the antisense primer was 5′-YTTYTTNGGN GGYTCNGGYTT-3′ (1,024-fold degeneracy). The PCR mixture (100 μl) contained 10 mM Tris-HCl (pH 8.3) plus 50 mM KCl, 1.5 mM MgC12, final concentrations of 0.2 mM for each deoxynucleoside triphosphate and 20 μM for each primer, 50 ng of C. immitis genomic DNA, and 2.5 U of Tag DNA polymerase (Sigma Chemical Co., St. Louis, Mo.). Thirty-five cycles were conducted for PCR amplification in the presence of the template genomic DNA under conditions previously reported (21). The PCR products were ligated into pZErO 2.1 (Invitrogen, Carlsbad, Calif.) by the TA-cloning method (2). The clones were subsequently screened by PCR with primers derived from nucleotide sequences in the multiple cloning site of the vector. Twenty selected clones (inserts of 0.2 to 1.2 kb) were sequenced by using an ABI Prism 310 genetic analyzer (Perkin-Elmer, Foster City, Calif.), and the nucleotide sequences were examined by using MacDNASIS Sequence Analysis Software (version 3.5; Hitachi, San Bruno, Calif.). A 0.7-kb insert was sequenced, translated, and shown to include the reported N-terminal and internal amino acid sequences of the native SOWgp82. The 0.7-kb insert was purified, labeled with [α-32P]dCTP (<3,000 Ci/mmol; ICN, Costa Mesa, Calif.) by using a Multiprimer DNA labeling system (Amersham, Cleveland, Ohio), and employed as a probe to screen the genomic library of C. immitis isolate C735 (45). Positive phages were selected and amplified, and DNA was extracted for restriction enzyme digestion and Southern hybridization (46) by using the same 0.7-kb PCR product as a probe. A 5.9-kb KpnI and a 3.6-kb XhoI genomic fragment each hybridized with the probe. The former was subsequently used for construction of a restriction map (see Fig. 7A). The 3.6-kb XhoI genomic fragment was isolated and subcloned into pZErO 2.1. Partial sequence analysis of the 5′ and 3′ ends of this insert suggested that it contained tandem repeats. The repeat sequences presented problems for primer design and conventional sequence analysis. To resolve this issue, the 3.6-kb genomic fragment was digested with PstI, which generated a 3.0-kb fragment. The plasmid vector contains a PstI restriction site upstream of the insert. The 3.0-kb fragment was isolated and subcloned into the PstI site of pZErO 2.1. The 0.6-kb fragment, which remained as part of the original plasmid, was sequenced as described above. To sequence the 3.0-kb fragment, a series of deletion clones were produced by using the Erase-a-Base System (Promega, Madison, Wis.). The linearized plasmids with inserts that ranged from 0.2 to 3.0 kb were religated and used to transform Escherichia coli DHα-competent cells. The clones were screened for inserts by PCR. Positive clones were purified by using a Qiagen Plasmid Purification System (Qiagen, Chatsworth, Calif.). Seventeen clones were selected and subjected to sequence analysis as described above.
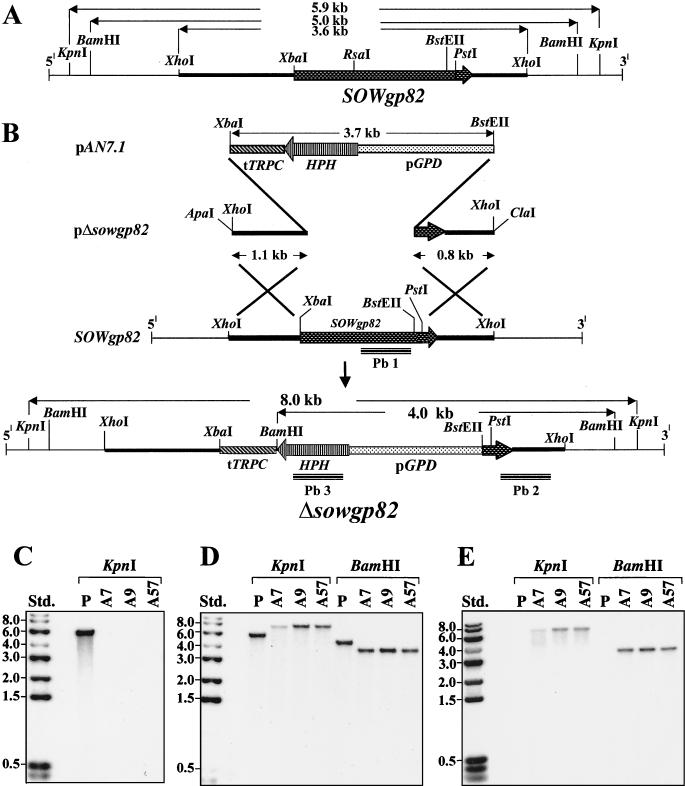
FIG. 7.
(A and B) Restriction map of 5.9-kb genomic fragment which contains the SOWgp82 gene (A) and disruption strategy showing plasmid construct and outcome of the pΔsowgp82 integration event (B). Homologous recombination of the plasmid construct with SOWgp82 by a double-crossover event resulted in replacement of the C. immitis gene with the HPH gene plus promoter (pGPD) and terminator (tTRPC) sequences. The probes used for Southern hybridization are labeled Pb1, Pb2, and Pb3. (C to E) Southern hybridization of genomic DNA with the Pb1 (C), Pb2 (D), and Pb3 (E) probes with restriction enzyme digests of both the parental (P) and transformant strains (A7, A9, and A57). The size of each band in panel C correlates with the predicted sizes in panels A and B, which provides evidence for single-site, homologous recombination and replacement of the SOWgp82 gene. Std., standards.
RACE.
Rapid amplification of cDNA ends (RACE) was used to resolve the location of the putative 5′ end of the untranslated region (UTR), the N-terminal methionine, and the poly(A) addition site(s) of the SOWgp82 gene (17, 21). The sequences of the PCR primers used for this procedure were based on the genomic sequence of the SOWgp82 gene, which has been deposited in GenBank (accession no. AF308873). The two gene-specific primers used for 5′ RACE were as follows: 5′-GAAATCAAACATAGAATGCAGCTG-3′ (nucleotides [nt] 3020 to 3043) and 5′-CCATAGGTATCGCAA TAACTGT-3′ (nt 1610 to 1589). For 3′ RACE, one of the two primers used was the synthesized oligo-d(T)17-adapter construct described by Frohman (14). The other was the gene-specific primer 5′-CAGTTCATACGCCAACTCAGTT-3′ (nt 2939 to 2960). The PCR conditions were the same as those used for amplification of the genomic fragment described above. The RACE products were separated by agarose gel electrophoresis (1.5% gel), isolated, subcloned into pZErO 2.1, and subjected to nucleotide sequence analysis as described above.
To confirm the locations of introns, template cDNA derived from the total RNA of 48-h-old parasitic-phase cultures was amplified with a sense primer, the sequence of which was based on the genomic sequence of SOWgp82 (5′-ACAGTTATTGCGATACCTATGG-3′ [nt 1589 to 1610]), and an antisense primer based on one of the two 5′ RACE primers (nt 3020 to 3043) reported above. The PCR conditions were the same as previously described. A 1.1-kb PCR product, which represented the SOWgp82 cDNA that spanned eight of the nine putative exons, was separated by agarose gel electrophoresis (1% gel), subcloned into pZErO 2.1, and sequenced as described above.
The same procedures with primers identical to those described above were conducted to obtain cDNA sequences of the SOWgp58 and SOWgp66 genes of the Silveira and C634 isolates (GenBank accession no. AF309491 and AF309490), respectively. The CLUSTAL W program (18) was used to align the three translated SOWgp sequences. The NCBI-BLAST and PSI-BLAST programs were used to search for protein sequences in the GenBank and SWISS-PROT databases with similarities to the translated sequences of the three C. immitis SOWgp genes (1). The PROSITE database was used to identify conserved motifs in SOWgp with homology to reported proteins (19), and the PSORT database was used for prediction of cellular localization sites of the glycoprotein (31).
Southern hybridization.
Isolation of intact chromosomal DNA and total genomic DNA of C. immitis isolates C735, C634, and Silveira were performed as previously described (21, 32). Southern hybridization was conducted at low stringency as reported elsewhere (46). A 1.5-kb PCR product of SOWgp82 was amplified from genomic DNA (sense primer, nt 1589 to 1610; antisense primer, nt 3043 to 3020), purified, labeled with [α-32P]dCTP, and used as a probe for the Southern hybridizations.
SELDI-TOF mass spectrometry.
The native 82- and 60-kDa SOWgp fractions of C. immitis isolate C735 were purified by electroelution of the protein bands from a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel after electrophoretic separation of the previously reported Triton X-114 (TX-114) extract of the crude spherule outer wall (SOW) (20). Surface-enhanced laser desorption/ionization-time of flight (SELDI-TOF) mass spectrometry was used to determine the molecular mass of the purified SOWgp fractions. Samples were applied to separate spots on H4 ProteinChip arrays (C-16 aliphatic chain hydrophobic surface) supplied by the manufacturer (Ciphergen Biosystems, Inc., Fremont, Calif.). To prevent cross-contamination between samples applied to the chip, a circle was drawn around each reaction spot on the chip surface by using a hydrophobic marker (Zymed Lab., Inc., San Francisco, Calif.). Each reaction spot was first activated with 2 μl of acetonitrile. Before the reactant was completely dry, a 2-μl sample (0.1 mg/ml) of the purified 82- or 60-kDa SOWgp in 0.1 M phosphate-buffered saline (PBS; pH 7.2) was applied to the reaction spot and allowed to air dry. The unbound material was removed by washing it three times with 5 μl of high-pressure liquid chromatography-grade water (Fisher Scientific, Pittsburgh, Pa.). The dried surface of each spot was finally reacted with 0.5 μl of a saturated solution which contained a patented energy absorbing molecule (EAM1; Ciphergen), which was solubilized in 50% acetonitrile plus 0.5% trifluoroacetic acid in distilled water. The protein chip was then subjected to mass analysis in a model PBSII laser desorption/ionization mass spectrometer (Ciphergen). The instrument was calibrated by using purified bovine serum albumin (BSA; the molecular mass of the major fraction is 66,410 Da) on protein chips provided by the PBSII manufacturer.
Northern hybridization.
Total RNA was isolated from 4-day mycelial cultures (saprobic-phase) and near-synchronous parasitic-phase cultures at 36, 72, 96, 120, and 144 h after inoculation with arthroconidia as described previously (21). Each RNA preparation was subjected to formaldehyde agarose gel electrophoresis (1% gel) and transferred to a nitrocellulose membrane (Hybond; Amersham, Arlington Heights, Ill.) as reported elsewhere (46). The 1.1-kb SOWgp82 cDNA described above was labeled with [α-32P]dCTP and used to hybridize with the membrane as previously described (46).
Expression of SOWgp.
Since it was difficult to obtain sufficient amounts of the native SOWgp for antiserum production and the adherence assays described below, we isolated the recombinant antigens (rSOWp82 and rSOWp58). The 1.1-kb SOWgp82 cDNA described above was subcloned into an EcoRI/XhoI site of the pET28b expression vector (Novagen, Madison, Wis.). Transformation of E. coli strain BL21(DE3) with the pET28b-SOWgp82 plasmid construct, induction of expression, purification, and amino acid sequence analysis of the recombinant protein were performed as previously reported (17). We also purified the 0.8-kb cDNA of SOWgp58 (Silveira isolate) and subcloned it into pET28b for expression of rSOWp58 by E. coli as described above. The isolation, digestion, subcloning, and expression methods were the same as described for SOWgp82.
Production of antiserum.
BALB/c mice (6 weeks old) were used to raise antiserum against the purified rSOWp82 by subcutaneous immunization by a standard protocol (20). The specificity of the antiserum was determined by immunoblot analysis. Preimmune mouse serum was used as a control.
Immunoblot analysis.
Detection of the native SOWgp82 glycoprotein in total homogenates of 4-day mycelial cultures and spherules from selected stages of the parasitic cell cycle was conducted by SDS-PAGE separation and immunoblot analysis (21) with the above murine anti-rSOWp82 serum. Total protein was extracted from each homogenate with 1% TX-114 (Sigma), and the concentration of protein in each sample was determined as reported earlier (20).
Microscopy.
Freshly isolated, presegmented spherules of C. immitis C735 (36- to 72-h cultures; see Fig. 5) were examined by immunofluorescence microscopy using the murine anti-rSOWp82 antibody described above. The immunofluorescence procedure was the same as that previously reported (20). In preparation for immunoelectron microscopy, in vitro-grown parasitic cells were first subjected to cryofixation by using a propane jet freezer as reported elsewhere (25). The spherules were then freeze substituted, embedded in LR White resin, thin sectioned, and stained with osmium tetroxide (OsO4) to enhance the visibility of lipoidal structures. The sections were incubated with the same primary murine antibody as described above, followed by treatment with goat anti-mouse immunoglobulin G (IgG) conjugated with colloidal gold (20 nm; Ted Pella, Inc., Redding, Calif.). The isolated, crude SOW (8) fraction of C. immitis was also prepared for immunoelectron microscopy by conventional fixation and resin-embedding procedures (38). Thin sections of SOW were incubated with serum from a human patient with confirmed C. immitis infection (complement fixation antibody titer of >1:64 [36]), followed by the addition of goat anti-human IgG conjugated with colloidal gold. Normal human serum was used as a control.
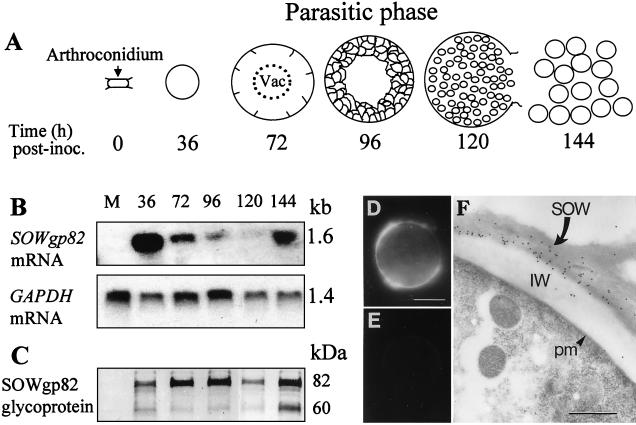
FIG. 5.
(A to C) Diagram of the parasitic cycle of C. immitis (A), Northern hybridization analysis of temporal expression of SOWgp and GAPDH genes (B), and corresponding immunoblot analysis of SOWgp82 production as detected by examination of detergent extracts of parasitic cell homogenates of isolate C735 (C). The developmental stages of first- and second-generation parasitic cells are identified by incubation times after inoculation of parasitic-phase cultures. (D and E) Immunolocalization of SOWgp in spherules (36- to 72-h stage of development) by immunofluorescence (D and E) and immunoelectron microscopy (F) with antiserum raised against the recombinant SOWp82 reveals that the antigen is presented at the parasitic cell surface. Control cells in panel E were incubated with secondary antibody-fluorescein isothiocyanate conjugate alone. IW, inner spherule wall; pm, plasma membrane. Bars: D and F, 10.0 and 0.5 μm, respectively.
Adherence assay.
The recombinant protein of the Silveira strain of C. immitis (rSOWp58) was tested for its ability to adhere to selected extracellular matrix (ECM) proteins (human plasma fibronectin [Boehringer Mannheim, Indianapolis, Ind.], mouse laminin [Sigma], and bovine dermal collagen type IV [ICN]). Purified rSOWp58 was biotin-labeled by incubation with sulfo-NHS-biotin (Pierce, Rockford, Ill.) according to the manufacturer's protocol (the stock concentration of labeled antigen was 4.5 μg/μl). The relative amount of rSOWp58 bound to different ECM proteins was determined by a microplate assay. The wells of microplates (Pro-Bind; 96-well flat-bottom plates; Becton Dickinson, Franklin Lakes, N.J.) were coated with either 50 μl of 1% BSA (Sigma) as a control or different concentrations of ECM proteins (1.3 to 20 μg/ml) in PBS (0.1 M [pH 7.4]) for 12 h at 4°C. The wells were then washed with PBS, blocked with 1% BSA (for 1 h at room temperature), and reacted with 50 μl of biotin-labeled rSOWp58 at a fixed concentration of 10 μg/ml in PBS containing 0.1% Tween 20 (PBST; Sigma) for 2 h at room temperature. The wells were washed three times with PBST, followed by incubation with 50 μl of peroxidase-conjugated streptavidin (diluted 1:1,000 in PBS; Boehringer Mannheim) for 1 h at room temperature, and then washed again as described above. Semiquantitative analysis of bound rSOWp58 was conducted by the addition of peroxidase substrate to the wells and determination of the optical density at 450 nm (OD450). These assays were performed in triplicate, and statistical analyses were conducted by using the Student's t test.
Deletion of the SOWgp82 gene.
The pΔsowgp82 plasmid was designed as a gene replacement vector (9) and constructed by using a 3.7-kb fragment of pAN7.1 that contained the hygromycin resistance gene (HPH [35]). The 3.6-kb genomic clone of SOWgp82 in pZErO 2.1 was digested with BstEII, followed by XbaI, which resulted in deletion of most of the coding region of the C. immitis gene (i.e., nt 1305 to 3039). The 3.7-kb pAN7.1 fragment was used to replace the deleted C. immitis gene fragment by ligation with the digested SOWgp82 plasmid to yield the pΔsowgp82 construct. The latter was amplified by transformed E. coli, isolated, and used for subsequent transformation of C. immitis isolate C735. Prior to transformation, the plasmid was linearized by ApaI/ClaI digestion and purified as reported (28). The transformation protocol and method used to isolate homokaryotic transformants were the same as those previously described (36). Confirmation of homologous recombination was based on results of Southern hybridization by using nucleotide probes derived from the HPH and targeted C. immitis genes. The HPH gene probe (i.e., probe 3 [see Fig. 7B]) was used to confirm single-site integration of the plasmid construct and consisted of a 600-bp PCR product generated by use of a sense primer (5′-ACAGCG TCTCCGACCTGATG-3′ [nt 2354 to 2373; pAN7.1]) and an antisense primer (5′-CCTCGCTCCAGTCAATGACC-3 [nt 2934 to 2953]) in the presence of pAN7.1 template DNA. Confirmation of homologous recombination and deletion of the coding region of SOWgp82 was performed by Southern hybridization by using a RsaI/BstEII digestion fragment of the SOWgp82 gene (nt 1414 to 2801) as a probe (i.e., probe 1). Confirmation that the selected transformant was homokaryotic was performed with a PstI/XhoI digestion fragment of SOWgp82 (nt 2969 to 3565) as a probe (i.e., probe 2). This latter nucleotide sequence was present in both the parental and mutant strains.
Generation of the revertant strain.
In order to select for transformants with the restored, wild-type SOWgp82 gene, we used the Δsowgp82 strain as the host and a plasmid which contained a second selection marker. We chose the pAN8.1 plasmid (kindly provided by J. Woods, University of Wisconsin at Madison) containing the BLE gene which encodes a protein that confers resistance to phleomycin. We demonstrated that growth of the saprobic phase of C. immitis isolate C735 was inhibited by phleomycin (Sigma) at a concentration of 1.0 μg/ml of glucose-yeast extract (GYE) medium. The pAN8.1 plasmid was first digested with EcoRI and XbaI to generate a 3.4-kb fragment. The DNA fragment was then cloned into the EcoRI/SpeI site of pZErO 2.1 that contained the previously described 3.6-kb XhoI restriction fragment of the wild-type SOWgp82 gene. This undigested plasmid construct (pSOWgpR [see Fig. 9]) was used to restore the wild-type SOWgp82 gene to the Δsowgp82 mutant host by employing the protoplast method described above in the gene knockout experiment. Transformants were selected on GYE agar plates that contained phleomycin at a concentration of 4 μg/ml of growth media. Homokaryotic revertants were isolated by serial transfer of subcultures to fresh agar media containing the same antibiotic (4 μg/ml). Southern hybridization and immunoblot analyses were conducted to confirm that the selected revertant strain (SOWgp82R) contained the wild-type gene, that it integrated into the chromosomal DNA at a single site by homologous recombination, and that the level of expression of the glycoprotein was comparable to the SOWgp expression of the parental isolate.
FIG. 9.
(A) Strategy used to restore wild-type SOWgp82 gene to the Δsowgp82 strain with a plasmid construct that contains the BLE gene which encodes resistance to phleomycin. (B) Results of Southern hybridization of restriction enzyme-digested genomic DNA of selected strains with the Pb2 probe (see Fig. 7B). Genomic DNA was obtained from the parental isolate (P), Δsowgp82 mutant (A9), and two putative revertant strains (R61 and R121). (C) SDS-PAGE separation and immunoblot analysis of total protein from parasitic cell homogenates of the same C. immitis strains as in panel B. The immunoblot was conducted with antiserum raised against the recombinant SOWp82. (D) Predicted outcome of the pSOWgpR integration event which yielded the R61 revertant strain (SOWgp82R).
Evaluation of the phenotype, adherence, and virulence of the Δsowgp82 mutant strain.
Loss of production of the SOWgp82 glycoprotein in total cell homogenates of the mutant strain and its presence in homogenates of the parental isolate and revertant strains were confirmed by immunoblot analysis with anti-rSOWp82 serum. Methods used for sample preparation and SDS-PAGE were as described above. Preparations of the isolated, crude SOW fraction from the parental and Δsowgp82 mutant strains were also examined by immunoelectron microscopy for presence of the SOWgp82 glycoprotein by using serum from a patient with confirmed coccidioidal infection as described above.
Evaluation of the relative adherence of intact, presegmented spherules isolated from 72-h parasitic-phase cultures of the parental and Δsowgp82 mutant strains were conducted in microplates by a competitive adherence inhibition assay. Cells from the parental strain were biotinylated, streptavidin-peroxidase labeled, and washed with PBST buffer to remove the unbound label. The wells of the microtiter plate were coated with 75 μg of fibronectin or laminin/ml. The labeled spherules of the parental strain (104 cells in 25 μl of PBS) were mixed with an equal volume of PBS that contained different numbers of unlabeled spherules from either the parental or the mutant strain (2.5 × 104 to 2.0 × 105 cells). The mixtures of cells were added to the precoated wells, and the plates were incubated at 37°C for 2 h. The wells were washed with PBST three times and then incubated for 2 min with the peroxidase substrate. The color intensity was determined by measuring the OD450. To control for the presence of endogenous peroxidase produced by the spherules, the cells used in this assay were first frozen at −20°C for 12 h and then tested for reactivity with the peroxidase substrate. The OD of the unlabeled cells from either the parental or mutant strain incubated with peroxidase substrate as described above was below the limit of detection by spectrophotometry. These assays were performed in triplicate, and statistical analyses were conducted by using the Student's t test.
To compare the virulence of the parental, mutant, and revertant strains, arthroconidia (70 viable cells) were obtained from GYE plate cultures of each grown for 30 days at 30°C. The arthroconidia were used to inoculate C57BL/10J mice (15 female mice per group, 8 weeks old) by the intranasal route as previously reported (27). The survival plots were subjected to Kaplan-Meier statistical analysis as previously described (36).
RESULTS
Structure of the SOWgp82 gene.
A 3.6-kb C. immitis clone isolated from the genomic DNA library was sequenced and shown to contain an open reading frame (ORF) of 1,266 bp. The translated sequence (422 amino acids [aa]) included the previously reported N-terminal and internal amino acid sequences of the native SOWgp82 (20). Analysis of the genomic sequence also revealed the presence of eight introns. The structure of the ORF and location of the eight introns and nine exons were confirmed by cDNA sequence analysis. The location of the N-terminal methionine (AUG) and the 5′ end of the upstream UTR were resolved by using the results of the 5′ RACE procedure. The 5′ UTR was shown to contain a putative TATA box located 103 bp upstream of the AUG. Results of 3′ RACE resolved the stop codon of the ORF and identified two poly(A) tail addition sites separated by 69 bp. An unusual feature of the nucleotide sequence of this gene is its organization into six tandem repeats. Each repeat contains a near-identical intron (54 to 56 bp long). The first and last of the six exon repeats translate 45 and 41 residues, respectively, whereas the others each translate a 47-aa sequence (Fig. 1A and B). Additional structural details of the SOWgp82 gene are provided under GenBank accession number AF308873.
FIG. 1.
Structure of the translated ORF of the SOWgp gene of isolate C735 (A), and alignment of the six proline- and aspartate-rich repeat sequences of the polypeptide (B). Underlined residues are sequence variants in the tandem repeats. Aligned cysteine and proline motifs are enclosed in boxes.
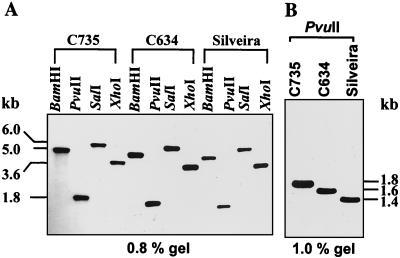
Southern hybridization.
Southern hybridization of electrophoretically separated chromosomal DNA suggested that the SOWgp gene is located on chromosome II (results not shown), in accordance with the nomenclature used for numbering coccidioidal chromosomes (32). Total genomic DNA preparations of C. immitis isolate C735 were separately digested with BamHI, PvuII, SalI, and XhoI, subjected to agarose gel electrophoresis (0.8 or 1.0% gels) and hybridized with the same labeled probe that was used for Southern hybridization with chromosomal DNA. Single bands were observed in each digest (5.0, 1.8, 6.0, and 3.6 kb, respectively), which were predicted on the basis of a restriction map of the genomic clone of the SOWgp82 gene. These results indicate that SOWgp82 is a single-copy gene. The same nucleotide probe and set of restriction enzymes were used for Southern hybridization with total genomic DNA digests derived from isolates C634 and Silveira (Fig. 2A and B). The results also revealed the presence of single bands for each restricted genomic fragment but showed that a clear difference exists in the size of the respective fragments from each isolate.
FIG. 2.
Southern hybridization of endonuclease-restricted genomic DNA obtained from three isolates of C. immitis (C735, C634, and Silveira) characterized by SOWgps with six, five, and four tandem repeats, respectively. The hybridization probe was a 1.5-kb PCR product of SOWgp82.
Protein sequence analysis and alignment.
The SOWgp82 gene encodes a protein with a predicted molecular size of 46.4 kDa and a pI of 4.2. Sequence analysis performed by PSORT showed that the first 20 aa at the N terminus had the characteristic of a signal peptide, with a positively charged residue (K2), followed by a hydrophobic core and a putative cleavage site between G20 and Y21 (Fig. 1A) (43). However, the N-terminal residue of the mature protein is G61, which was previously determined by sequence analysis of the purified, native SOWgp82 glycoprotein (20). A propeptide (aa 21 to 60; Fig. 1A) was deduced on the basis of this N-terminal sequence of the native glycoprotein. The C terminus of the translated SOWgp82 contains a predicted glycosylphosphatidylinositol (GPI) signal sequence with a putative anchor site at residue G399 (http://members.nbci.com/_XMCM/jkronegg/unige/GPI-anchor/index_en.html [11]).
Examination of the amino acid composition of the deduced, full-length SOWgp82 indicated that it is a proline- and aspartate-rich protein with 16.6% proline (as mol%), 14.7% aspartic acid, 11.6% tyrosine, 10.2% glycine, and 9.2% lysine. Each amino acid repeat described above (Fig. 1A and B) is enriched in proline (20.4%), aspartate (19.7%), tyrosine (13.1%), lysine (10.9%), and glycine (10.9%). There are four cysteine residues at identical positions in each repeat, except for the fourth repeat, which contains only three cysteines (Fig. 1B). Each repeat also contains a polyproline motif (PPPPP). No N-glycosylation sites were predicted in the translated SOWgp82 sequence. However, based on positive periodic acid-Schiff (PAS) staining of the native SOWgp82 (20), we concluded that the cell surface polypeptide is linked to sugar residues. The presence of multiple serines and threonines suggest that the protein may be O glycosylated. Glycosylation of the native SOWgp82 may also be associated with the GPI anchor complex (11). The results of our BLAST searches of the GenBank and SWISS-PROT databases for homologous sequences revealed that only low similarity (≤20%) exists between SOWgp82 and sequences of other reported proline-rich proteins.
Alignment of the translated SOWgp genes of three isolates of C. immitis showed sequence identity in the N- and C-terminal regions but differences in the number of 47-aa repeats (Fig. 3). The difference in the predicted molecular size of the SOWgps correlates with the number of conserved repeat sequences. However, the predicted molecular size of the translated, mature SOWgp (minus the signal peptide and propeptide) of each isolate and the estimated molecular size of the secreted, mature glycoprotein based on SDS-PAGE are incongruent (Table 1). The disparity is highest in SOWgp82, which contains the largest number of repeats and, therefore, the highest proline content. Proline-rich proteins typically demonstrate slow migration in SDS-PAGE gels, presumably due to conformational peculiarities that persist even after exposure to the reducing conditions of sample preparation (44).
FIG. 3.
Amino acid sequence alignment of translated SOWgp genes of C. immitis isolates Silveira (SOWgp58), C634 (SOWgp66), and C735 (SOWgp82). An asterisk indicates a nonconserved substitution of residues; a filled circle indicates a conserved substitution as defined by Higgins and Sharp (18).
TABLE 1.
Summary of differences between SOWgps of three clinical isolates of C. immitis
| Antigen | Isolate | Sizes (kDa)a ± SD | No. of aa | No. of repeats |
|---|---|---|---|---|
| SOWgp58 | Silveira | (58/29.1/30.7) ± 0.3 | 328 | 4 |
| SOWgp66 | C634 | (66/34.6/37.7) ± 0.3 | 375 | 5 |
| SOWgp82 | C735 | (82/39.5/43.8) ± 0.3 | 422 | 6 |
Sizes are presented as a/b/c, where a is an estimate of the molecular size of native glycoprotein based on mobility in SDS-PAGE gels under reducing conditions; b is the molecular size of SOWgp deduced from the translated gene sequence of the predicted, mature protein; and c is the molecular size of the native SOWgp as determined by SELDI-TOF mass spectrometry. The molecular sizes are mean values of three separate determinations ± the standard deviations.
Molecular size of the native SOWgp determined by SELDI-TOF mass spectrometry.
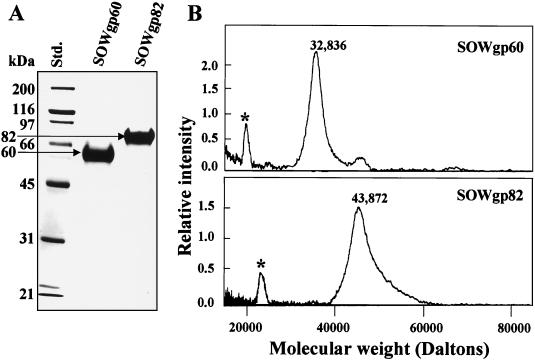
SDS-PAGE gel separations of the TX-114-extracted, crude SOW fraction of strain C735 typically revealed two Coomassie blue-stained bands at 60 and 82 kDa (Fig. 4A [20]). The 60-kDa band was usually less prominent than the upper band but was clearly visible in SDS-PAGE gel separations of SOW obtained from first- and second-generation parasitic-phase cultures which contained young presegmented spherules (20). The N-terminal amino acid sequences of the purified, native SOWgp82 and SOWgp60 are identical, and it was suggested that the 60-kDa glycoprotein is a product of proteolysis of the C terminus of SOWgp82 (20). In the present study, we attempted to clarify the disparity between the molecular size of the native SOWgp fractions of isolate C735 based on SDS-PAGE and the predicted size of the mature protein based on its translated gene sequence. The 60- and 82-kDa glycoproteins were each purified as previously described (Fig. 4A [20]), electroeluted from SDS-PAGE gels, and subjected to SELDI-TOF mass spectrometry to determine their actual molecular size (Fig. 4B). The results confirmed that the SDS-PAGE estimates of molecular sizes of the native, mature proline-rich proteins are markedly inflated. The actual molecular size of the secreted SOWgp82 is 43.8 kDa. This value is similar to the predicted size of the mature protein (i.e., 39.5 kDa [Table 1]). SOWgp60 was shown to have an actual molecular size of 32.8 kDa, indicating that proteolysis in the C-terminal region of SOWgp82 results in the release of an ca. 10-kDa peptide. Since cleavage of this C-terminal peptide would result in loss of the GPI anchor motif, the previously reported positive PAS reaction with the mature 60-kDa fraction (20) is most likely due to putative upstream sites of O glycosylation.
FIG. 4.
Molecular sizes of SOWgp fractions of isolate C735 as estimated by SDS-PAGE (A) and SELDI-TOF mass spectrometry (B). The asterisks in panel B locate low intensity peaks, which represent double-charged molecules.
Expression and immunolocalization of SOWgp82.
Total RNA was isolated from saprobic- and parasitic-phase cultures of C. immitis isolate C735, separated by formaldehyde agarose gel electrophoresis, and hybridized with the labeled 1.1-kb cDNA of SOWgp82. Total RNA (4 μg) was extracted from mycelial cultures grown for 4 days and from each of five different developmental stages of in vitro-grown parasitic cells (36, 72, 96, 120, and 144 h after arthroconidium inoculation [Fig. 5A ]). The Northern blot analysis in Fig. 5B showed that the predicted 1.6-kb cDNA band of SOWgp82 mRNA was present only in the parasitic phase. Comparison of levels of SOWgp82 gene expression in the parasitic cycle to the expression of the constitutive glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene of C. immitis (Fig. 5B) (20) suggested that production of the highest amounts of the SOWgp82 transcript correlated with the early stages of isotropic growth of first- and second-generation, presegmented spherules. On the other hand, the amount of SOWgp82 mRNA sharply decreased to an almost undetectable level during the intervening stages of development.
Parallel studies of total protein were conducted by immunoblot analysis to compare levels of production of the SOWgp82 glycoprotein in TX-114 extracts of homogenates of mycelia and parasitic cells (Fig. 5C). Murine antiserum raised against the purified rSOWp82 revealed a single band in immunoblots of SDS-PAGE separations of the E. coli homogenates (not shown). To confirm that the recombinant protein was encoded by the SOWgp82 gene, the gel-purified rSOWp82 was subjected to Lys-C digestion and N-terminal sequence analysis (17). The amino acid sequence of a selected peptide of the digest (AASSGSTDRPASTPV) was identical to aa 377 to 391 of the translated SOWgp82 gene (Fig. 3). In the immunoblot of different stages of C. immitis development (Fig. 5C), the same amount of total protein (20 μg) of each homogenate was applied to the SDS-PAGE gel as previously reported (21). The antiserum failed to recognize any component of the mycelial homogenate but recognized both the 82- and 60-kDa fractions of SOWgp82 in the parasitic cell preparations. Results of immunoblot analysis of the same developmental stages of the parasitic cycle as those described above suggested that the amount of SOWgp82 glycoprotein was highest during the early stages of spherule development in both the first and the second generations (36 to 96 h and 144 h, respectively) but decreased sharply at the time of endosporulation (120 h).
The same antiserum raised against the rSOWp82 was used for immunolocalization of the native glycoprotein in presegmented spherules by immunofluorescence (Fig. 5D and E) and immunoelectron microscopy (Fig. 5F). In both cases, the glycoprotein was found at the cell surface as patches of concentrated antigen (Fig. 5D and F). C. immitis spherules which were incubated with secondary antibody alone showed no label (Fig. 5E).
Adherence of SOWgp to ECM.
The amount of recombinant SOWp produced by E. coli transformed with the pET-SOWgp58 plasmid construct was ∼100-fold higher than the amount of bacterium-expressed rSOWp82. We cannot explain this difference in the level of expression of the two recombinant proteins. Because high sequence homology exists between rSOWp82 and rSOWp58, except for the number of 47-aa repeats (Fig. 3), we decided to use rSOWp58 for the adherence assays (Fig. 6). The results show that the labeled recombinant protein binds most avidly to laminin and significantly less so to fibronectin and collagen. A linear relationship exists between the amount of bound rSOWp58 and the concentrations of fibronectin and laminin used to test adherence. Our intent in this assay was to demonstrate that SOWgp is a putative adhesin rather than to present a detailed evaluation of the adherence properties of the cell surface molecule.
FIG. 6.
Adherence of biotin-labeled recombinant SOWp58 to mammalian ECM proteins in a microtiter plate assay. Bound protein was detected spectrophometrically by secondary reaction with peroxidase-conjugated streptavidin plus substrate. BSA was used as a control. The bars represent mean values ± the standard deviation of three separate experiments. The difference in relative binding of rSOWp58 to laminin versus fibronectin was statistically significant (P = 0.0011).
Transformation and isolation of the Δsowgp82 strain.
The transformation plasmid construct, pΔsowgp82, was designed to integrate into C. immitis chromosomal DNA by homologous recombination at two crossover sites (Fig. 7A and B). Transformation of 5 × 107 protoplasts with 5 μg of linearized DNA derived from ApaI/ClaI digestion of the plasmid construct yielded 80 to 100 hygromycin-resistant C. immitis colonies. A selection of 39 putative transformants were initially screened by PCR for deletion of the SOWgp82 gene fragment and integration of the plasmid into chromosomal DNA of the pathogen at a single site (36). Of the 39 colonies examined, 7 were identified as candidate, homokaryotic transformants in which the SOWgp82 gene fragment was replaced by homologous recombination with pΔsowgp82.
Three putative transformants (A7, A9, and A57) were selected from the seven colonies identified above and used for Southern hybridization with three different probes (probes Pb1 to Pb3; Fig. 7B). A restriction map was constructed for the 5.9-kb genomic fragment, which included the full-length SOWgp82 gene plus the 5′ and 3′ flanking regions (Fig. 7A). A restriction map was also constructed for the hypothetical structure of the chromosomal fragment which contained the integrated pΔsowgp82 plasmid (Fig. 7B). The results of hybridization of KpnI and BamHI digests of genomic DNA from the parental strain (P) and three selected transformants (A7, A9, and A57) with Pb1 (Fig. 7C), Pb2 (Fig. 7D), and Pb3 (Fig. 7E) provide evidence that homologous recombination occurred at a single locus in A7, A9, and A57. Pb2 and Pb3 hybridized weakly with bands in the KpnI digest but not the BamHI digest of genomic DNA of transformant A7 (Fig. 7D and E). Nevertheless, the single bands of predicted size in the Southern hybridizations of all three selected transformants indicate that the SOWgp82 gene fragment in these homokaryotic strains had been deleted. The A9 transformant was selected for further phenotypic analysis, and this strain was designated Δsowgp82.
Loss of SOWgp production by the Δsowgp82 transformant.
Total cell homogenates of 72-h parasitic-phase cultures (see Fig. 5A) of the parental and Δsowgp82 strains were subjected to SDS-PAGE gel separation as described above and then to immunoblot analysis with the murine serum raised against rSOWp82 (Fig. 8A). The parental strain showed the characteristic 82- and 60-kDa bands of isolate C735, whereas the antiserum failed to recognize any components of the cell homogenate derived from the Δsowgp82 strain. Additional evidence that the transformant does not express the SOWgp82 glycoprotein is presented in Fig. 8B and C. Serum obtained from a patient with confirmed coccidioidal infection (36) was reacted with thin sections of the crude SOW fraction isolated from 72-h parasitic-phase cultures of the parental (Fig. 8B) and the Δsowgp82 strains (Fig. 8C). In both cases, the electron micrographs reveal the typical membranous and amorphous material that comprises the SOW complex (7, 8). In Fig. 8B, the immunogold particles conjugated with anti-human IgG are bound primarily to the membranous components of SOW derived from the parental strain. On the other hand, no specific binding of immunogold particles to SOW of the transformant was visible (Fig. 8C).
FIG. 8.
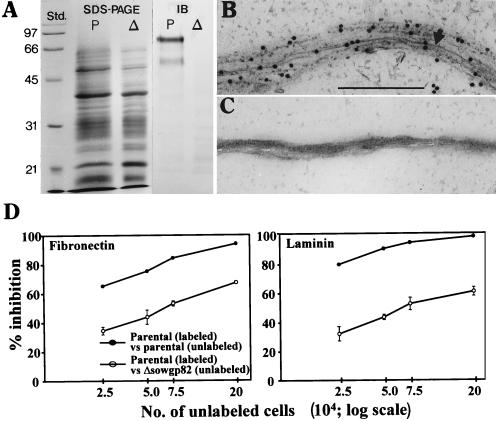
Biochemical, structural, and functional analysis of the Δsowgp82 mutant strain. (A) SDS-PAGE and corresponding immunoblot (IB) analysis of expression of native SOWgp82 in parasitic cell homogenates of the parental (P) and Δsowgp82 (Δ) strains. (B and C) Immunoelectron microscopy of isolated SOW from parental (B) and Δsowgp82 (C) strains with sera from a patient with confirmed coccidioidal infection. Bar (panel B), 0.5 μm. (D) Results of competitive inhibition adherence assay with biotin-streptavidin-labeled parental spherules (72 h) incubated with different concentrations of either unlabeled parental or Δsowgp82 spherules (72 h) in microtiter wells coated with fibronectin or laminin. The datum points represent mean values ± the standard deviation of three separate assays.
Reduced adherence of Δsowgp82-derived presegmented spherules to ECM.
Presegmented spherules isolated from 72-h cultures of the parasitic phase of the parental strain and the Δsowgp82 transformant were also tested for their ability to bind to fibronectin and laminin in a competitive adherence inhibition microplate assay (Fig. 8D). Unlabeled parental cells added to labeled parental cells at a ratio of 20:1 showed nearly complete inhibition of adherence of intact cells to the ECM proteins. Unlabeled spherules isolated from the mutant strain, on the other hand, failed to completely inhibit binding of the labeled parental cells and showed less of an inhibitory effect on binding of the parental cells to laminin than to fibronectin. These data are in agreement with the results presented in Fig. 6, which showed that purified recombinant protein (rSOWp58) binds more avidly to laminin than fibronectin. Results of this competitive inhibition assay suggest that absence of the SOWgp82 from the surface of presegmented spherules of the mutant strain significantly reduces the ability of these parasitic cells to bind to the ECM proteins.
Transformation and isolation of the SOWgp82R revertant.
The transformation plasmid construct, pSOWgpR, successfully integrated into C. immitis chromosomal DNA by homologous recombination at a single crossover site, as illustrated in Fig. 9A. Transformation of 5 × 107 protoplasts with 5 μg of undigested plasmid DNA yielded 15 phleomycin-resistant C. immitis colonies. The putative transformants were tested for their ability to grow in GYE agar in the presence of both hygromycin (100 μg/ml) and phleomycin (4 μg/ml). All of these colonies were screened by PCR for presence of the full-length SOWgp82 gene. The parental isolate, the Δsowgp82 mutant strain, and two selected revertants (R61 and R121) were examined by Southern hybridization (Fig. 9B) with a labeled probe (Pb2) described above (Fig. 7B). The results of hybridization of BamHI, KpnI, and XhoI digests of genomic DNA from the parental (P), mutant (A9), and two putative revertants (R61 and R121) provide evidence that the revertant strain R61 is homokaryotic and contains a single copy of the wild-type SOWgp82 gene which integrated into the chromosomal DNA by homologous recombination. The other revertant (R121) also contained the wild-type gene, but it integrated at an ectopic site. Immunoblot analysis of total protein preparations of parasitic cell homogenates from each of the four strains were conducted with antiserum raised against the recombinant SOWp82 as described above (Fig. 9C). The results show comparable amounts of SOWgp82 present in the cell homogenates of the parental (P) and revertant (R61) strains, and no protein detected by the anti-rSOWp82 antibody in the mutant strain (A9). The ectopically integrated wild-type gene in the second revertant (R121) expressed the glycoprotein but at a markedly reduced level. The revertant strain R61 was chosen for further study and was designated SOWgp82R. A predicted restriction map of the integrated pSOWgpR plasmid is shown in Fig. 9D. The results of Southern hybridization suggest that the plasmid integrated by a single crossing-over event at the tTRPC locus (Fig. 9A).
Comparison of the virulence of the parental, mutant, and revertant strains.
Equal numbers (n = 70) of arthroconidia isolated from the parental, Δsowgp82, and SOWgp82R strains were used to separately inoculate three groups of C57BL/10J mice by the intranasal route (Fig. 10). Mice challenged with the inoculum of the C735 parental or revertant strain showed a high mortality rate beginning on day 14 postinoculation. All animals in these two groups had died by day 21. In contrast, mice inoculated with the Δsowgp82 mutant strain showed a slower rate of mortality, and 58% of the animals survived beyond 40 days postchallenge. The difference in survival plots for mice challenged with the mutant versus the parental strain was statistically significant (P = 0.0019).
FIG. 10.
Survival plots of C57BL/10J mice challenged intranasally with equal numbers of arthroconidia isolated from the parental, Δsowgp82 mutant, or SOWgp82R revertant strain.
DISCUSSION
We have previously described a membrane-like SOW fraction of C. immitis that is released into the culture medium from the surface of presegmented parasitic cells (8). This material can be easily isolated from liquid shake cultures of the parasitic phase by differential centrifugation and has been subjected to compositional analysis and immunoassays (7). Phospholipids are prominent components of this cell surface fraction (8), a finding which is in agreement with the pioneering work of Tarbet and Breslau (41), who first reported that the walls of spherules are rich in “lipid complexes.” TX-114 extraction of the isolated SOW, followed by separation of the detergent and aqueous phases, yielded a water-soluble fraction composed of a single glycoprotein (SOWgp) which was detected in Coomassie blue-stained SDS-PAGE gels (20). The molecular size of this glycoprotein varies between C. immitis isolates. However, the N-terminal amino acid sequence of each purified SOWgp is identical, and each is reactive with sera from patients with coccidioidal infection (20). The isolated SOWgp fraction has also been shown to elicit an in vitro proliferative response of human peripheral blood monocytic cells obtained from individuals who are skin test reactive to C. immitis antigen (20). Antiserum raised against SOWgp and used for immunolocalization of the antigen in mycelial and parasitic-phase preparations revealed that the glycoprotein is expressed only on the surface of presegmented spherules (20). The possibility that this immunodominant, parasitic phase-specific antigen has a significant influence on the course of coccidioidal infection prompted us to clone the SOWgp gene and explore the potential functions of the expressed glycoprotein during fungus-host interaction.
The translated SOWgp gene of isolate C735 predicted a 39.5-kDa protein composed of a signal peptide and propeptide, six tandemly repeated proline- and aspartic acid-rich motifs, and a GPI anchor signal consensus sequence. The three isolates of C. immitis examined here (C735, C634, and Silveira) were shown to contain six, five, and four copies, respectively, of this repeat sequence. The Southern hybridization data showed that the difference in the number of repeats is not due to alternative splicing but is instead the result of genetic polymorphism.
No N-glycosylation sites were predicted from the deduced sequence. However, earlier studies of the native SOWgp showed that it reacts with PAS stain (20). A hydropathic profile of the deduced SOWgp protein, based on a Kyte-Doolittle plot (26; data not shown), revealed hydrophobic regions at the N and C termini that correspond to the secretory and GPI anchor signal sequences, respectively. The intervening region, composed primarily of 47-aa tandem repeats, is hydrophilic. This hydropathic profile is characteristic of GPI-linked proteins that are anchored to the outer leaflet of the plasmalemma (11). However, immunoelectron microscopic examinations of thin sections of presegmented spherules did not show SOWgp associated with the plasma membrane. Alternatively, SOWgp could be localized to the cell surface in a manner comparable to GPI-linked mannoproteins of yeast, which have been shown to be covalently bound to cell wall polysaccharides (22). However, SOWgp was found only in association with the SOW complex at the cell surface, and our extraction procedure released the glycoprotein from the complex as a water-soluble fraction of the detergent-aqueous-phase partition (20). These data suggest that SOWgp is not covalently linked to other components of the SOW complex. However, we cannot rule out the possibility that some SOWgp remained bound to the SOW layer. Results of transmission electron microscopic studies of the crude SOW fraction have revealed that this membrane-like material has a tripartite structure in cross-section that is comparable to the ultrastructure of biomembranes (8). Isolated GPI-anchored proteins of mammalian cells have been reported to reintegrate with cell surface membranes when these proteins are added to in vitro-grown cells (29). It is possible that the SOWgp of C. immitis is transported in exocytotic vesicles to the spherule surface, where it integrates into the putative, amphipathic phospholipid layer of the SOW complex. The presence of cysteine residues at conserved locations in the tandem repeats of the hydrophilic domain of SOWgp may permit the formation of regularly spaced disulfide bonds. In the case of the Candida albicans HWP1 protein, it was suggested that disulfide bonds may be functionally important in establishment of a conformation for intermolecular association of proteins at the hyphal surface (39). In fact, proline-rich motifs such as those in the tandem repeats of SOWgp (i.e., PPPPP) have been shown to be responsible for both untramolecular and intermolecular interactions of proteins (4, 23).
The high proline content of proteins typically leads to contradictory data on molecular size based on estimates derived from the deduced protein sequence, on the one hand, and SDS-PAGE, on the other (34). Although the relatively high serine and threonine content of the C-terminal region of SOWgp may be associated with some degree of O glycosylation, it does not account for the wide discrepancy in estimates of molecular size of the glycoprotein homologs from the three isolates examined in this study (Table 1). SELDI-TOF mass spectrometry has proved to be a valuable tool for characterization of posttranslationally modified proteins whose molecular masses cannot be accurately predicted from sequence data alone (30). The SELDI-TOF determination of molecular size of the native SOWgp82 of isolate C735 was 43.8 kDa, whereas the molecular size of the deduced, mature protein was 39.5 kDa. On this basis we suggest that the polypeptide is not highly glycosylated. Nevertheless, the sugar moiety may contribute to immunoreactivity and adhesion, and future studies will address these possibilities. SDS-PAGE separations of the native SOWgp from the C735 isolate consistently showed two bands with estimated molecular sizes of 82 and 60 kDa. The latter is probably a product of proteolysis, and its actual molecular size determined by SELDI-TOF analysis is 32.9 kDa. We used this same mass spectrometric method to compare the actual molecular sizes of the native, mature glycoproteins to the sequence-deduced sizes of the cloned SOWgps from C. immitis isolates C634 (37.7 versus 34.6 kDa, respectively) and Silveira (30.7 versus 29.1 kDa).
Recent studies in our laboratory have focused on C. immitis genes which demonstrate differential expression during the parasitic cycle. The rationale for these investigations is that the results may provide clues about mechanisms of morphogenetic control (21) and may simultaneously contribute to our search for potential drug targets, as well as immunoreactive and protective antigens that can be used in a vaccine against coccidioidomycosis (N. Delgado, J.-J. Yu, and G. T. Cole, Abstr. 101th Gen. Meet. Am. Soc. Microbiol., abstr. F-45, p. 363, 2001). In the present study we confirmed our earlier suggestion that SOWgp expression is parasitic phase specific. The results of both Northern hybridization and immunoblot analyses support this conclusion. SOWgp is produced during isotropic growth and early maturation of spherules but sharply decreases in expression during the endosporulation phase. Whether the decrease in specific mRNA is due to activation of a regulatory element or is the result of a precipitous loss of labile message is unknown. We recently purified a metalloproteinase (unpublished results) from the crude SOW fraction isolated from first-generation parasitic-phase cultures during the stage of endospore differentiation when the SOWgp content of spherules has markedly decreased compared to spherules in the isotropic growth phase (Fig. 5C). However, we have not yet determined whether the cell surface glycoprotein is specifically digested by this metalloproteinase. Immunolocalization of SOWgp with antibody raised against the recombinant protein revealed that the native glycoprotein is present only in the outer membranous and amorphous layer of the spherule wall and not in the inner wall layer. The patches of high and low density of SOWgp revealed by immunofluorescence correlated with the variable thickness of the SOW complex shown in thin sections.
Little is specifically known about the influence of SOW or SOWgp on host phagocyte interaction with parasitic cells of C. immitis. We have performed preliminary experiments (unpublished results) to test the ability of biotin-labeled recombinant SOWgp to bind to macrophages of a murine cell line (IC-21) by using a previously reported method (12). No adherence of the glycoprotein to macrophage membrane preparations was observed. An isolated, unrelated 36-kDa macrophage-binding protein of C. immitis was used as a positive control (G. T. Cole and C. Fradin, Abstr. 101th Gen. Meet. Am. Soc. Microbiol., abstr. F-13, p. 356, 2001). The results of earlier reports on interactions of polymorphonuclear neutrophils (PMNs) with parasitic cells of C. immitis suggested that the SOW complex may actually impede the ability of host cells to kill C. immitis. Frey and Drutz (13) showed that in vitro phagocytic and fungicidal activities of PMNs declined during C. immitis maturation from arthroconidia to young, presegmented spherules. These authors observed an absence of adequate PMN-spherule interaction and attributed this to production of an extracellular, fibrillar matrix that appeared to restrict intimate contact between the host and parasitic cells. This fibrillar matrix is synonymous with the SOW layer. It is possible, therefore, that the SOW complex provides protection to the pathogen against phagocytic and fungicidal activities of the host innate immune system.
Although SOWgp may not bind to the surface of macrophages, we have demonstrated here that the recombinant protein and intact, presegmented spherules adhere to host ECM proteins (laminin > fibronectin > collagen). Laminin and fibronectin are candidate ligands which could mediate adherence of the respiratory fungal pathogen to host tissues, influence colonization, and permit initiation of infection (16, 42). Host epithelial damage, which may result in exposure of the subepithelial basement membrane, is known to increase accessibility to host ligands, including laminin, fibronectin, and type IV collagen (16). In order to more critically evaluate the significance of SOWgp interaction with ECM proteins, we performed an SOWgp gene-targeted replacement experiment by using the transformation system which has recently been developed for C. immitis (36). Our previous targeted gene knockout approach utilized a disruption protocol (36). In the present study we report the first successful gene replacement strategy (9) applied to C. immitis. The yield of transformants with confirmed homologous recombination by this method was comparable to that of the disruption procedure. The transformants showed normal mycelial and parasitic-phase growth, and presegmented spherules demonstrated characteristic release of the SOW fraction from their cell surface in liquid culture. However, the isolated and extracted SOW from the Δsowgp82 strain lacked the SOWgp component, as determined by immunoblot analysis. A surprising discovery was that SOW from the mutant was not reactive with sera from patients with confirmed, disseminated coccidioidomycosis, suggesting that the SOWgp glycoprotein is the sole component of this parasitic cell surface layer that is recognized by patient antibody. To test whether loss of the SOWgp gene product influenced adherence to ECM proteins, a competitive inhibition assay was conducted with intact, presegmented spherules. The results confirmed that the absence of the glycoprotein component of SOW results in ca. 30 to 50% reduction in the ability of parasitic cells of the mutant strain to bind to fibronectin and laminin. However, the data also suggested that additional macromolecules exist on the surface of spherules that can bind to these same ligands. Studies are under way with SOW isolated the from the Δsowgp82 mutant strain to determine which additional components of this cell surface fraction show affinity for ECM ligands.
To evaluate the effect of deletion of the SOWgp gene on host response, a comparative survival experiment was conducted in which mice were challenged intranasally with an equal inoculum of arthroconidia obtained from either the parental or Δsowgp82 mutant strain. A revertant strain in which the deleted SOWgp gene was restored to the disruptant was also tested in the survival experiment. This is the first report of successful generation of a revertant strain of C. immitis by homologous recombination using two separate selective markers (HPH and BLE). The results of the challenge experiment demonstrated that the mutant had significantly decreased in virulence and that the difference between the three survival curves was most evident between 16 and 21 days postchallenge. It is not known whether reduced virulence of the mutant is due to decreased efficiency of binding to host tissue, loss of a B-cell dominant surface antigen which may influence the course of immune response to the pathogen, or both. Parasitic cell surface adhesins isolated from Blastomyces dermatitidis (BAD1 [5]) and Paracoccidioides brasiliensis (gp43 [3]) have also been reported to elicit both strong humoral and cellular immune responses during the course of the respective mycoses. As in coccidioidal infection, activation of the T helper 1 (Th1) rather than the Th2 subset of T cells appears to be essential for immunoprotection against these fungal diseases (20). All three antigens (BAD1, gp43, and SOWgp) contain immunodominant B-cell epitopes (24, 37). High titers of antibody to gp43 in patients infected with P. brasiliensis were correlated with T-cell hyporesponsiveness (3). These authors suggested that patient Th2 response to the immunodominant gp43 antigen may have compromised the ability of the host to control P. brasiliensis infection. SOWgp is a structurally complex antigen associated with a still undefined cell surface matrix (SOW). We suggest that SOWgp and other components of the SOW layer may play important roles both in adherence to host tissue and modulation of the immune response to C. immitis infection.
Acknowledgments
This investigation was supported by Public Health Service grant AI19149 from the National Institute of Allergy and Infectious Diseases.
Editor: T. R. Kozel
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Wiley, New York, N.Y.
- 3.Benard, G., M. J. Mendes-Giannini, M. Juvenale, E. T. Miranda, and A. J. Duarte. 1997. Immunosuppression in paracoccidioidomycosis: T-cell hyporesponsiveness to two Paracoccidioides brasiliensis glycoproteins that elicit strong humoral response. J. Infect. Dis. 175:1263-1267. [DOI] [PubMed] [Google Scholar]
- 4.Brady, L. J., D. G. Cvitkovitch, C. M. Geric, M. D. Addison, J. C. Joyce, P. J. Crowley, and A. S. Bleiweis. 1998. Deletion of the central proline-rich repeat domain results in altered antigenicity and lack of surface expression of the Streptococcus mutans P1 adhesin molecule. Infect. Immun. 66:4274-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandhorst, T. T., M. Wüthrich, T. Warner, and B. Klein. 1999. Targeted gene disruption reveals an adhesin indispensable for pathogenicity by Blastomyces dermatitidis. J. Exp. Med. 189:1207-1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, G. T., and C.-Y. Hung. 2001. The parasitic cell wall of Coccidioides immitis. Med. Mycol. 39(Suppl. 1):31-40. [PubMed] [Google Scholar]
- 7.Cole, G. T., T. N. Kirkland, M. Franco, S. Zhu, L. Yuan, S. H. Sun, and V. M. Hearn. 1988. Immunoreactivity of a surface wall fraction produced by spherules of Coccidioides immitis. Infect. Immun. 56:2695-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cole, G. T., K. R. Seshan, M. Franco, E. Bukownik, S. H. Sun, and V. M. Hearn. 1988. Isolation and morphology of an immunoreactive outer wall fraction produced by spherules of Coccidioides immitis. Infect. Immun. 56:2686-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.d'Enfert, C., G. Weidner, P. C. Mol, and A. A. Brakhage. 1999. Transformation systems of Aspergillus fumigatus, p. 146-166. In A. A. Brakhage and J. B. Schmidt (ed.), Aspergillus fumigatus: contribution to microbiology, vol. 2. Kruger Press, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 10.Drutz, D. J., and M. Huppert. 1983. Coccidioidomycosis: factors affecting the host-parasite interaction. J. Infect. Dis. 147:372-390. [DOI] [PubMed] [Google Scholar]
- 11.Eisenhaber, B., P. Bork, and F. Eisenhaber. 1998. Sequence properties of GPI-anchored proteins near the ω-site: constraints for the polypeptide binding site of the putative transamidase. Protein Eng. 11:1155-1161. [DOI] [PubMed] [Google Scholar]
- 12.Fradin, C., D. Poulain, and T. Jouault. 2000. Beta-1,2-linked oligomannosides from Candida albicans bind to a 32-kilodalton macrophage membrane protein homologous to the mammalian lectin galectin-3. Infect. Immun. 68:4391-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frey, C. L., and D. J. Drutz. 1986. Influence of fungal surface components on the interaction of Coccidioides immitis with polymorphonuclear neutrophils. J. Infect. Dis. 153:933-943. [DOI] [PubMed] [Google Scholar]
- 14.Frohman, M. A. 1990. RACE: rapid amplification of cDNA ends, p. 28-38. In M. A. Innis, D. H. Gelfund, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.
- 15.Galgiani, J. N. 1999. Coccidioidomycosis: a regional disease of national importance: rethinking approaches for control. Ann. Intern. Med. 130:293-300. [DOI] [PubMed] [Google Scholar]
- 16.Gil, M. L., M. C. Peñalver, J. L. Lopez-Ribot, J. E. O'Connor, and J. P. Martinez. 1996. Binding of extracellular matrix proteins to Aspergillus fumigatus conidia. Infect. Immun. 64:5239-5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guevara-Olvera, L., C.-Y. Hung, J.-J. Yu, and G. T. Cole. 2000. Sequence, expression and functional analysis of the Coccidioides immitis ODC (ornithine decarboxylase) gene. Gene 242:437-448. [DOI] [PubMed] [Google Scholar]
- 18.Higgins, D. G., and P. M. Sharp. 1988. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene 73:237-244. [DOI] [PubMed] [Google Scholar]
- 19.Hofmann, K., P. Bucher, L. Falquet, and A. Bairoch. 1999. The PROSITE database: its status in 1999. Nucleic Acids Res. 27:215-218., [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung, C.-Y., N. M. Ampel, L. Christian, K. R. Seshan, and G. T. Cole. 2000. A major cell surface antigen of Coccidioides immitis which elicits both humoral and cellular immune responses. Infect. Immun. 68:584-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hung, C.-Y., J.-J. Yu, P. F. Lehmann, and G. T. Cole. 2001. Cloning and expression of the gene which encodes a tube precipitin antigen and wall-associated β-glucosidase of Coccidioides immitis. Infect. Immun. 69:2211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kapteyn, J. C., L. L. Hoyer, J. E. Hecht, W. H. Muller, A. Andel, A. J. Verkleij, M. Makarow, H. Van Den Ende, and F. M. Klis. 2000. The cell wall architecture of Candida albicans wild-type cells and cell wall-defective cells. Mol. Microbiol. 35:601-611. [DOI] [PubMed] [Google Scholar]
- 23.Kay, B. K., M. P. Williamson, and M. Sudol. 2000. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 44:231-241. [PubMed] [Google Scholar]
- 24.Klein, B. S., and S. L. Newman. 1996. Role of cell surface molecules on Blastomyces dermatitidis in host-pathogen interactions. Trends Microbiol. 4:246-251. [DOI] [PubMed] [Google Scholar]
- 25.Kruse, D., and G. T. Cole. 1992. A seroreactive 120-kilodalton β-1,3-glucanase of Coccidioides immitis which may participate in spherule morphogenesis. Infect. Immun. 60:4350-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, R. M., A. C. Huston, and P. D. Hoeprich. 1977. Reproducible method for induction of pulmonary coccidioidomycosis in mice. J. Infect. Dis. 135:117-119. [DOI] [PubMed] [Google Scholar]
- 28.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Medof, M. E., S. Nagajan, and M. L. Tykocinski. 1996. Cell-surface engineering with GPI-anchored proteins. FASEB J. 10:574-586. [DOI] [PubMed] [Google Scholar]
- 30.Merchant, M., and S. R. Weinberger. 2000. Recent advances in surface-enhanced laser desorption/ionization-time-of-flight mass spectrometry. Electrophoresis 21:1164-1167. [DOI] [PubMed] [Google Scholar]
- 31.Nakai, K., and M. Kanehisa. 1992. A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14:897-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan, S., and G. T. Cole. 1992. Electrophoretic karyotypes of clinical isolates of Coccidioides immitis. Infect. Immun. 60:4872-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perfect, S. E., R. J. O'Connell, E. F. Green, C. Doering-Saad, and J. R. Green. 1998. Expression cloning of a fungal proline-rich glycoprotein specific to the biotrophic interface formed in the Colletotrichum-bean interaction. Plant J. 15:273-279. [DOI] [PubMed] [Google Scholar]
- 34.Proft, T., H. Hilbert, G. Layh-Schmitt, and R. Herrmann. 1995. The proline-rich P65 protein of Mycoplasma pneumoniae is a component of the Triton X-100-insoluble fraction and exhibits size polymorphism in strains M129 and FH. J. Bacteriol. 177:3370-3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Punt, P. J., R. P. Oliver, M. A. Dingemanse, P. H. Pouwels, and C. A. M. J. J. van den Hondel. 1987. Transformation of Aspergillus based on the hygromycin B resistance marker from Escherichia coli. Gene 5:117-124. [DOI] [PubMed] [Google Scholar]
- 36.Reichard, U., C.-Y. Hung, P. W. Thomas, and G. T. Cole. 2000. Disruption of the gene which encodes a serodiagnostic antigen and chitinase of the human fungal pathogen Coccidioides immitis. Infect. Immun. 68:5830-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saraiva, E. C. O., A. Altermani, M. F. Franco, C. S. Unterkircher, and Z. P. Camargo. 1996. Paracoccidioides brasiliensis-gp 43 used as paracoccidioidin. J. Med. Vet. Mycol. 34:155-161. [DOI] [PubMed] [Google Scholar]
- 38.Seshan, K. R., and G. T. Cole. 1994. Structural studies in Coccidioides immitis, p. 265-273. In B. Maresca and G. S. Kobayashi (ed.), Molecular biology of pathogenic fungi. Telos Press, New York, N.Y.
- 39.Staab, J. F., S. D. Bradway, P. L. Fidel, and P. Sundstrom. 1999. Adhesive and mammalian transglutaminase substrate properties of Candida albicans Hwp1. Science 283:1535-1538. [DOI] [PubMed] [Google Scholar]
- 40.Sundstrom, P. 1999. Adhesins in Candida albicans. Curr. Opin. Microbiol. 2:353-357. [DOI] [PubMed] [Google Scholar]
- 41.Tarbet, J. E., and A. M. Breslau. 1953. Histochemical investigation of the spherule of Coccidioides immitis in relation to host reaction. J. Infect. Dis. 72:183-190. [DOI] [PubMed] [Google Scholar]
- 42.Vicentini, A. P., J. Z. Moraes, J.-L. Gesztesi, M. F. Franco, W. De Souza, and J. D. Lopes. 1997. Laminin-binding epitope on gp43 from Paracoccidioides brasiliensis is recognized by a monoclonal antibody raised against Staphylococcus aureus laminin receptor. J. Med. Vet. Mycol. 35:37-43. [PubMed] [Google Scholar]
- 43.von Heijne, G. 1986. Towards a comparative anatomy of N-terminal topogenic protein sequences. J. Mol. Biol. 189:239-242. [DOI] [PubMed] [Google Scholar]
- 44.Williamson, M. P. 1994. The structure and function of proline-rich regions in proteins. Biochem. J. 297:249-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wyckoff, E. E., E. J. Pishko, T. N. Kirkland, and G. T. Cole. 1995. Cloning and expression of a gene encoding a T-cell reactive protein from Coccidioides immitis: homology to 4-hydroxyphenylpyruvate deoxygenase and the mammalian F antigen. Gene 161:107-111. [DOI] [PubMed] [Google Scholar]
- 46.Yu, J.-J., S. L. Smithson, P. W. Thomas, T. N. Kirkland, and G. T. Cole. 1997. Isolation and characterization of the urease gene (URE) from the pathogenic fungus Coccidioides immitis. Gene 198:387-391. [DOI] [PubMed] [Google Scholar]
- 47.Zimmermann, C. R., S. M. Johnson, G. W. Martens, A. G. White, B. L. Zimmer, and D. Pappagianis. 1998. Protection against lethal murine coccidioidomycosis by a soluble vaccine from spherules. Infect. Immun. 66:2342-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]