Abstract
The response of the intracellular pathogen Rhodococcus equi to H2O2 treatment, a situation potentially encountered after the oxidative burst of alveolar macrophages, was analyzed. Compared to other bacteria, including Deinococcus radiodurans, R. equi showed exceptionally high resistance to this stress. A proteomic approach showed that four polypeptides present in the wild-type strain (85F) are missing in the plasmid-cured strain 85F(P-), and by using a DNA macroarray, we identified two plasmid-encoded vap genes, vapA and vapG, whose expression was highly induced by H2O2 treatment. Whereas the transcript size of vapA was compatible with a monocistronic mRNA, the transcript of vapG was considerably longer. Rapid amplification of cDNA ends PCRs showed that the transcriptional start sites of the two operons were 69 and 269 nucleotides (nt) upstream of the start codon, respectively. Analysis of these leader sequences revealed the presence of a small open reading frame named podG, which encodes a sequence of 55 amino acids preceded by a putative ribosome binding site sequence in the vapG transcript. Taking this result into account, the untranslated leader of the podG/vapG operon is 87 nt. Alignment of this sequence with the leader sequences of vapA and vapD, genes previously shown to be induced by acid, revealed significant homologies. Since our results showed that vapA, vapD, and vapG are genes highly induced by macrophage-related stresses, their gene products may, within the Vap protein family, play a dominant role inside these phagocytic cells and may be the most promising candidates for vaccination strategies.
Rhodococcus equi is a gram-positive, facultative intracellular pathogen which can cause severe pneumonia or occasionally septic arthritis or osteomyelitis in young foals (14). In the last decade, an increase in cases of pneumonia caused by R. equi in immunocompromised people, especially individuals with AIDS or those undergoing immunosuppressive therapy, has been reported (8, 27). The organism is thought to be delivered to the lungs via inhalation and grows primarily, if not exclusively, within host macrophages (18, 24). R. equi strains isolated from pneumonic foals typically contain a large plasmid of about 85 to 90 kb with a 27.5-kb pathogenicity island containing seven vap genes (named vap for virulence-associated protein) (32, 33). Plasmid-cured isogenic mutants of virulent strains lose their ability to survive in alveolar macrophages and fail to induce pneumonia in foals (15, 17).
To survive within alveolar macrophages, the bacteria must adapt to their intracellular environment including the stresses they encounter in a host cell armed with a powerful array of antimicrobial defenses (25). The involvement of stress-induced proteins in the pathogenesis of several bacterial pathogens has been demonstrated. For example, the heat shock proteins GroEL and DnaK are overexpressed inside macrophages for intracellular pathogens such as Brucella abortus and Mycobacterium tuberculosis (28, 41). Several studies on pathogenic bacteria have demonstrated that they have adapted to the intracellular environment of macrophages by altering protein synthesis from multiple regulons (16). It is speculated that the enhanced expression of stress proteins may give intracellular bacteria a selective advantage in their ability to survive in phagocytes, resulting in their multiplication and dissemination throughout the host. Clearly, the ability to withstand a stressing environment is an important factor in the virulence of intracellular bacteria. For this reason, identifying and characterizing stress-induced proteins is potentially of great importance in understanding the pathogenesis of R. equi.
Little is known about the gene products required for survival, growth, and pathogenesis of R. equi in the intracellular environment of phagocytic cells. There is evidence from other bacterial systems indicating that intracellular pathogens respond to macrophage phagocytosis by expressing several stress proteins which are also induced by oxidative stress in vitro (1, 22). Indeed, concomitant with phagocytosis, professional phagocytes produce an oxidative burst, resulting in a number of toxic by-products. These reactive oxygen intermediates include singlet oxygen (·O2), superoxide anions (O2−), hydrogen peroxide (H2O2), and the hydroxyl radical (.OH), which are part of the bactericidal defense mechanisms employed by phagocytic cells. An oxidative stress can also be a stimulus for induction of virulence genes responsible for survival or multiplication of bacteria within phagocytes. For example, in Yersinia enterocolitica, an insertional mutation of gsrA (named gsr for global stress requirement) resulted in the inability of the organism to survive within macrophages and greatly increased susceptibility to oxidative stress (40).
Virtually nothing is known about the role of the megaplasmid or more precisely the vap genes in the pathogenesis of R. equi infections. Nevertheless, we have shown in a previous study that acid pH, one of the signals encountered by the bacterium inside phagosomes, induced expression of a set of proteins, including proteins encoded by two vap genes, vapA and vapD (5). In the present study, we showed that vap genes were also highly induced by H2O2, evidence that vap gene products may play important roles in the survival and growth of R. equi within macrophages.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
R. equi 85F, provided by the Institut de Pathologie du Cheval (Association Française de Sécurité Sanitaire des Aliments [AFSSA]-Dozulé, Dozulé, France), and its isogenic plasmid-cured strain, 85F(P-) (4), were used for all experiments. Strain 85F, isolated from pneumonic foals, possesses a 85-kb virulence plasmid named p85F (5). Cultures were grown in M17 broth medium (36) supplemented with 0.5% glucose (GM17) and incubated at 37°C under shaking (90 rpm). Dilutions were performed in physiological solution (0.9% NaCl).
Escherichia coli XL1 Blue (Stratagene, La Jolla, Calif.) and Enterococcus faecalis JH2-2 (20) were used as reference bacterial strains in the hydrogen peroxide challenge assay. These organisms were cultured under the same conditions as R. equi.
Hydrogen peroxide challenge assay.
Cultures were grown to mid-exponential growth phase in GM17 medium. Cells were harvested by centrifugation, and the pellets were resuspended in an equal volume of physiological solution containing H2O2 at different concentrations. Survival of bacteria was monitored over 60 min of incubation at 37°C. Viable counts of bacteria were performed at specific times by serially diluting samples and plating them onto GM17 agar plates. Colonies were counted after 24 h (E. coli) or 48 h at 37°C. Survival was defined as the ratio of CFU present after the challenge treatment to CFU at time zero. In all experiments, each value was the average of duplicate plates, and all experiments were performed in triplicate.
Extraction of membrane and cytoplasmic proteins.
Bacteria were grown to mid-exponential growth phase in 100 ml of GM17 broth medium at 37°C. Cells were harvested by centrifugation (10 min, 3,000 × g, 4°C) and washed in a solution containing 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1% β-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, and 100 μg of chloramphenicol per ml. Membrane and cytoplasmic proteins were extracted as described previously (5).
2DGE.
High-resolution two-dimensional gel electrophoresis (2DGE) was performed by the method of O'Farrell (26) with the modifications described by Lopez et al. (23), using the Investigator 2D-Electrophoresis system (Millipore, Bedford, Mass.). After the second-dimensional gel electrophoresis was performed, the 14% polyacrylamide gels were silver stained (Millipore). All experiments were performed at least three times.
Western blot analysis.
After centrifugation of the culture (10 min, 3,000 × g, 20°C), the cell pellet was solubilized in sodium dodecyl sulfate (SDS) buffer (62.5 mM Tris-HCl [pH 6.8], 10% [vol/vol] glycerol, 2% SDS, 5% 2-mercaptoethanol, 0.02% bromophenol blue) and boiled for 10 min. The undissolved material was sedimented by centrifugation at 13,000 × g for 3 min. SDS-polyacrylamide gel electrophoresis was performed by the method of Laemmli (21) with a vertical slab gel containing 14% polyacrylamide (Bio-Rad Mini-Protean II system). After electrophoresis, proteins were transferred onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore) by electroblotting (Milliblot-Graphite Electroblotter; Millipore) according to the manufacturer's instructions. Immunoblot analysis was performed with an enhanced chemiluminescence Western blot analysis system (Amersham Pharmacia Biotech). Monoclonal antibody against VapA (provided by S. Taouji, AFSSA-Dozulé) was used for immunoblotting procedures.
Total RNA extraction from R. equi and preparation of 32P-labeled total first-strand cDNA.
Total RNA of R. equi was isolated with the RNeasy Midi Kit (Qiagen) as instructed by the manufacturer. Total first-strand cDNA was synthesized exactly by the method of Chuang et al. (7).
Construction of plasmid gene library.
Using the sequence of the virulence plasmid p103 (accession number AF116907) (33), primer pairs were determined to generate specific fragments of each open reading frame (ORF) present in the pathogenicity island of the virulence plasmid p85F (Table 1). The PCR fragments, obtained by using Ready To Go PCR beads (Amersham Pharmacia Biotech), were purified with the QIAquick kit (Qiagen) and sequenced by the dideoxy chain termination method with the ABI prism sequencing system (PE Biosystem). A 100-ng sample of DNA of each PCR product was dotted onto a Hybond-N+ membrane (Amersham Pharmacia Biotech) according to the manufacturer's instructions.
TABLE 1.
Primers used in PCR experiments
| ORF | Primer | Sequence | Orientationa |
|---|---|---|---|
| ORF1 | Orf1A | 5′-CATCGGTTGGAGTCATCGC-3′ | + |
| f2 | 5′-GTAGGCCAAGTTGGAGTTGCG-3′ | − | |
| vapG | G1 | 5′-CTTCTCCAGGACTGCTGGC | − |
| G2 | 5′-CACACGAGACATGGTCGG-3′ | + | |
| ORF3 | Orf3A | 5′-AGATATCGCACAGGGTCCG-3′ | − |
| Orf3B | 5′-GTCCTGACCAACTCTCGCC-3′ | + | |
| ORF4 | LysR1 | 5′-GACCGAACTCCACTCAGCC-3′ | + |
| LysR2 | 5′-CGTACCTGACGCCTTCGG-3′ | − | |
| ORF5 | BCMP1 | 5′-GCAGGCAGCAATGTATGCG-3′ | + |
| BCMP2 | 5′-CCTGCCGAACAATTTCGGC-3′ | − | |
| vapH | H1 | 5′-GACGAGACGAGAGGGGTGC-3′ | + |
| H2 | 5′-TCTTCGCTTGGACTACGC-3′ | − | |
| ORF7 | Orf7A | 5′-CGACCTGCTTTCGAATCCG-3′ | + |
| Orf7B | 5′-GTCTTCAAACTTTGAACCG-3′ | − | |
| ORF8 | TwoC1 | 5′-GATGAAGTGCTCAGATCGG-3′ | + |
| TwoC2 | 5′-CTGACGTGTACAGTGTCCG-3′ | − | |
| ORF9 + ORF10 | Orf9A | 5′-CTTGGACGCCTGATCTCGC-3′ | + |
| Orf10B | 5′-ACAGCAGAACATCTGGCGC-3′ | − | |
| ORF11 | Orf11A | 5′-TCCAGACGTGTCTGGACCG-3′ | + |
| Orf11B | 5′-CGCAGCGGTTATCAATCGC-3′ | − | |
| vapA | Rho1 | 5′-GACTCTTCACAAGACGGT-3′ | + |
| Rho2 | 5′-TAGGCGTTGTGCCAGCTA-3′ | − | |
| ORF13 | Orf13A | 5′-TCGCAACAGGATTGGTGCC-3′ | + |
| Orf13B | 5′-GGAGCCGCTCAAGAATCGC-3′ | − | |
| vapC | C3 | 5′-CAATGTAGTCGCTCCGTCCGGC-3′ | + |
| C4 | 5′-GCAATTCATCCCGCGCGAGCG-3′ | − | |
| vapD | D3 | 5′-CTCGCGGTGGCTGTGATCGC-3′ | + |
| D4 | 5′-CCCTTGCAAGGACTTGTTCC-3′ | − | |
| ORF16 | Orf16A | 5′-CGTCACAGCGATCCATCCG-3′ | + |
| Orf16B | 5′-GTCGGGGACCTAACTGCGC-3′ | − | |
| ORF17 | Orf17A | 5′-GCCGTTCCCGGTTCGCGGT-3′ | − |
| Orf17B | 5′-TCGTATGACGGGATAACGC-3′ | + | |
| ORF18 | Orf18A | 5′-GTTCACCGTCCTGGCAGGC-3′ | − |
| Orf18B | 5′-AGGACCGCGACCATGAGCG-3′ | + | |
| vapE | E3 | 5′-CTGACTTATAGTAGCTGCTCC-3′ | + |
| E4 | 5′-GTGCCTCCTCCGATGCCCACC-3′ | − | |
| vapF | F1 | 5′-GTGAGTGATAGAATATGCC-3′ | + |
| F2 | 5′-GCTTCTGCCAATCATTGCG-3′ | − | |
| ORF21 | Orf21A | 5′-GTCCGTAACCGCTTAAGCC-3′ | − |
| Orf21B | 5′-CAGCGGTTGTCGACAGCCG-3′ | + |
+, forward primers; −, reverse primers.
Analysis of mRNA transcription by Northern blotting.
For Northern blot studies, total RNA preparations were treated with DNase before precipitation. After electrophoresis of exactly 10 μg of RNA of R. equi, Northern blots were prepared using Hybond-N+ membranes and standard procedures (29). For quantification of the relative intensity of the hybridizing bands in the Northern blots, rRNA bands observed after ethidium bromide staining of gels were used as an internal standard for each sample. For this purpose, the stained 16S and 23S rRNA bands were scanned and quantified by densitometry with the Optiquant Image Analysis software (Packard Instrument Company, Canberra, Australia). The sizes of the transcripts were estimated by comparing band mobility of standards in an RNA ladder (0.56 to 9.4 kb) (Amersham Pharmacia Biotech). PCRs were performed using Ready To Go PCR beads (Amersham Pharmacia Biotech) to generate specific fragments of vapA and vapG genes using primer pairs Rho1-Rho2 and G2-G1, respectively (Table 1). The PCR fragments were then purified with the QIAquick kit (Qiagen) and used as templates to generate specific single-strand probes by elongation with the reverse primers (200 pmol) in a reaction mixture (100 μl) containing 2 mM (each) dGTP, dCTP, and dTTP, 1.5 mM MgCl2, 1 × PCR buffer (Amersham Pharmacia Biotech), 5 U of Taq DNA polymerase (Amersham Pharmacia Biotech), and 20 μCi of [α-32P]dATP (Amersham Pharmacia Biotech). Reactions were run for 10 cycles. Prehybridization and hybridization of membrane-bound RNA with single-strand DNA probes were performed at 55°C in a solution containing 0.25 M NaH2PO4, 0.25 M NaHPO4, and 5% SDS.
Mapping of the transcriptional start site.
In order to determine the transcriptional start site, a 5′→3′ rapid amplification of cDNA ends (RACE) (Roche Molecular Biochemicals) was performed. Briefly, after reverse transcription with primers G3 (5′-GAGCTCCCGCTCTTCGC-3′), 47C (5′-GTCGTACTGCTGCTCTTGCC-3′), and Dtr3 (5′-CGTAACCAAGTCGTTCGC-3′), for vapG, vapA, and vapD, respectively, a 3′ dA tail was added to the CDNAs and then PCR amplified with an oligo(dT) primer and a nested primer (G4 [5′-CAACGAGCGTTGCCGCC-3′], Atr1 [5′-GCAGCCGCAGCTACGGCTG-3′], and Dtr1 [5′-CCTTCTTATCCTCAGC-3′] for vapG, -A and -D, respectively). The resulting products were subsequently sequenced by the dideoxy chain termination method with the ABI prism sequencing system (PE Biosystems) using primers G8 (5′-GCGAATACCTCCGAACGG-3′), Rho1c (5′-ACCGTCTTGTGAAGAGTC-3′), and Dtr2 (5′-CCATTGAGACCGTTGCGA-3′) for vapG, vapA, and vapD, respectively.
General molecular methods.
DNA and amino acid sequences were analyzed using Mac Vector software (Kodak, Scientific Imaging Systems), and database searches were performed using the BLAST software (2). Other standard techniques were performed as described by Sambrook et al. (29).
RESULTS
Sensitivity of R. equi to H2O2 stress.
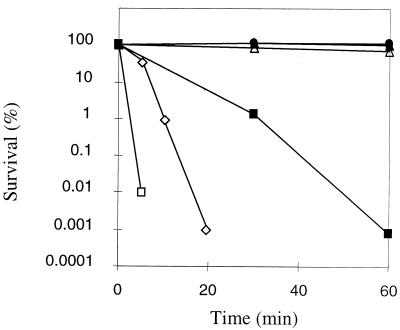
We first examined the survival of R. equi strain 85F and its isogenic plasmid-cured strain, 85F(P-), in the presence of increasing concentrations of H2O2. Survival of the bacteria was monitored over 60 min of incubation in physiological water (0.9% NaCl) in the presence of 10, 50, 100, and 150 mM H2O2 (Fig. 1). Surprisingly, the viability of R. equi was not affected with challenge doses as high as 100 mM. Only with the highest concentration tested (150 mM) was a decrease in cultivability observed. Indeed, the survival dropped by approximately 5 orders of magnitude after a 60-min exposure. In comparison to these results, two other bacteria (E. coli and E. faecalis) tested against the highest dose (150 mM H2O2) were significantly more sensitive than R. equi (Fig. 1). Only the results obtained with the parental strain 85F are shown in Fig. 1, but a similar result was obtained with the plasmid-cured strain 85F(P-) (data not shown). This shows that plasmid-encoded proteins do not play a role in H2O2 resistance of R. equi.
FIG. 1.
Effect of H2O2 treatments on R. equi 85F survival. Cultures were grown to mid-exponential growth phase and either unstressed (•) or treated with H2O2 at concentrations of 10 mM (○), 50 mM (▴), 100 mM (▵), and 150 mM (▪) for 60 min. The survival of two reference bacteria, E. coli (□) and E. faecalis (◊) in the presence of the highest H2O2 dose (150 mM) after 5, 10, and 20 min, determined under the same conditions, are also shown. The values are the averages of results from three independent experiments. Comparable results were obtained with the plasmid-cured strain.
Modification of protein expression by hydrogen peroxide treatment.
Membrane and cytoplasmic protein extracts from H2O2-stressed and unstressed cells of strains 85F and 85F(P-) were subjected to 2DGE to determine which polypeptides showed altered expression under oxidative stress.
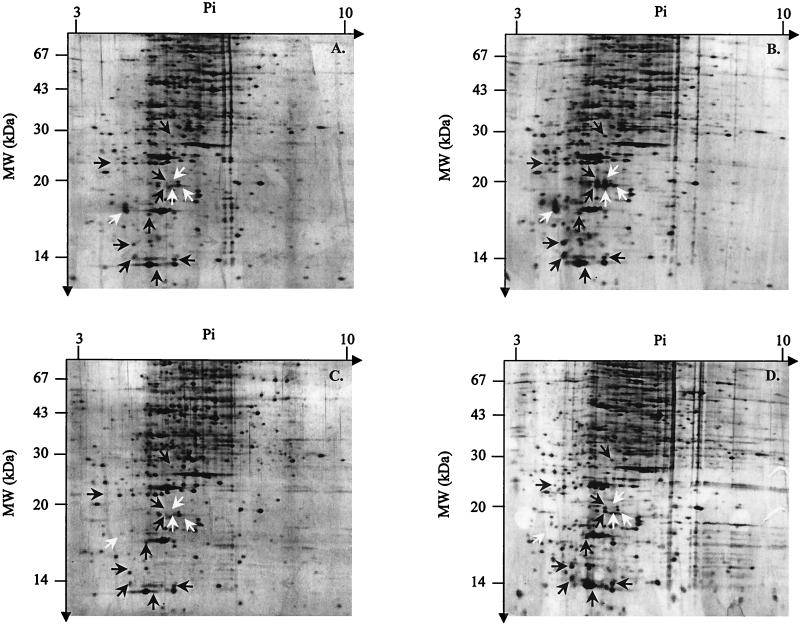
Whereas no differences were observed with the membrane protein fractions (data not shown), analysis of spectra obtained from cytoplasmic proteins revealed that 13 OSP polypeptides (named OSP for oxidative stress protein) were overexpressed in response to H2O2 in strain 85F (Fig. 2A and B). Among the OSPs, nine were also induced in strain 85F(P-), but interestingly, four were absent in the plasmid-cured strain (Fig. 2C and D). Using preparative 2DGE, we tried to identify some of the induced proteins by N-terminal microsequencing. Unfortunately, by increasing the protein concentration, the quality of separation of the polypeptides significantly decreased (horizontal smears) and the proteins could not be isolated in sufficient quantity for the Edman reaction.
FIG. 2.
Silver-stained two-dimensional gels of cytoplasmic proteins from the R. equi strains 85F (A and B) and 85F(P-) (C and D). The protein patterns of untreated (A and C) and H2O2-treated (10 mM for 45 min) (B and D) cultures are shown. Molecular mass (MW) is shown on the y axis, and pI is shown the x axis. Proteins overexpressed in strains 85F and 85F(P-) (black arrows) and polypeptides whose expression is specifically induced in strain 85F (white arrows) are indicated. Only reproducible changes observed from at least three individual experiments are indicated.
Are plasmid-encoded genes regulated in response to oxidative stress?
As described above, four proteins showing induced expression in response to H2O2 were present only in strain 85F. However, the proteomic approach did not allow us to determine whether the proteins were encoded by plasmid genes and regulated by oxidative stress or whether the genes were located on the bacterial chromosome and under the control of a plasmid regulator. Thus, we used a strategy of differential dot blot hybridization to identify plasmid-encoded genes potentially upregulated by hydrogen peroxide.
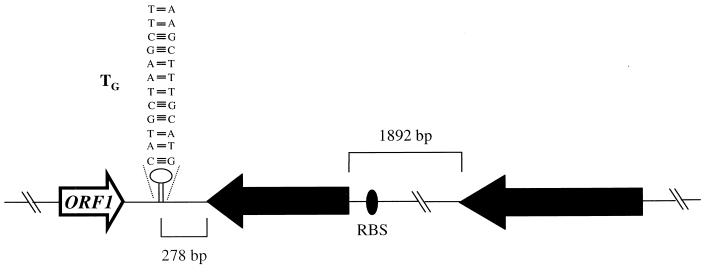
The size of the pathogenicity island of the virulence plasmid p85F was estimated to be approximately 27.5 kb containing 21 ORFs (33) (accession number AF116907). Entire or internal fragments of each of these ORFs were amplified using specific primer pairs (Materials and Methods). The resulting PCR products were sequenced and compared to proteins in the databases. All of the PCR products showed 100% identity with the sequence of the homologous virulence plasmid of R. equi 103 (accession number AF116907). Finally, the PCR products were dotted on a nylon membrane and hybridized with 32P-labeled cDNA isolated from either naive cells or cells exposed to 10 mM H2O2. Compared to the results obtained with cDNA prepared from nonexposed cells, a strong signal was observed for two ORFs corresponding to vapA and vapG (data not shown). The genomic context of the vapA gene of R. equi has already been described (5, 35). The surrounding regions of the vapG gene of R. equi 85F were sequenced and analyzed. The structural organization of vapG is shown in Fig. 3. The environment of vapG also showed 100% identity to that of the virulence plasmid p103 (accession number AF116907). vapG is an ORF of 519 bp initiated by the start codon GTG which encodes a 172-amino-acid protein (33) with a calculated molecular mass of 17.4 kDa and a pI of 4.78. A putative ribosome binding site (RBS) sequence (GAGAGG) is located 8 nucleotides (nt) upstream of the start codon which corresponds to the optimal spacing (7 to 9 bp) determined by Vellanoweth and Rabinowith (38). An inverted repeat (IR) located 278 bp downstream of vapG (ΔG value of −27 kcal · mol−1; determined by the method of Tinoco et al. [37]) may function as a Rho-independent transcription terminator (Fig. 3).
FIG. 3.
Structural organization of the genetic environment of the vapG gene. Large arrows represent the ORFs, and their orientation shows the transcriptional direction. The distances between vapG and ORF3 and the putative Rho-independent terminator (TG) located downstream of the vapG stop codon are shown.
Upregulation of vap genes after exposure to H2O2.
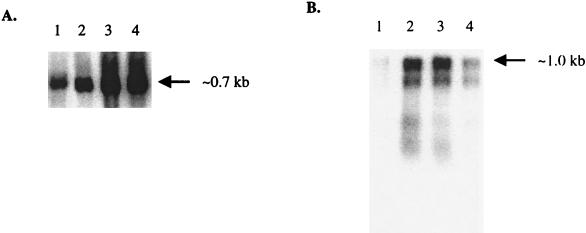
To confirm that the expression of the vapA and vapG genes was regulated by H2O2 and to determine transcript sizes, transcriptional analysis by Northern blot hybridization was performed. Using a vapA-specific probe, a single transcript with a size of approximately 700 nt (Fig. 4A), which corresponds to the expected size for vapA mRNA (5), was obtained. Figure 4A shows that the signal increased by increasing H2O2 concentrations. With 25 and 50 mM H2O2, the induction factors (i.e., the ratio between the amount of transcripts in H2O2-treated and nontreated cells) were 4.5 and 4.2, respectively. This induction was confirmed on the protein level using a VapA-specific antibody. Figure 5 shows an increase in the amount of VapA after 15- and 30-min exposure to 25 mM H2O2 (lanes 2 and 3, respectively), compared with that in nonexposed cells (lane 1). It is interesting that after 45 min of exposure, VapA was only weakly detectable (lane 4).
FIG. 4.
Expression analysis of the R. equi 85F vap genes. Northern blot analysis of vapA (A) and vapG (B) under oxidative stress conditions. Total RNA was isolated from strain 85F after 10-min incubation without H2O2 (lane 1) and with H2O2 at concentrations of 10, 25, and 50 mM (lanes 2 to 4, respectively). Hybridization was done using vapA- and vapG-specific probes, respectively. Only the relevant parts of autoradiograms are shown.
FIG. 5.
Western blot analysis of VapA. Total proteins were extracted after 15 min without H2O2 (lane 1) or 15, 30, and 45 min in the presence of 25 mM H2O2 (lanes 2 to 4, respectively). In all cases, only the relevant parts of the autoradiograms are shown.
Northern blot hybridization with a vapG-specific probe showed a unique transcript of about 1.0 kb (Fig. 4B). This transcript was approximately twice as long as the vapG ORF (Fig. 3). As described for vapA, the amount of transcripts dramatically increased in the presence of H2O2. In contrast, maximal induction was detected at concentrations ranging from 10 to 25 mM with induction factors of 6.75 and 6.2, respectively (Fig. 4B, lanes 2 and 3, respectively). The induction factor dropped to 2.2 at 50 mM (Fig. 4B, lane 4).
Mapping of the transcriptional start site of vapA, vapG, and vapD.
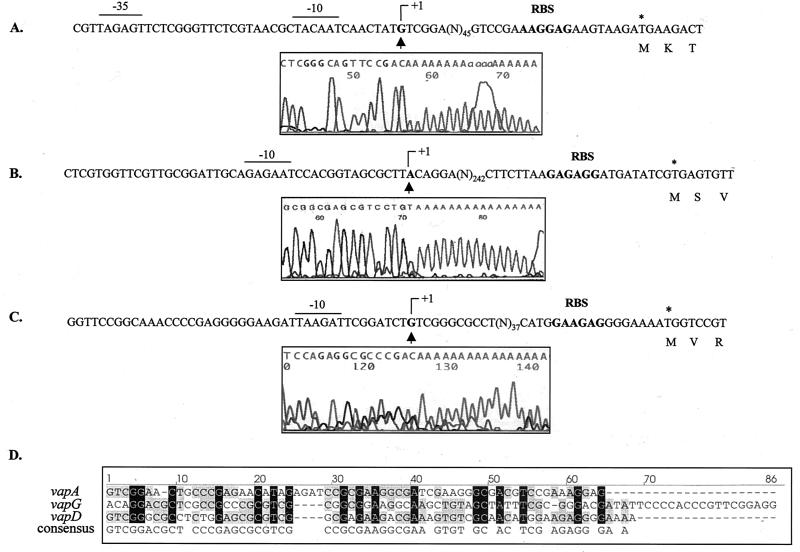
To locate the region involved in the initiation of transcription of vapA and vapG, 5′ RACE PCR experiments were performed with total RNA extracted from cells treated with 25 mM H2O2 for 10 min. For each gene, a unique specific band was observed after electrophoresis of the 5′ RACE PCR product. Surprisingly, the transcriptional start site (TSS) (G) of the vapA mRNA was 69 nt upstream of the start codon (Fig. 6A) and was separated from the putative −10 box (TACAAT) by 7 nt. The calculated size for the mRNA from the transcriptional initiation nucleotide (+1) to the putative Rho-independent terminator was 670 nt, which is in accordance with the Northern blot hybridization result (Fig. 4A). Concerning vapG, the TSS of the vapG mRNA was 269 nt upstream of the start codon (Fig. 6B). Fourteen nucleotides upstream of the TSS (A), we found a putative −10 box (GAGAAT). On the other hand, no sequence homology with a consensus −35 box could be identified (3). The calculated size for the mRNA from the transcriptional initiation nucleotide to the putative Rho-independent terminator was 1,067 nt, which is consistent with the Northern blot hybridization result (Fig. 4B).
FIG. 6.
Sequences of the vapA (A), vapG (B), and vapD (C) promoter regions. The transcriptional initiation nucleotides (+1), the putative −10 and −35 boxes, and the RBS sequences are indicated. Electrophoretograms obtained from 5′ RACE PCR experiments are shown. The sequences were obtained using Rho1c, G8, and Dtr2 primers for vapA, vapG, and vapD, respectively, and cDNA from 5′ dA-tailed RNA using RACE kit (Roche Molecular Biochemicals). The TSS (G, A, and G for vapA, vapG, and vapD, respectively) are indicated above. (D) Alignment of the spacers between the transcriptional initiation nucleotide and the RBS sequence of the vap genes. Gaps introduced to maximize alignment are indicated by the dashes.
We recently showed that like vapA, vapD, another vap gene of the virulence plasmid, is highly induced by acid stress (5). Interestingly, like vapD, the TSS is separated by a 63-bp spacer from the translational initiation region (TIR) (Fig. 6C). Furthermore, the leader sequence from the vapA transcript shows 42 and 54% identity with vapG and vapD transcripts, and the identity between vapD and vapG is 53% (Fig. 6D).
Analysis of the leader sequence of the vapG transcript.
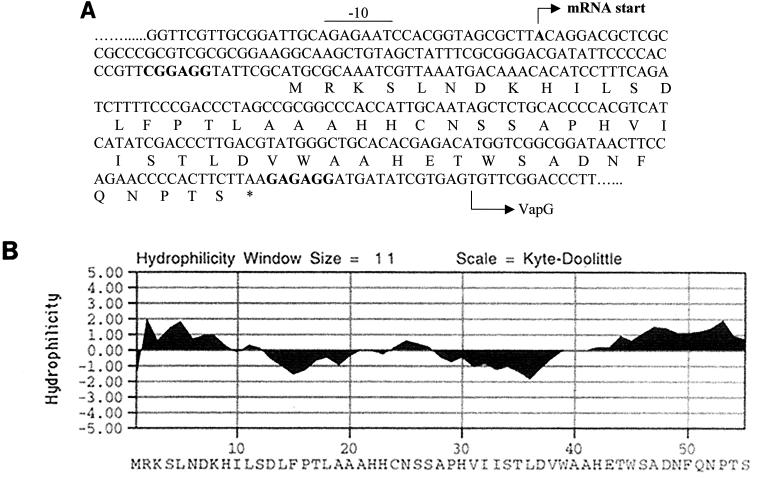
The hypothesis that there is another ORF upstream of vapG could explain the extreme distance observed between the TSS and TIR of this gene. Indeed, analysis of the leader sequence revealed the presence of a small ORF of 165 bp starting with an ATG codon and encoding a hypothetical 55-amino-acid protein (Fig. 7A). In addition, this ORF is preceded by a putative RBS sequence (CGGAGG) and may code for a polypeptide with a calculated molecular mass of 6 kDa and a pI of 6.15. This ORF, named podG, is separated from the TSS by 87 nt, with a genetic organization comparable to that observed for vapA and vapD. No inverted repeats were found downstream of the stop codon TAA, which is separated by 14 bp from the initiation codon of vapG. These results suggest that vapG and podG could form a bicistronic operon. Hydrophobicity analysis showed that PodG is composed of a central hydrophobic region surrounded by hydrophilic parts (Fig. 7B). However, searches of databases revealed no significant homology with other proteins.
FIG. 7.
(A) Spacer region of vapG with the amino acid sequence of the putative ORF (podG). The TSS of the vapG mRNA, determined by 5′ RACE PCR, is indicated by the arrow. Putative RBS sequences for vapG and podG are shown in boldface type. A putative −10 box is indicated. (B) Hydrophilicity plot of PodG obtained using MacVector software. Positive and negative values on the Kyte-Doolittle scale correspond to hydrophilic and hydrophobic domains, respectively.
DISCUSSION
We show in this report that, in comparison to other bacteria, R. equi is extremely resistant to hydrogen peroxide stress. In preliminary experiments, in which cells were treated with concentrations of H2O2 ranging from 10 to 200 mM in GM17 medium, no effects on bacterial survival were observed (data not shown), whereas survival of Enterococcus faecalis, known to be particularly resistant to numerous stresses (12), decreased by 3 and 5 orders of magnitude after 30 or 20 min of exposure to 45 mM H2O2 (13) or 150 mM H2O2 (Fig. 1), respectively. Even Deinococcus radiodurans (a bacterium known to be highly resistant to oxidative stress) is more sensitive to H2O2 treatment. Indeed, its survival dropped by 5 orders of magnitude after a 60-min exposure to 80 mM H2O2 (39). As a control, to determine that the resistance of R. equi was not due to a protective effect of the rich medium, experiments with cells resuspended in physiological water were performed. Under these conditions, R. equi conserved its high resistance to hydrogen peroxide. Indeed, 0.1% of R. equi cells were still cultivable after a 40-min exposure to 150 mM H2O2. In contrast to R. equi, an approximately 15-fold-lower concentration of this chemical oxidant was sufficient to decrease survival of Nocardia asteroides, a pathogenic bacterium described as highly resistant to oxidative metabolites (11). In regards to these results, it was evident that the resistance of R. equi to hydrogen peroxide was exceptional and may be the basis of survival of the bacterium inside macrophages.
It has been shown for many pathogens that H2O2-inducible polypeptides were involved in virulence (1, 22, 40). Relative to other bacteria like E. coli or Salmonella enterica serovar Typhimurium (10), the response to H2O2 in R. equi is characterized by fewer changes in the protein pattern. Indeed, only 13 OSP proteins showed enhanced synthesis, whereas more than 30 polypeptides are induced above the basal level in E. coli and S. enterica serovar Typhimurium (10). Likewise, saprophytic and pathogenic mycobacteria exhibit a limited response to H2O2. In fact, after H2O2 treatment, there were changes in the relative abundance of approximately 12 and only 3 proteins in Mycobacterium smegmatis and Mycobacterium avium, respectively (30). Fewer changes in the protein pattern compared to other bacteria had already been observed in R. equi exposed to acidic pH (5). For these reasons, it appears that R. equi possesses very efficient housekeeping defense mechanisms to resist the antimicrobial attack of macrophages.
Only four OSP proteins were present in the wild-type strain, evidence that they are polypeptides encoded either by the virulence plasmid p85F or by the chromosome and under the control of a positive regulator, such as the transcriptional regulator belonging to the LysR family or the two-component response regulator encoded by the megaplasmid (33). By differential dot blot hybridization, we showed that vapA and vapG were highly induced by H2O2. The products of these genes may correspond to two of the four OSPs present only in the wild-type strain. This supposition is supported by the fact that three of these OSPs mapped in a molecular weight and pI region compatible with the characteristics of VapA and VapG. However, VapA is induced by acid pH (5) and H2O2 stresses, but there is no overlap between OSPs and those induced by acid pH. It is therefore unlikely that VapA is one of the proteins present only in the wild-type strain.
By Western blotting, we showed that maximal expression of VapA appeared after 15 min in 25 mM H2O2 but was at the limit of detection after 45 min. VapC, VapD, and VapE were shown to be secreted into the culture medium, but VapA was undetected outside the cells (6). Thus, the decrease of the protein amount after 45 min of exposure to H2O2 could be explained by its instability rather than its secretion.
It has been suggested that expression of VapA, -C, -D, and -E is under coordinated regulation, since they are all expressed at 37°C but not at 30°C (6, 34). However, our Northern blot analysis revealed that vapA, -D, and -G were only weakly expressed at 37°C and that they were mainly stress induced. More interestingly, the stresses triggering this induction are acid pH and H2O2 treatment, which correspond to situations potentially encountered by R. equi inside macrophages. In contrast, an osmotic shock does not induce expression of the corresponding genes (5; also data not shown). Furthermore, whereas vapG and vapD were induced by oxidative or acid stress, vapA responds to both treatments, showing that even these stress-inducible vap genes seem to be regulated differently. Since expression of the other vap genes does not appear to be induced by the two treatments, one may speculate that within the Vap protein family, VapA, VapD, and VapG play a dominant role inside macrophages.
Due to the fact that VapC, -D, and -E are secreted, whereas VapA is not, it has been suggested that the former Vap proteins could be useful in immunization strategies to prevent rhodococcal pneumonia (6). In contrast to VapC and VapE, expression of VapD may be highly induced inside macrophages (5) and may be the most promising candidate for such vaccination studies. In this respect, VapG is another interesting candidate. Like other Vap proteins with the exception of VapF, a signal peptidase I (A-X-A) site is present in VapG (33), but at present, it is not known whether this protein is secreted.
The mapping of the transcriptional start sites of vapA, -D, and -G (if we consider the presence of podG) showed that they were separated by an unusually long sequence from the translation initiation region. In the closely related mycobacteria and other actinomycetes, several promoters have been mapped (9, 31), including those of virulence genes such as mtrA, an essential response regulator in M. tuberculosis induced upon entry into macrophages (42). In the majority of mapped promoters, the TSS was in close proximity to the TIR. At present, only one exception has been reported; the exception is the hspX gene of M. tuberculosis where the TSS was separated by 32 nt from the ATG codon (19). However, the leader sequences present in the vap transcripts were approximately two to three times longer. It will be interesting to determine whether this is a general characteristic or whether it is specific to the stress-induced subgroup of vap genes. If it is specific to the stress-induced subgroup, the leader sequence could be involved in the upregulation of VapA, -D, and -G by acid pH and/or H2O2. In this context, an attractive hypothesis is the eventual implication of the regulators present in the pathogenicity island of the megaplasmid in this regulation.
Acknowledgments
We thank A. Rincé, J.-C. Giard, J.-M. Laplace, and V. Pichereau for helpful discussions and M. Dabrowski-Coton for proofreading the manuscript.
This work was supported by the Conseil Régional de Basse-Normandie (France) and the AFSSA (France).
Editor: V. J. DiRita
REFERENCES
- 1.Abshire, K. Z., and F. C. Neidhardt. 1993. Analysis of proteins synthesized by Salmonella typhimurium during growth within a host macrophage. J. Bacteriol. 175:3734-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Andersen, S. J., S. Quan, B. Gowan, and E. R. Dabbs. 1997. Monooxygenase-like sequence of a Rhodococcus equi gene conferring increased resistance to rifampin by inactivating this antibiotic. Antimicrob. Agents Chemother. 41:218-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benoit, S., S. Taouji, A. Benachour, and A. Hartke. 2000. Resistance of Rhodococcus equi to acid pH. Int. J. Food Microbiol. 55:295-298. [DOI] [PubMed] [Google Scholar]
- 5.Benoit, S., A. Benachour, S. Taouji, Y. Auffray, and A. Hartke. 2001. Induction of vap genes encoded by the virulence plasmid of Rhodococcus equi during acid tolerance response. Res. Microbiol. 152:439-449. [DOI] [PubMed]
- 6.Byrne, B. A., J. F. Prescott, G. H. Palmer, S. Takai, V. M. Nicholson, D. C. Alperin, and S. A. Hines. 2001. Virulence plasmid of Rhodococcus equi contains inducible gene family encoding secreted proteins. Infect. Immun. 69:650-656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuang, S. E., D. L. Daniels, and F. R. Blattner. 1993. Global regulation of gene expression in Escherichia coli. J. Bacteriol. 175:2026-2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Donisi, A., M. G. Suardi, S. Casari, M. Longo, G. P. Cadeo, and G. Carosi. 1996. Rhodococcus equi infection in HIV-infected patients. AIDS 10:359-362. [DOI] [PubMed] [Google Scholar]
- 9.Dussurget, O., J. Timm, M. Gomez, B. Gold, S. Yu, S. Z. Sabol, R. K. Holmes, W. R. Jacobs, and I. Smith. 1999. Transcriptional control of the iron-responsive fxbA gene by the mycobacterial regulator IdeR. J. Bacteriol. 181:3402-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Filice, G. A. 1983. Resistance of Nocardia asteroides to oxygen-dependent killing by neutrophils. J. Infect. Dis. 148:861-867. [DOI] [PubMed] [Google Scholar]
- 12.Flahaut, S., P. Boutibonnes, and Y. Auffray. 1997. Les entérocoques dans l'environnement proche de l'homme. Can. J. Microbiol. 43:699-708. [PubMed] [Google Scholar]
- 13.Flahaut, S., J. M. Laplace, J. Frère, and Y. Auffray. 1998. The oxidative stress response in Enterococcus faecalis: relationship between H2O2 tolerance and H2O2 stress proteins. Lett. Appl. Microbiol. 26:259-264. [DOI] [PubMed] [Google Scholar]
- 14.Giguere, S., and J. F. Prescott. 1997. Clinical manifestations, diagnosis, treatment, and prevention of Rhodococcus equi infections in foals. Vet. Microbiol. 56:313-334. [DOI] [PubMed] [Google Scholar]
- 15.Giguere, S., M. K. Hondalus, J. A. Yager, P. Darrah, D. M. Mosser, and J. F. Prescott. 1999. Role of the 85-kilobase plasmid and plasmid-encoded virulence-associated protein A in intracellular survival and virulence of Rhodococcus equi. Infect. Immun. 67:3548-3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Handfield, M., and R. C. Levesque. 1999. Strategies for isolation of in vivo expressed genes from bacteria. FEMS Microbiol. Rev. 23:69-91. [DOI] [PubMed] [Google Scholar]
- 17.Hondalus, M. K., and D. M. Mosser. 1994. Survival and replication of Rhodococcus equi in macrophages. Infect. Immun. 62:4167-4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hondalus, M. K. 1997. Pathogenesis and virulence of Rhodococcus equi. Vet. Microbiol. 56:257-268. [DOI] [PubMed] [Google Scholar]
- 19.Hu, Y., and A. R. M. Coates. 1999. Transcription of the stationary-phase-associated hspX gene of Mycobacterium tuberculosis is inversely related to synthesis of the 16-kilodalton protein. J. Bacteriol. 181:1380-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacob, A. E., and S. J. Hobbs. 1974. Conjugal transfer of plasmid-borne multiple antibiotic resistance in Streptococcus faecalis var. zymogenes. J. Bacteriol. 117:360-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Lee, B. Y., and M. A. Horwitz. 1995. Identification of macrophage and stress-induced proteins of Mycobacterium tuberculosis. J. Clin. Investig. 96:245-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez, M. F., W. F. Patton, B. L. Utterback, N. Chung-Welch, P. Barry, W. M. Skea, and R. P. Cambria. 1991. Effect of various detergents on protein migration in the second dimension of two-dimensional gels. Anal. Biochem. 199:109-113. [DOI] [PubMed] [Google Scholar]
- 24.Martens, R. J., R. A. Fiske, and H. W. Renshaw. 1982. Experimental subacute foal pneumonia induced by aerosol administration of Corynebacterium equi. Equine Vet. J. 14:111-116. [DOI] [PubMed] [Google Scholar]
- 25.Nathan, C. F. 1983. Mechanisms of macrophage antimicrobial activity. Trans. R. Soc. Trop. Med. Hyg. 77:620-630. [DOI] [PubMed] [Google Scholar]
- 26.O'Farrell, P. H. 1975. High resolution two-dimensional electrophoresis of proteins. J. Biol. Chem. 250:4007-4021. [PMC free article] [PubMed] [Google Scholar]
- 27.Prescott, J. F. 1991. Rhodococcus equi: an animal and human pathogen. Clin. Microbiol. Rev. 4:20-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rafie-Kolpin, M., R. C. Essenberg, and J. H. Wyckoff III. 1996. Identification and comparison of macrophage-induced proteins and proteins induced under various stress conditions in Brucella abortus. Infect. Immun. 64:5274-5283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Sherman, D. R., P. J. Sabo, M. J. Hickey, T. M. Arain, G. G. Mahairas, Y. Yuan, C. E. Barry III, and C. K. Stover. 1995. Disparate responses to oxidative stress in saprophytic and pathogenic mycobacteria. Proc. Natl. Acad. Sci. USA 92:6625-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strohl, W. R. 1992. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 20:961-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takai, S., Y. Watanabe, T. Ikeda, T. Ozawa, S. Matsukura, Y. Tamada, S. Tsubaki, and T. Sekizaki. 1993. Virulence-associated plasmids in Rhodococcus equi. J. Clin. Microbiol. 31:1726-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takai, S., S. A. Hines, T. Sekizaki, V. M. Nicholson, D. A. Alperin, M. Osaki, D. Takamatsu, M. Nakamura, K. Suzuki, N. Ogino, T. Kakuda, H. Dan, and J. F. Prescott. 2000. DNA sequence and comparison of virulence plasmids from Rhodococcus equi ATCC 33701 and 103. Infect. Immun. 68:6840-6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai, S., N. Fukanega, K. Kamisawa, Y. Imai, Y. Sasaki, and S. Tsubaki. 1996. Expression of virulence-associated antigens of Rhodococcus equi is regulated by temperature and pH. Microbiol. Immunol. 40:591-594. [DOI] [PubMed] [Google Scholar]
- 35.Tan, C., J. F. Prescott, M. C. Patterson, and V. M. Nicholson. 1995. Molecular characterization of a lipid-modified virulence-associated protein of Rhodococcus equi and its potential in protective immunity. Can. J. Vet. Res. 59:51-59. [PMC free article] [PubMed] [Google Scholar]
- 36.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tinoco, I., P. N. Borer, and B. Dengler. 1973. Improved estimation of secondary structure in ribonucleic acids. Nature New Biol. 246:40-41. [DOI] [PubMed] [Google Scholar]
- 38.Vellanoweth, R. L., and J. C. Rabinowith. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escher-ichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]
- 39.Wang, P., and H. E. Schellhorn. 1995. Induction of resistance to hydrogen peroxide and radiation in Deinococcus radiodurans. Can. J. Microbiol. 41:170-176. [DOI] [PubMed] [Google Scholar]
- 40.Yamamoto, T., T. Hanawa, S. Ogata, and S. Kamiya. 1996. Identification and characterization of the Yersinia enterocolitica gsrA gene, which protectively responds to intracellular stress induced by macrophage phagocytosis and to extracellular environmental stress. Infect. Immun. 64:2980-2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Young, D. B., A. Mehlert, V. Bal, P. Mendez-Samperio, J. Ivanyi, and J. R. Lamb. 1988. Stress proteins and the immune response to mycobacteria-antigens as virulence factors? Antonie Leeuwenhoek 54:431-439. [DOI] [PubMed] [Google Scholar]
- 42.Zahrt, T. C., and V. Deretic. 2000. An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182:3832-3838. [DOI] [PMC free article] [PubMed] [Google Scholar]