Abstract
Spirochetes, including Treponema denticola, are implicated in the pathogenesis of periodontal disease. Because T. denticola lacks lipopolysaccharides that serve as targets for human β-defensin (hβD) binding, we postulated that T. denticola would resist killing by hβD. We showed that T. denticola is resistant to hβD-1 and -2. Protease inhibitors did not enhance killing of T. denticola by hβD-2, suggesting that degradation of hβD-2 by treponemal proteases is not a major factor in T. denticola resistance.
Periodontal disease is a destructive inflammatory condition resulting in tooth loss; severe disease requires clinical intervention and affects 6 million Americans (17, 31). Periodontal disease is linked to cardiovascular disease, as well as delivery of preterm, low-birth-weight infants, increasing its recognition as a public health concern (19, 22, 24). Oral spirochetes, including Treponema denticola, are implicated in the pathogenesis of periodontal disease (2, 6, 15), and spirochetes comprise 40% of the microflora found in diseased sites (16).
Microorganisms induce a variety of responses from epithelial cells, including the expression of antimicrobial peptides, such as β-defensins. These small, cationic peptides interact with negatively charged cell wall components of bacteria and fungi, disrupting membrane integrity (9). Human β-defensin 1 (hβD-1) is expressed constitutively by gingival epithelial cells, while hβD-2 expression is induced in response to periodontal microorganisms such as Fusobacterium nucleatum (12, 13). Some antimicrobial peptides have significant in vitro bactericidal activity against periodontal pathogens, including Porphyromonas gingivalis, Actinobacillus actinomycetemcomitans, and Eikenella corrodens (20, 21); the role of antimicrobial peptides in controlling periodontal pathogens in vivo is unknown.
Defensins are known to interact strongly with lipopolysaccharide (LPS) due to its negative charge. T. denticola lacks a traditional LPS, as do many other spirochetes (25). Further, T. denticola is resistant to polymyxin B, a cationic peptide antibiotic that interacts with LPS (1), at more than 1,000 μg/ml. These facts prompted us to investigate whether T. denticola is resistant to human β-defensins.
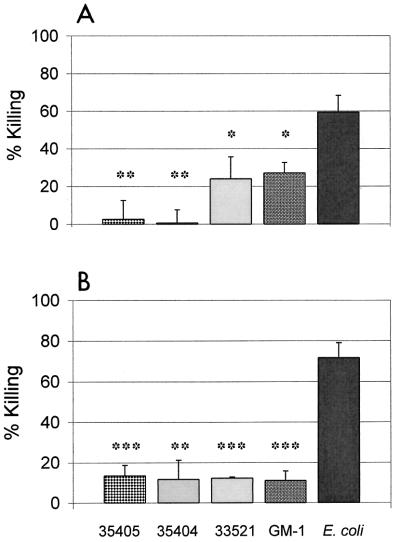
T. denticola strains ATCC 35405, ATCC 35404, ATCC 33521, and GM-1 (30) were obtained from Pamela Braham (University of Washington) and maintained as previously described (4). Escherichia coli strain DH5α was obtained from Patricia Totten (University of Washington) and maintained in Luria-Bertani (LB) medium at 37°C. Four-day, late-logarithmic-phase cultures of T. denticola and overnight cultures of E. coli were centrifuged at 10,000 × g for 10 min at 20°C. Bacteria were washed once and resuspended in phosphate-buffered saline to yield a final concentration of 107 organisms/ml. Bacteria with or without hβD-1 or -2 (Peptides International, Louisville, Ky., and United Biochemical Research, Inc., Seattle, Wash.) were added to duplicate wells of a 96-well polypropylene plate (Costar) at physiologically relevant concentrations ranging from 0.01 to 100 μg/ml and incubated at 37°C for 4 h (18, 26). Motile treponemes were enumerated by dark-field microscopy. E. coli cells were diluted in distilled H2O and plated on LB agar overnight to enumerate CFU. The percentage of killing was calculated as follows: 100 − [(number of viable treated organisms/number of viable untreated organisms) × 100]. All T. denticola strains tested were significantly less susceptible to killing by either defensin than was E. coli (Fig. 1). There was no statistical difference in the number of live treponemes after incubation with or without peptide (P > 0.05, Student's two-tailed t test assuming unequal variances). Similar results were seen with 3-day, mid-logarithmic-phase T. denticola ATCC 35404, and no increase in killing of T. denticola was seen even after 24 h of exposure to 100 μg of hβD per ml (data not shown). The different susceptibilities of various strains of T. denticola to hβD-1 may influence their relative abilities to colonize the oral cavity (13). No strain-to-strain differences in susceptibility were seen with hβD-2. These data demonstrate that, unlike E. coli, T. denticola is able to resist killing by human β-defensins.
FIG. 1.
Sensitivity of T. denticola and E. coli to human β-defensins. T. denticola strains representing three serotypes (ATCC 35405, ATCC 33521, and ATCC 35404), as well as one clinical isolate (GM-1), remain motile in the presence of 10 μg of hβD-1 (A) and hβD-2 (B) per ml. ∗, P < 0.05, ∗∗, P < 0.01, and ∗∗∗, P < 0.001, compared to killing of E. coli. Percent killing was calculated as described in the text. Data are means ± SEM from multiple 4-h experiments. P values were determined by Student's t test for two samples, assuming unequal variances.
Because loss of spirochetal motility does not always correlate with loss of viability, we tested metabolic activity of defensin-exposed T. denticola using Alamar Blue, a dye that is reduced in the presence of actively metabolizing organisms (28). This method has been used successfully to monitor antimicrobial activity against Mycobacterium tuberculosis (8). As there was little difference in hβD-2 sensitivity among strains, T. denticola ATCC 35404 was used for all experiments. Organisms (final concentration, 107/ml) were incubated with 10 μg of hβD-2 per ml or 0.1% sodium dodecyl sulfate for 4 h at 37°C and 5% CO2. A 1/10 volume of Alamar Blue (Biosource, Camarillo, Calif.) was then added, and the bacteria were incubated for an additional 20 h. E. coli was used as a control for hβD-2 activity. Optical density for each well was read on a Dynatech colorimetric plate reader at 570 and 600 nm. Percent reduction was calculated as described previously (8). Results from four experiments confirm that T. denticola ATCC 35404 remains metabolically active after incubation with hβD-2. In a representative experiment, incubation in medium alone showed 72.06% ± 10.01% (mean ± standard error of the mean [SEM]) reduction, while incubation with 10 μg of hβD-2 per ml resulted in 71.57% ± 9.88% reduction. Dead bacteria yielded 12.91% ± 1.49% reduction. These results confirm that under these assay conditions, T. denticola remains metabolically active after treatment with hβD-2.
The precise mechanism of killing by β-defensins is unknown. Research with other cationic peptides suggests that secondary targets, such as cytochromes or components of DNA synthesis, may be necessary for killing (14, 29). To determine the ability of T. denticola to replicate following exposure to hβD-2, T. denticola ATCC 35404 cultures at 107 organisms/ml were incubated with and without 10 μg of hβD-2 per ml in GM-1 medium at 37°C and 5% CO2 for 4 h. Cultures were diluted serially in semisolid GM-1 medium (with 0.5% Noble agar and 0.5% gelatin) and incubated anaerobically at 37°C for 1 to 2 weeks (5). E. coli incubated in GM-1 medium was used as a control for hβD-2 activity and was quantitated by serial dilution on LB agar. Two independent experiments demonstrate that T. denticola was not killed by hβD-2 under these assay conditions: untreated treponemes produced 7.2 × 106 ± 4.9 × 106 CFU/ml, while treponemes incubated in the presence of peptides produced 1.8 × 107 ± 1.4 × 107 CFU/ml (P = 0.55, Student's two-tailed t test assuming unequal variances). Further, colony size for the hβD-2-treated treponemes was equivalent to that for untreated treponemes, suggesting that there was no sublethal effect on growth. E. coli incubated with hβD-2 in this medium was killed as readily as in phosphate-buffered saline, suggesting that the GM-1 medium itself did not inhibit the activity of hβD-2 (data not shown).
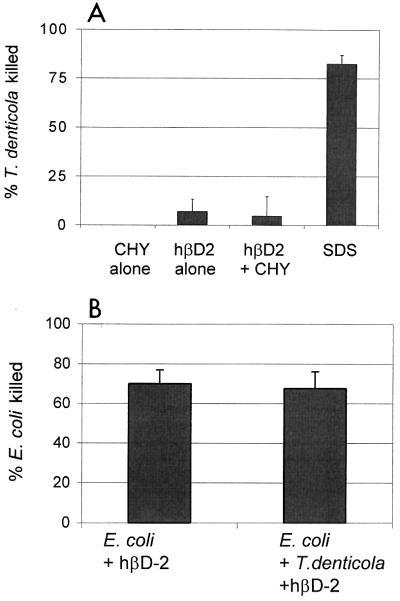
One reason for T. denticola's resistance to β-defensins may be that the peptides are degraded or inactivated. The proteases of T. denticola are known to degrade several host proteins and bioactive peptides (3, 7, 10, 11, 23, 27). To test the hypothesis that T. denticola can degrade hβD-2, thus preventing its activity, we examined the effect of protease inhibitors on survival of T. denticola exposed to hβD-2. T. denticola ATCC 35404 and 10 μg of hβD-2 per ml were incubated in the presence or absence of final concentrations of 100 μM chymostatin (Sigma Chemicals, St. Louis, Mo.) at 37°C and 5% CO2 for 4 h. Viable bacteria were enumerated by dark-field microscopy (T. denticola) or plate counts (E. coli). Killing of T. denticola was not enhanced in the presence of chymostatin, which inhibits the major outer membrane-associated protease of T. denticola, dentilisin (Fig. 2A). Similar results were obtained with other strains of T. denticola and with aprotinin (Sigma) and Complete protease inhibitor cocktail (Roche Molecular Biochemicals, Indianapolis, Ind.) (data not shown). The activity of the protease inhibitors was confirmed by measuring the degradation of recombinant interleukin 8 (R&D Systems, Minneapolis, Minn.) by T. denticola in the presence or absence of the protease inhibitors; chymostatin effectively inhibited >90% of interleukin 8 degradation by T. denticola (data not shown). These results suggest that resistance of T. denticola to hβD-2 is not due to proteolytic destruction of defensin peptides. To examine the possibility that hβD-2 is inactivated by T. denticola by an independent method, coculture killing assays were performed. If T. denticola inactivates hβD-2 by proteolytic degradation, then the presence of T. denticola should protect E. coli from killing by hβD-2 in these experiments. Killing of E. coli by 10 μg of hβD-2 per ml was unaffected by the presence of equal numbers of T. denticola, confirming that T. denticola does not inactivate hβD-2 or decrease its biological activity for sensitive organisms (Fig. 2B). These results suggest that mechanisms other than degradation by protease are responsible for resistance to defensins.
FIG. 2.
T. denticola proteases are not responsible for resistance to hβD-2. (A) T. denticola ATCC 35404 incubated in the presence or absence of 100 μM chymostatin (CHY) remained resistant to 10 μg of hβD-2 per ml over a 4-h period. (B) E. coli was incubated in the presence of 10 μg of hβD-2 per ml for 4 h, with or without equal numbers of T. denticola ATCC 35404. Data are means ± SEM from five (A) and three (B) experiments. SDS, sodium dodecyl sulfate.
In conclusion, we have demonstrated that T. denticola is resistant to killing by human β-defensins, that this resistance exists across strains, and that this resistance is not due to inactivation of the defensins by treponemal proteases. Spirochetes are frequently seen in close association with the epithelium, and resistance to the elevated concentrations of β-defensins at this interface may explain the preponderance of spirochetes observed at those sites in periodontal lesions. The resistance of T. denticola to β-defensins may confer a survival advantage for T. denticola, allowing colonization and persistence in the periodontal pocket.
ADDENDUM IN PROOF
The resistance to human β-defensin 2 by Treponema denticola tested in low-sodium (10 μM) buffer was equal to that seen in phosphate-buffered saline.
Acknowledgments
We thank Richard Darveau and Brian Bainbridge for helpful discussions.
C.A.B. was supported by NRSA Institutional Research training grant T32 DE 07063.
Editor: V. J. DiRita
REFERENCES
- 1.Abramson, I. J., and R. M. Smibert. 1971. Bactericidal activity of antimicrobial agents for treponemes. Br. J. Vener. Dis. 47:413-418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armitage, G. C., W. R. Dickinson, R. S. Jenderseck, S. M. Levine, and D. W. Chambers. 1982. Relationship between the percentage of subgingival spirochetes and the severity of periodontal disease. J. Periodontol. 53:550-556. [DOI] [PubMed] [Google Scholar]
- 3.Beausejour, A., N. Deslauriers, and D. Grenier. 1997. Activation of the interleukin-1β precursor by Treponema denticola: a potential role in chronic inflammatory periodontal diseases. Infect. Immun. 65:3199-3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blakemore, R. P., and E. Canale-Parola. 1976. Arginine catabolism by Treponema denticola. J. Bacteriol. 128:616-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, E. C., A. De Ciccio, R. McLaughlin, A. Klitorinos, and R. Siboo. 1997. An inexpensive solid medium for obtaining colony-forming units of oral spirochetes. Oral Microbiol. Immunol. 12:372-376. [DOI] [PubMed] [Google Scholar]
- 6.Darveau, R. P., A. Tanner, and R. C. Page. 2000. The microbial challenge in periodontitis. Periodontology 14:12-32. [DOI] [PubMed] [Google Scholar]
- 7.Deng, Q. D., Y. Han, X. Xia, and H. K. Kuramitsu. 2001. Effects of the oral spirochete Treponema denticola on interleukin-8 expression from epithelial cells. Oral Microbiol. Immunol. 16:185-187. [DOI] [PubMed] [Google Scholar]
- 8.Franzblau, S. G., R. S. Witzig, J. C. McLaughlin, P. Torres, G. Madico, A. Hernandez, M. T. Degnan, M. B. Cook, V. K. Quenzer, R. M. Ferguson, and R. H. Gilman. 1998. Rapid, low-technology MIC determination with clinical Mycobacterium tuberculosis isolates by using the microplate Alamar Blue assay. J. Clin. Microbiol. 36:362-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ganz, T. 2001. Antimicrobial proteins and peptides in host defense. Semin. Respir. Infect. 16:4-10. [DOI] [PubMed] [Google Scholar]
- 10.Grenier, D. 1996. Degradation of host protease inhibitors and activation of plasminogen by proteolytic enzymes from Porphyromonas gingivalis and Treponema denticola. Microbiology 142:955-961. [DOI] [PubMed] [Google Scholar]
- 11.Grenier, D., and D. Mayrand. 2001. Cleavage of human immunoglobulin G by Treponema denticola. Anaerobe 7:1-4. [Google Scholar]
- 12.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lichtenstein, A. K., T. Ganz, T. M. Nguyen, M. E. Selsted, and R. I. Lehrer. 1988. Mechanism of target cytolysis by peptide defensins. Target cell metabolic activities, possibly involving endocytosis, are crucial for expression of cytotoxicity. J. Immunol. 140:2686-2694. [PubMed] [Google Scholar]
- 15.Loesche, W. J. 1988. The role of spirochetes in periodontal disease. Adv. Dent. Res. 2:275-283. [DOI] [PubMed] [Google Scholar]
- 16.Loesche, W. J. 1988. The spirochetes, p. 228-236. In M. G. Newman and R. J. Nisengard (ed.), Oral microbiology and immunology. Saunders, Philadelphia, Pa.
- 17.Loesche, W. J., and N. S. Grossman. 2001. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin. Microbiol. Rev. 14:727-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray, Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meyer, D. H., and P. M. Fives-Taylor. 1998. Oral pathogens: from dental plaque to cardiac disease. Curr. Opin. Microbiol. 1:88-95. [DOI] [PubMed] [Google Scholar]
- 20.Miyasaki, K. T., A. L. Bodeau, M. E. Selsted, T. Ganz, and R. I. Lehrer. 1990. Killing of oral, gram-negative, facultative bacteria by the rabbit defensin, NP-1. Oral Microbiol. Immunol. 5:315-319. [DOI] [PubMed] [Google Scholar]
- 21.Miyasaki, K. T., R. Iofel, A. Oren, T. Huynh, and R. I. Lehrer. 1998. Killing of Fusobacterium nucleatum, Porphyromonas gingivalis and Prevotella intermedia by protegrins. J. Periodontal Res. 33:91-98. [DOI] [PubMed] [Google Scholar]
- 22.Offenbacher, S., H. L. Jared, P. G. O'Reilly, S. R. Wells, G. E. Salvi, H. P. Lawrence, S. S. Socransky, and J. D. Beck. 1998. Potential pathogenic mechanisms of periodontitis associated pregnancy complications. Ann. Periodontol. 3:233-250. [DOI] [PubMed] [Google Scholar]
- 23.Pederson, E. D., B. L. Lamberts, and I. L. Shklair. 1988. Susceptibility of fibronectin to degradation by various gram-negative and gram-positive oral micro-organisms. Microbios 53:83-90. [PubMed] [Google Scholar]
- 24.Satcher, D. S. 2000. Linkages with general health, p. 95-127. In Oral health in America: a report of the surgeon general. U.S. Department of Health and Human Services, National Institute of Dental and Craniofacial Research, National Institutes of Health, Rockville, Md.
- 25.Schultz, C. P., V. Wolf, R. Lange, E. Mertens, J. Wecke, D. Naumann, and U. Zahringer. 1998. Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola. Functioning permeation barrier without lipopolysaccharides. J. Biol. Chem. 273:15661-15666. [DOI] [PubMed] [Google Scholar]
- 26.Shi, J., G. Zhang, H. Wu, C. Ross, F. Blecha, and T. Ganz. 1999. Porcine epithelial beta-defensin 1 is expressed in the dorsal tongue at antimicrobial concentrations. Infect. Immun. 67:3121-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorsa, T., T. Ingman, K. Suomalainen, M. Haapasalo, Y. T. Konttinen, O. Lindy, H. Saari, and V. J. Uitto. 1992. Identification of proteases from periodontopathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect. Immun. 60:4491-4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stapleton, J. T., L. V. Stamm, and P. J. Bassford, Jr. 1985. Potential for development of antibiotic resistance in pathogenic treponemes. Rev. Infect. Dis. 7(Suppl. 2):S314-S317. [DOI] [PubMed] [Google Scholar]
- 29.Wachinger, M., A. Kleinschmidt, D. Winder, N. von Pechmann, A. Ludvigsen, M. Neumann, R. Holle, B. Salmons, V. Erfle, and R. Brack-Werner. 1998. Antimicrobial peptides melittin and cecropin inhibit replication of human immunodeficiency virus 1 by suppressing viral gene expression. J. Gen. Virol. 79:731-740. [DOI] [PubMed] [Google Scholar]
- 30.Weinberg, A., and S. C. Holt. 1990. Interaction of Treponema denticola TD-4, GM-1, and MS25 with human gingival fibroblasts. Infect. Immun. 58:1720-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zambon, J. J. 1990. Microbiology of periodontal disease, p. 147-160. In R. J. Genco, H. M. Goldman, and D. W. Cohen (ed.), Contemporary periodontics. Mosby, St. Louis, Mo.