Abstract
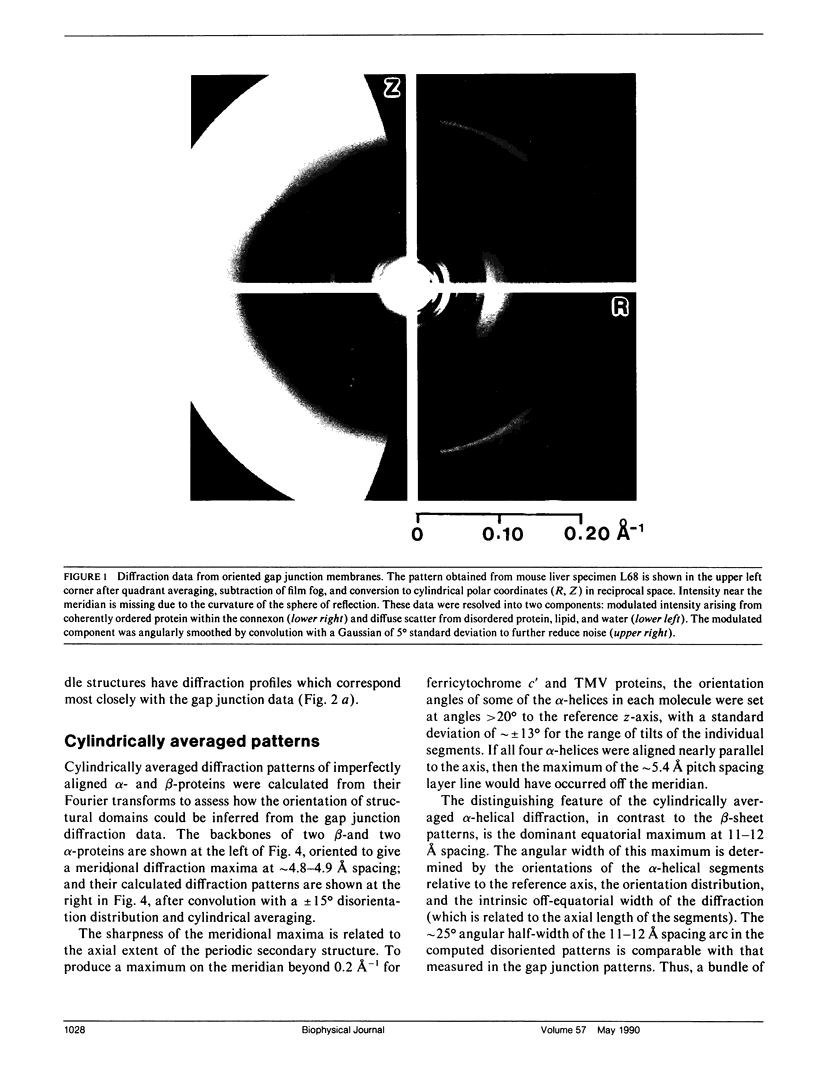
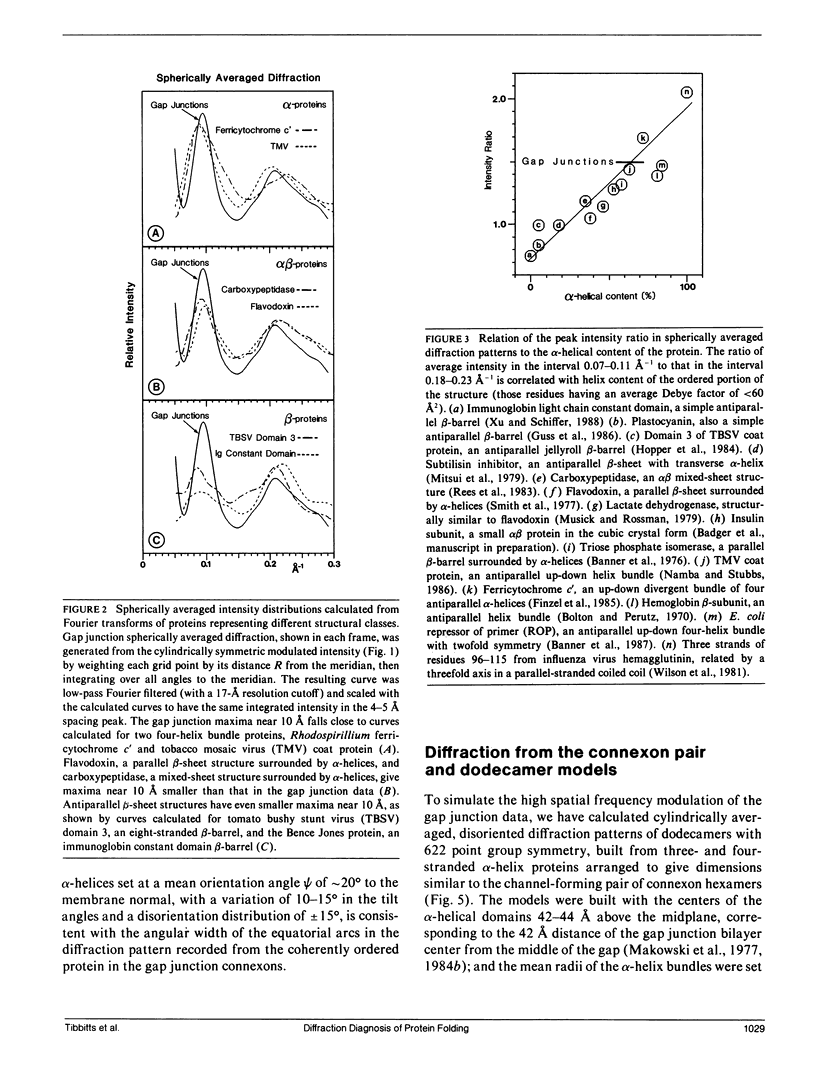
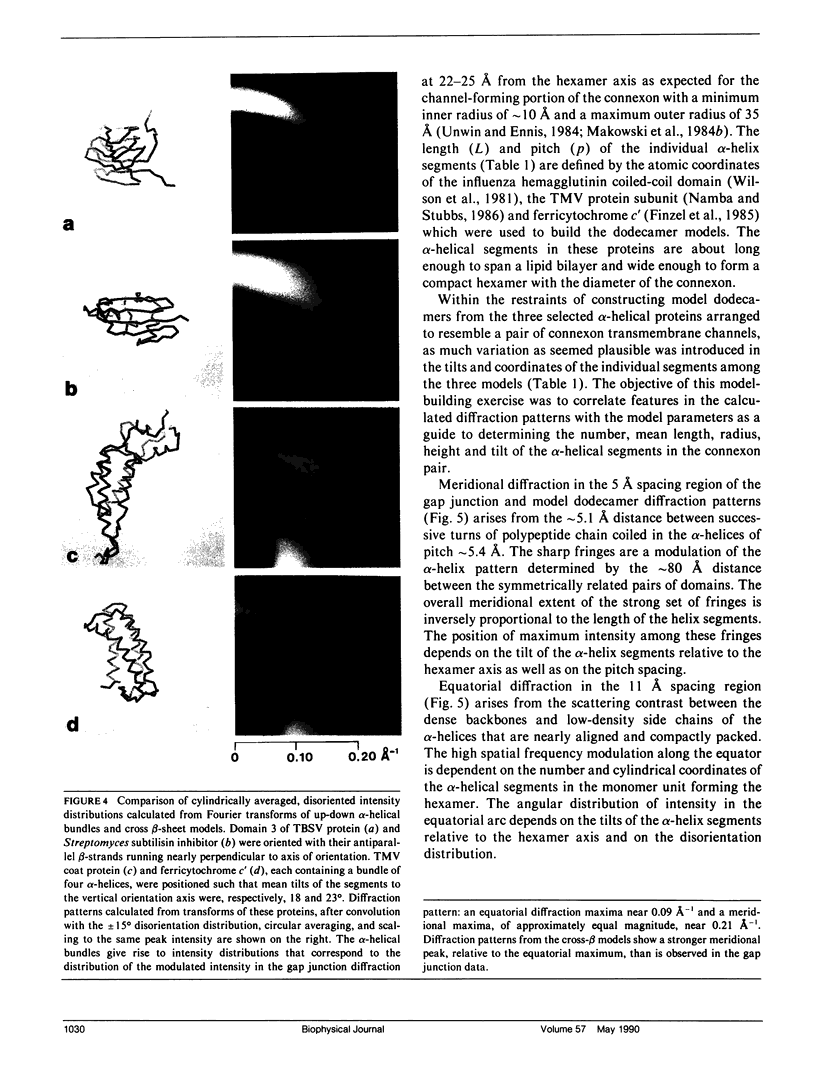
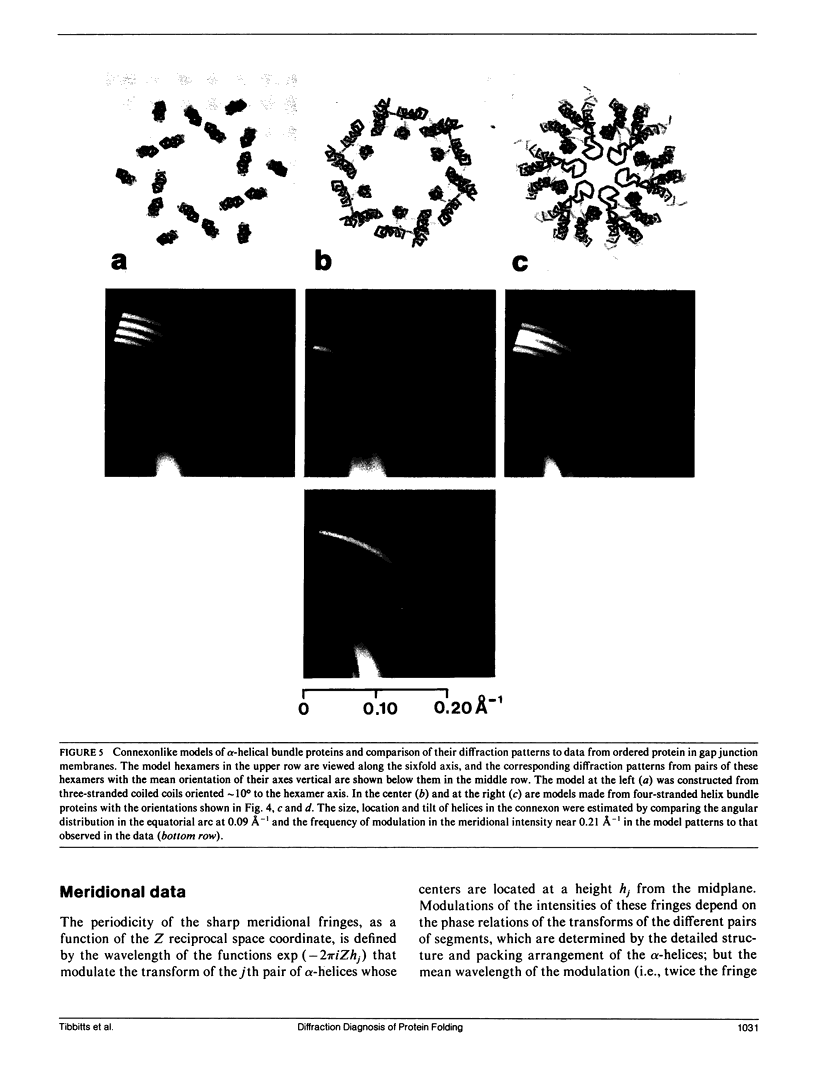
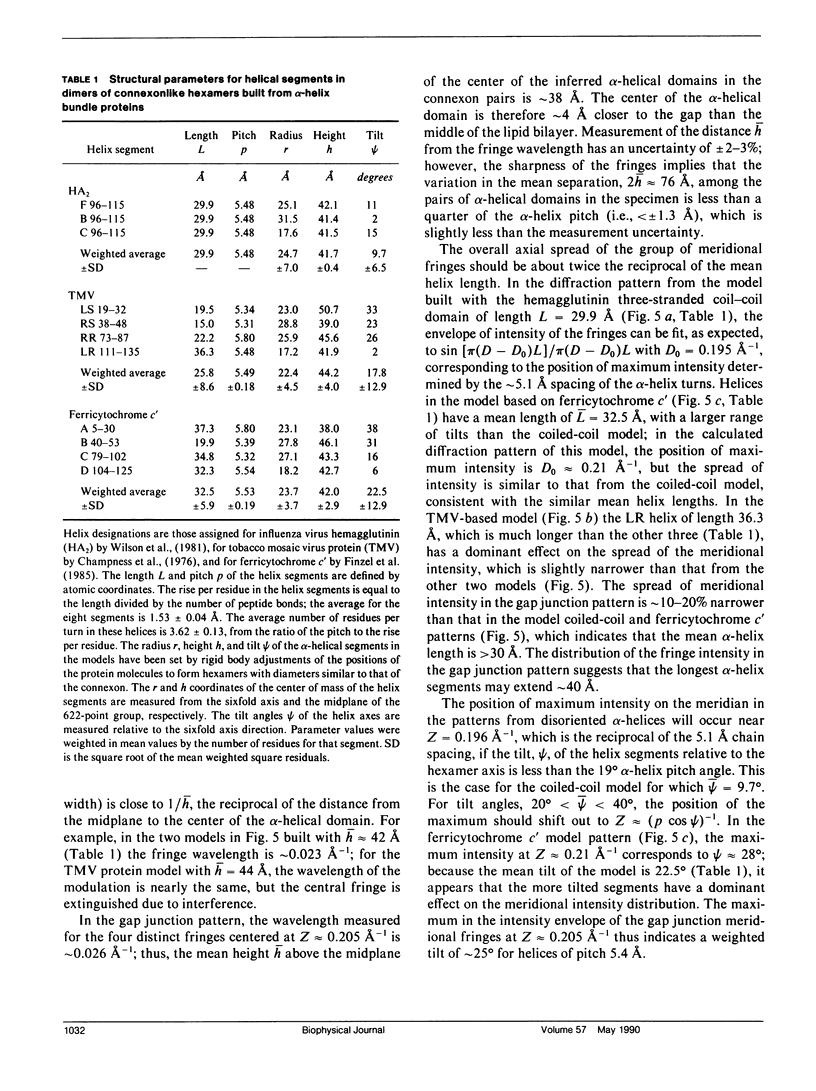
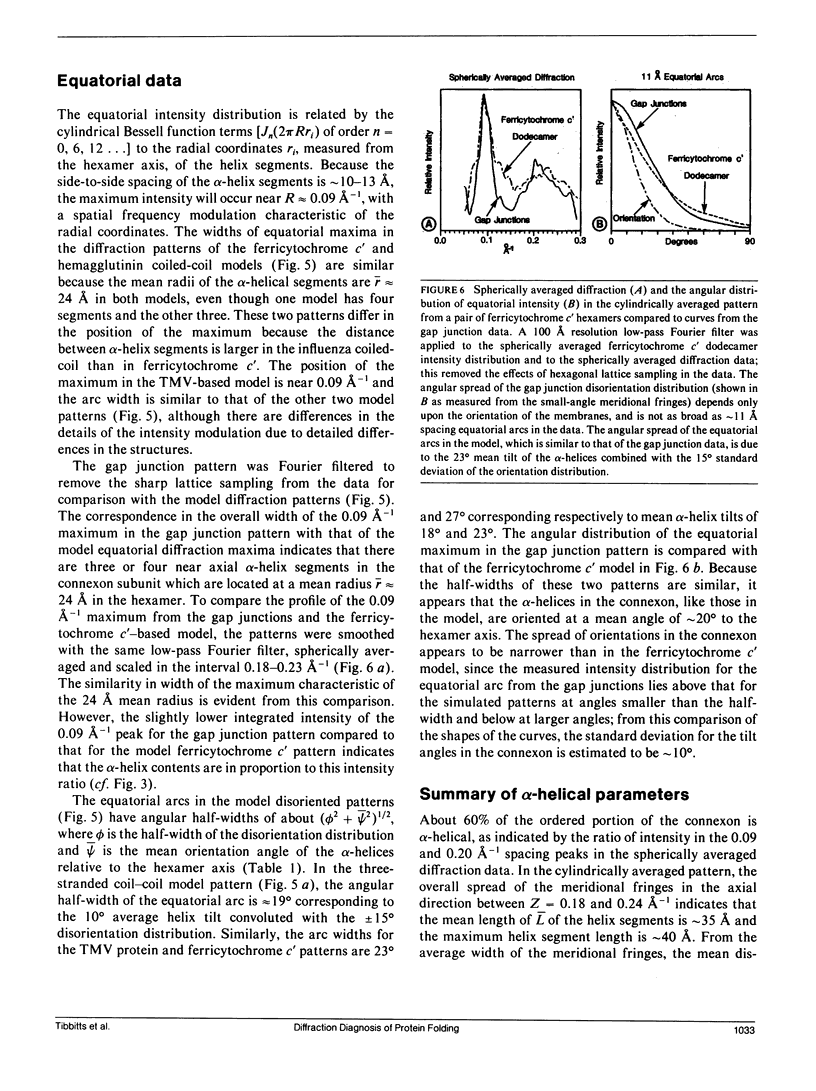
To diagnose the regular polypeptide conformation in gap junction membranes, the x-ray intensities diffracted from oriented specimens have been separated into a modulated component due to the coherently ordered portion of the channel-forming pairs of connexon hexamers and a diffuse component due to the disordered parts. The spherically averaged ordered protein diffraction, in the resolution range 15-4 A, was compared with intensity curves calculated from the Fourier transforms of proteins representative of the major tertiary structural classes. From this comparison the alpha-helical content of the ordered portion of the connexon was estimated to be approximately 60%. Calculation of cylindrically averaged patterns for oriented distributions of alpha-helical and beta-sheet proteins demonstrated that the ratio of the modulated diffracted intensity near 5 A spacing on the meridian and 10 A spacing on the equator observed from the gap junctions can be accounted for by alpha-helical segments inclined relative to the connexon axis. Model dimers of connexonlike hexamers were constructed from alpha-helix bundle proteins to correlate features in the calculated diffraction patterns with the model parameters. On the basis of these correlations, the ordered gap junction diffraction data indicate that alpha-helical segments centered at 38 A from the midplane of the gap have a mean radial location approximately 24 A from the hexamer axis, and an axial projected length of approximately 35 A. Thus, these alpha-helical segments traverse the hydrocarbon core of the lipid bilayer, as expected for the four hydrophobic sequences of the connexin molecule.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banner D. W., Bloomer A. c., Petsko G. A., Phillips D. C., Wilson I. A. Atomic coordinates for triose phosphate isomerase from chicken muscle. Biochem Biophys Res Commun. 1976 Sep 7;72(1):146–155. doi: 10.1016/0006-291x(76)90972-4. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Beyer E. C., Paul D. L., Goodenough D. A. Connexin43: a protein from rat heart homologous to a gap junction protein from liver. J Cell Biol. 1987 Dec;105(6 Pt 1):2621–2629. doi: 10.1083/jcb.105.6.2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton W., Perutz M. F. Three dimensional fourier synthesis of horse deoxyhaemoglobin at 2.8 Angstrom units resolution. Nature. 1970 Nov 7;228(5271):551–552. doi: 10.1038/228551a0. [DOI] [PubMed] [Google Scholar]
- Caspar D. L., Goodenough D. A., Makowski L., Phillips W. C. Gap junction structures. I. Correlated electron microscopy and x-ray diffraction. J Cell Biol. 1977 Aug;74(2):605–628. doi: 10.1083/jcb.74.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champness J. N., Bloomer A. C., Bricogne G., Butler P. G., Klug A. The structure of the protein disk of tobacco mosaic virus to 5A resolution. Nature. 1976 Jan 1;259(5538):20–24. doi: 10.1038/259020a0. [DOI] [PubMed] [Google Scholar]
- Chothia C. Structural invariants in protein folding. Nature. 1975 Mar 27;254(5498):304–308. doi: 10.1038/254304a0. [DOI] [PubMed] [Google Scholar]
- Fallon R. F., Goodenough D. A. Five-hour half-life of mouse liver gap-junction protein. J Cell Biol. 1981 Aug;90(2):521–526. doi: 10.1083/jcb.90.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzel B. C., Weber P. C., Hardman K. D., Salemme F. R. Structure of ferricytochrome c' from Rhodospirillum molischianum at 1.67 A resolution. J Mol Biol. 1985 Dec 5;186(3):627–643. doi: 10.1016/0022-2836(85)90135-4. [DOI] [PubMed] [Google Scholar]
- Gimlich R. L., Kumar N. M., Gilula N. B. Sequence and developmental expression of mRNA coding for a gap junction protein in Xenopus. J Cell Biol. 1988 Sep;107(3):1065–1073. doi: 10.1083/jcb.107.3.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough D. A., Paul D. L., Jesaitis L. Topological distribution of two connexin32 antigenic sites in intact and split rodent hepatocyte gap junctions. J Cell Biol. 1988 Nov;107(5):1817–1824. doi: 10.1083/jcb.107.5.1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guss J. M., Harrowell P. R., Murata M., Norris V. A., Freeman H. C. Crystal structure analyses of reduced (CuI) poplar plastocyanin at six pH values. J Mol Biol. 1986 Nov 20;192(2):361–387. doi: 10.1016/0022-2836(86)90371-2. [DOI] [PubMed] [Google Scholar]
- Henderson R. The structure of the purple membrane from Halobacterium hallobium: analysis of the X-ray diffraction pattern. J Mol Biol. 1975 Apr 5;93(2):123–138. doi: 10.1016/0022-2836(75)90123-0. [DOI] [PubMed] [Google Scholar]
- Henderson R., Unwin P. N. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975 Sep 4;257(5521):28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- Hopper P., Harrison S. C., Sauer R. T. Structure of tomato bushy stunt virus. V. Coat protein sequence determination and its structural implications. J Mol Biol. 1984 Aug 25;177(4):701–713. doi: 10.1016/0022-2836(84)90045-7. [DOI] [PubMed] [Google Scholar]
- Kumar N. M., Gilula N. B. Cloning and characterization of human and rat liver cDNAs coding for a gap junction protein. J Cell Biol. 1986 Sep;103(3):767–776. doi: 10.1083/jcb.103.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Goodenough D. A., Phillips W. C. Gap Junction Structures: III. The Effect of Variations in the Isolation Procedure. Biophys J. 1982 Jan;37(1):189–191. doi: 10.1016/S0006-3495(82)84663-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Marvin D. A. Filamentous bacteriophage Pf1 structure determined at 7A resolution by refinement of models for the alpha-helical subunit. J Mol Biol. 1980 Jun 25;140(2):149–181. doi: 10.1016/0022-2836(80)90101-1. [DOI] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Baker T. S., Goodenough D. A. Gap junction structures. VI. Variation and conservation in connexon conformation and packing. Biophys J. 1984 Jan;45(1):208–218. doi: 10.1016/S0006-3495(84)84149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Goodenough D. A. Gap junction structures. II. Analysis of the x-ray diffraction data. J Cell Biol. 1977 Aug;74(2):629–645. doi: 10.1083/jcb.74.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Caspar D. L., Phillips W. C., Goodenough D. A. Gap junction structures. V. Structural chemistry inferred from X-ray diffraction measurements on sucrose accessibility and trypsin susceptibility. J Mol Biol. 1984 Apr 15;174(3):449–481. doi: 10.1016/0022-2836(84)90331-0. [DOI] [PubMed] [Google Scholar]
- Marvin D. A. X-ray diffraction and electron microscope studies on the structure of the small filamentous bacteriophage fd. J Mol Biol. 1966 Jan;15(1):8–17. doi: 10.1016/s0022-2836(66)80205-x. [DOI] [PubMed] [Google Scholar]
- Milks L. C., Kumar N. M., Houghten R., Unwin N., Gilula N. B. Topology of the 32-kd liver gap junction protein determined by site-directed antibody localizations. EMBO J. 1988 Oct;7(10):2967–2975. doi: 10.1002/j.1460-2075.1988.tb03159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsui Y., Satow Y., Watanabe Y., Iitaka Y. Crystal structure of a bacterial protein proteinase inhibitor (Streptomyces subtilisin inhibitor) at 2.6 A resolution. J Mol Biol. 1979 Jul 15;131(4):697–724. doi: 10.1016/0022-2836(79)90198-0. [DOI] [PubMed] [Google Scholar]
- Musick W. D., Rossmann M. G. The structure of mouse testicular lactate dehydrogenase isoenzyme C4 at 2.9 A resolution. J Biol Chem. 1979 Aug 25;254(16):7611–7620. doi: 10.2210/pdb1ldx/pdb. [DOI] [PubMed] [Google Scholar]
- Namba K., Stubbs G. Structure of tobacco mosaic virus at 3.6 A resolution: implications for assembly. Science. 1986 Mar 21;231(4744):1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- PAULING L., COREY R. B. Atomic coordinates and structure factors for two helical configurations of polypeptide chains. Proc Natl Acad Sci U S A. 1951 May;37(5):235–240. doi: 10.1073/pnas.37.5.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PAULING L., COREY R. B. The pleated sheet, a new layer configuration of polypeptide chains. Proc Natl Acad Sci U S A. 1951 May;37(5):251–256. doi: 10.1073/pnas.37.5.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D. L. Molecular cloning of cDNA for rat liver gap junction protein. J Cell Biol. 1986 Jul;103(1):123–134. doi: 10.1083/jcb.103.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RILEY D. P., ARNDT U. W. X-ray scattering by some native and denatured proteins in the solid state. Proc R Soc Lond B Biol Sci. 1953 Mar 11;141(902):93–97. doi: 10.1098/rspb.1953.0025. [DOI] [PubMed] [Google Scholar]
- Rees D. C., Lewis M., Lipscomb W. N. Refined crystal structure of carboxypeptidase A at 1.54 A resolution. J Mol Biol. 1983 Aug 5;168(2):367–387. doi: 10.1016/s0022-2836(83)80024-2. [DOI] [PubMed] [Google Scholar]
- Richards F. M. The interpretation of protein structures: total volume, group volume distributions and packing density. J Mol Biol. 1974 Jan 5;82(1):1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- Richardson J. S., Richardson D. C. Interpretation of electron density maps. Methods Enzymol. 1985;115:189–206. doi: 10.1016/0076-6879(85)15016-0. [DOI] [PubMed] [Google Scholar]
- Richardson J. S. The anatomy and taxonomy of protein structure. Adv Protein Chem. 1981;34:167–339. doi: 10.1016/s0065-3233(08)60520-3. [DOI] [PubMed] [Google Scholar]
- Smith W. W., Burnett R. M., Darling G. D., Ludwig M. L. Structure of the semiquinone form of flavodoxin from Clostridum MP. Extension of 1.8 A resolution and some comparisons with the oxidized state. J Mol Biol. 1977 Nov 25;117(1):195–225. doi: 10.1016/0022-2836(77)90031-6. [DOI] [PubMed] [Google Scholar]
- Sosinsky G. E., Jésior J. C., Caspar D. L., Goodenough D. A. Gap junction structures. VIII. Membrane cross-sections. Biophys J. 1988 May;53(5):709–722. doi: 10.1016/S0006-3495(88)83152-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unwin N. Is there a common design for cell membrane channels? Nature. 1986 Sep 4;323(6083):12–13. doi: 10.1038/323012a0. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Ennis P. D. Two configurations of a channel-forming membrane protein. Nature. 1984 Feb 16;307(5952):609–613. doi: 10.1038/307609a0. [DOI] [PubMed] [Google Scholar]
- Unwin P. N., Zampighi G. Structure of the junction between communicating cells. Nature. 1980 Feb 7;283(5747):545–549. doi: 10.1038/283545a0. [DOI] [PubMed] [Google Scholar]
- Wilson I. A., Skehel J. J., Wiley D. C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981 Jan 29;289(5796):366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]