Abstract
Borrelia burgdorferi differentially expresses many of the OspE/F/Elp paralogs during tick feeding. These findings, combined with the recent report that stable B. burgdorferi infection of mammals occurs only after 53 h of tick attachment, prompted us to further analyze the expression of the OspE/F/Elp paralogs during this critical period of transmission. Indirect immunofluorescence analysis revealed that OspE, p21, ElpB1, ElpB2, and OspF/BbK2.11 are expressed in the salivary glands of ticks allowed to feed on mice for 53 to 58 h. Interestingly, many of the spirochetes in the salivary glands that expressed abundant amounts of these antigens were negative for OspA and OspC. Although prior reports have indicated that OspE/F/Elp orthologs are surface exposed, none of the individual lipoproteins or combinations of the lipoproteins protected mice from challenge infections. To examine why these apparently surface-exposed lipoproteins were not protective, we analyzed their genetic stability during infection and their cellular locations after cultivation in vitro and within dialysis membrane chambers, mimicking a mammalian host-adapted state. Combined restriction fragment length polymorphism and nucleotide sequence analyses revealed that the genes encoding these lipoproteins are stable for at least 8 months postinfection. Interestingly, cellular localization experiments revealed that while all of these proteins can be surface localized, there were significant populations of spirochetes that expressed these lipoproteins only in the periplasm. Furthermore, host-specific signals were found to alter the expression patterns and final cellular location of these lipoproteins. The combined data revealed a remarkable heterogeneity in populations of B. burgdorferi during tick transmission and mammalian infection. The diversity is generated not only by temporal changes in antigen expression but also by modulation of the surface lipoproteins during infection. The ability to regulate the temporal and spatial expression patterns of lipoproteins throughout infection likely contributes to persistent infection of mammals by B. burgdorferi.
Lyme disease, the most common arthropod-borne infection in the United States, is a multisystemic infectious disease caused by the spirochete Borrelia burgdorferi (31). If the disease is left untreated, the spirochete can survive in a mammalian host for extended periods of time, which can lead to dermatitis, carditis, arthritis, and neuritis (42). B. burgdorferi is typically transmitted to mammals by hard ticks of the genus Ixodes (24). During tick feeding, B. burgdorferi travels from the tick midgut to the hemolymph and salivary glands before being deposited at the host dermis (4, 14, 37). During this feeding, migration, and transmission process, numerous borrelial lipoproteins have been shown to be differentially upregulated (e.g., OspC, OspE, and OspF) (20, 41, 47) or downregulated (e.g., OspA, OspB, and lp6.6) (1, 11, 22, 28, 40). Additionally, several other lipoproteins have been identified which appear to be expressed exclusively in the mammalian host environment; these lipoproteins include pG, BbK2.10, ElpA1, and p21 (3, 20, 48, 50). Many of the differentially expressed proteins are encoded by members of plasmid-encoded gene families (1, 3, 20, 34, 47, 48, 50). Among these, the OspE paralogs, OspF paralogs, and Elp paralogs, all of which are encoded on circular 32- or 18-kb plasmids (2, 7), have been identified in all borrelial strains studied to date, including the B31 strain, in which they have been termed Erps (OspE/F-related proteins) (8, 9, 17). Recent studies by other workers and by us have revealed that the lipoproteins encoded by members of the three gene families are induced by temperature and/or mammal-specific factors (1, 3, 20, 44, 47). Furthermore, it also has been reported that many of the OspE, OspF, and Elp orthologs are surface exposed in B. burgdorferi (16, 23), suggesting that they play a role in host-pathogen interactions during mammalian infection.
Ohnishi et al. (30) recently reported that OspA and OspC are not expressed by a majority (∼80%) of organisms that are in the salivary glands of ticks that have fed on animals for more than 53 h, a time period that coincides with the time required for stable infection of mammals. While we recently reported that many of the B. burgdorferi strain 297 OspE/F/Elp paralogs are upregulated in tick midguts during feeding (20), we did not assess the expression of these lipoproteins in the salivary glands of ticks during the feeding process. To extend our prior studies, we analyzed the expression patterns of the OspE/F/Elp paralogs as spirochetes migrated from the tick midgut to the salivary glands during the transmission process. Here, we present data which show that many of the OspE/F/Elp paralogs are expressed by spirochetes in salivary glands of ticks allowed to feed on mammals for 53 to 58 h, which indicates that these paralogs may play a role in the transmission process. Furthermore, our combined findings indicate that antigenically diverse subsets of spirochetes can be generated during infection by changes in both the temporal (i.e., when a protein is expressed) and spatial (i.e., where a protein is localized) expression of lipoproteins. Both of these regulatory mechanisms likely contribute to the overall parasitic strategy of B. burgdorferi and aid in maintaining persistent infections.
MATERIALS AND METHODS
Bacterial strains.
Virulent B. burgdorferi strain 297, originally isolated from the cerebrospinal fluid of a Lyme disease patient (43), was cultivated in BSK-H medium (Sigma Chemical Co., St. Louis, Mo.) supplemented with 6% rabbit serum (Pel-Freeze Biologicals, Rogers, Ark.). Spirochetes were passaged no more than three times before experimental manipulations were performed. For temperature shift experiments, organisms were cultivated at 23°C to the mid-logarithmic phase (approximately 5 × 106 to 1 × 107 organisms per ml), and 1,000 spirochetes per ml were then transferred to medium prewarmed to 37°C. To obtain B. burgdorferi in a mammalian host-adapted state, organisms were cultivated in dialysis membrane chambers (DMCs) implanted into rat peritoneal cavities as described previously (1). PCR amplification with specific primer pairs was used to ensure that borrelial cultures and infected ticks contained all seven different plasmids which harbor the nine ospE/F/elp paralogs as described previously (20).
To generate isogenic clones of B. burgdorferi, ear punch biopsies were cultured in medium containing rifampin (50 μg/ml) and amphotericin B (25 μg/ml). Recovered spirochetes were then subsurface plated in medium containing 0.7% high-strength, analytical grade agarose (Bio-Rad, Hercules, Calif.). The plates were incubated at 34°C with 5% CO2 in a humidified chamber until colonies were visible. Well-isolated colonies were carefully picked and cultivated in medium for further analyses. Electrocompetent Escherichia coli strain DH5α (Gibco/BRL Life Technologies, Gaithersburg, Md.) was used for transformation experiments; all E. coli clones and transformants were grown using tryptone-yeast agar or broth supplemented with the appropriate antibiotics.
Immunological reagents.
Glutathione S-transferase (GST) fusion constructs, recombinant proteins, and antisera were generated as described previously (1, 3, 12, 20, 22). To eliminate cross-reactivity between the closely related OspE and p21 proteins, 1 ml of each polyclonal rat antiserum was incubated overnight at 4°C with 500 μg of the appropriate cross-reactive GST fusion protein attached to a glutathione-agarose bead matrix (Amersham Pharmacia, Piscataway, N.J.). Cross-reactive antibodies adsorbed to the beads were separated by centrifugation at 4,000 × g for 10 min. The resulting supernatants were tested for specificity as previously described (20). The same procedure also was performed to adsorb out cross-reactivity between the highly homologous OspF and BbK2.11 proteins; however, multiple rounds of the adsorption procedure were not sufficient to completely eliminate all cross-reactivity. Therefore, the antibodies used to analyze the expression patterns of OspF and BbK2.11 are indicated below as OspF/BbK2.11 since the antiserum recognized both proteins. All other antibodies were affinity purified as previously described (20). Monoclonal antibodies directed against FlaB (1H6-33) and OspA (14D2-27) have been described previously (1).
Protection experiments.
Groups of five C3H/HeJ mice were actively immunized as described previously (19) with either (i) all nine of the OspE-related, OspF-related, and Elp lipoproteins, (ii) the OspE-related proteins (i.e., OspE and p21), (iii) the OspF-related proteins (i.e., OspF, BbK2.10, and BbK2.11), (iv) the Elp proteins (i.e., ElpA1, ElpA2, ElpB1, and ElpB2), or (v) each of the nine lipoproteins individually. Serum from each mouse was collected via tail bleeding, and specific titers were determined by an enzyme-linked immunosorbent assay (ELISA) as previously described (20). Antibody titers were determined to be greater than 1:100,000 before protection experiments were performed. Tick infection, rearing, and infestation were performed as described previously (51). Prior to infestation of mice, ticks infected with spirochetes were confirmed to contain all seven plasmids encoding the ospE-related, ospF-related, and elp genes by using the primer pairs and PCR parameters described previously (20). Mice were assessed for infection by cultivating ear punch biopsies taken 2 weeks after infestation in media containing rifampin (50 μg/ml) and amphotericin B (25 μg/ml). All mice were subjected to tail bleeding for subsequent immunoblot and ELISA analyses.
Amplification, restriction fragment length polymorphism (RFLP), and sequence analysis of the ospE-related, ospF-related, and elp genes.
The ospE/elpB1 and p21/elpB2 operons, ospF-related genes, and elp genes were amplified from 20 to 40 clonal isolates generated from each group of mice that were actively immunized with the corresponding OspE-related, OspF-related, or Elp protein by using the primers listed in Table 1. The ospE/F/elp paralogs also were amplified from 20 to 40 different clonal isolates recovered from mice 8 months postinfection. The B. burgdorferi strain 297 ospA gene (GenBank accession number X85442) and vlsE gene (GenBank accession number AY052626) were amplified from 10 isogenic clones from each group of immunized mice and from 20 clones from mice infected for 8 months by using the primers listed in Table 1. PCR amplifications were performed in 40-μl reaction mixtures by following the Expand High Fidelity PCR system protocol (Roche Molecular Biochemicals, Mannheim, Germany) and using 2 μl of DNA isolated from each isogenic clone as the template. PCR amplifications were performed as follows: 94°C for 5 min, 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min, and then a final extension at 72°C for 7 min. Five microliters of each amplification reaction mixture was then subjected to agarose gel electrophoresis and stained with ethidium bromide to visualize amplicons for size verification. The resulting amplicons were cleaned with a QiaQuick PCR purification kit (Qiagen, Valencia, Calif.) and then digested with 10 U of Tru9I (Promega, Madison, Wis.) before they were separated in a 4% MetaPhor agarose gel (Biowhittaker Molecular Applications, Rockland, Maine). For sequence analysis, ospE/elpB1, p21/elpB2, ospF, bbk2.10, bbk2.11, elpA1, and elpA2 amplicons generated from isogenic clones derived from mice immunized with each corresponding protein and from clones isolated from mice infected for 8 months were ligated into the pCR4Blunt-TOPO vector (Invitrogen, San Diego, Calif.). Nucleotide sequencing was performed by using an ABI model 373A automated DNA sequencer and PRISM ready reaction DyeDeoxy terminators (Applied Biosystems Inc., Foster City, Calif.) as described previously (20).
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence (5′ to 3′) | Description |
|---|---|---|
| ospA-5′ | ATGAAAAAATATTTATTGGGAATAG | PCR, bases 1 to 25 (plus strand) |
| ospA-3′ | TTTTAAAGCGTTTTTAATTTCATCA | PCR, bases 795 to 819 (minus strand) |
| vlsE-5′ | TTTCAAGTGCAATTTTTTTAGGAGC | PCR, bases 11 to 35 (plus strand) |
| vlsE-3′ | CCTTAGCCATTCCCCGCAGAAC | PCR, bases 904 to 925 (minus strand) |
| ospE-5′ | TGTAAAAATCATACTTTATATGATGG | PCR, bases 58 to 83 (plus strand) |
| elpB1-3′ | CTTCTTCATCATAATTATCCTCACTACCAG | PCR, bases 1079 to 1108 (minus strand) |
| p21-5′ | TGCAAAATTCATACTTCATATGATGA | PCR, bases 58 to 83 (plus strand) |
| elpB2-3′ | ATATAAATTACTATCCTCGAT | PCR, bases 1198 to 1218 (minus strand) |
| ospF-5′ | GCAACTAGTAAAGATTTAGAAGGGG | PCR, bases 70 to 94 (plus strand) |
| ospF-3′ | CTTTTTTGACTTCTCCATTATTTTCTATAG | PCR, bases 656 to 685 (minus strand) |
| bbk2.10-5′ | GCAAGTGGTGAAGATATAAAACAAAAT | PCR, bases 70 to 96 (plus strand) |
| bbk2.10-3′ | GTGGCTCTGCTAGCTTTTTAAGCTC | PCR, bases 577 to 601 (minus strand) |
| bbk2.11-5′ | GCAAGTGGTGAAAATCTAAAAAATT | PCR, bases 70 to 94 (plus strand) |
| bbk2.11-3′ | ATTATAATCTTTAGATTCTTTCTCTAAC | PCR, bases 663 to 690 (minus strand) |
| elpA1-5′ | GAATAAAGATAGTAAGGATTTAAAAAGCGC | PCR, bases 75 to 104 (plus strand) |
| elpA1-3′ | CTGGTTCTTCTAGTTCTTCAATAAG | PCR, bases 877 to 901 (minus strand) |
| elpA2-5′ | ATAAAGATTTAAAACAAAATGTAAAAGAAC | PCR, bases 77 to 106 (plus strand) |
| elpA2-3′ | TTCACAATCAACATCATCCG | PCR, bases 1037 to 1056 (minus strand) |
Proteinase K accessibility experiments.
Mid-logarithmic-phase cultures of temperature-shifted or DMC-cultivated spirochetes were gently pelleted by centrifugation for 5 min at 4,000 × g, washed once in phosphate-buffered saline (PBS) containing 10 mM magnesium chloride, and resuspended in the same buffer at a concentration of 1 × 109 organisms per ml. Aliquots of washed cells were removed and digested at room temperature with between 40 and 200 μg of proteinase K (Sigma Chemical Co.) per ml for 30 or 120 min prior to addition of phenylmethylsulfonyl fluoride (Sigma Chemical Co.) at a final concentration of 1.6 mg/ml to stop the proteinase digestion. A separate aliquot of organisms also was incubated for 120 min without proteinase K as a control. For some experiments, organisms were preincubated for 15 min with 0.05% Triton X-100 prior to the addition of proteinase K to permeabilize the spirochetal outer membrane. After proteinase digestion, the spirochetes were pelleted by centrifugation for 10 min at 10,000 × g before they were boiled for 10 min in final sample buffer (62.5 μM Tris-HCl [pH 6.8], 10% [vol/vol] glycerol, 5% [vol/vol] 2-mercaptoethanol, 5% sodium dodecyl sulfate, 0.001% bromphenol blue). Equivalent amounts of whole-cell lysates were then subjected to polyacrylamide gel electrophoresis through 2.4% polyacrylamide stacking and 12.5% polyacrylamide resolving gels. The gels were then transferred electrophoretically to 0.2-μm-pore-size nitrocellulose membranes (Schleicher and Schuell, Keene, N.H.) for immunoblot analysis. The membranes were blocked for 2 h in PBS-0.02% Tween containing 5% fetal bovine serum before 1:500 dilutions of affinity-purified rat polyclonal antiserum or 1:50 dilutions of mouse monoclonal antiserum were added and the preparations were incubated for 1 h at room temperature. The membranes were then washed, 1:5,000 dilutions of horseradish peroxidase-conjugated goat anti-rat or anti-mouse immunoglobulin G (IgG) (heavy and light chain specific) secondary antibodies (Zymed, San Francisco, Calif.) were added, and the preparations were incubated for 45 min at room temperature. The membranes were again washed before chemilumigrams were developed by using the Enhanced Chemiluminescence Plus Western blot detection system according to the manufacturer's instructions (Amersham Pharmacia).
Indirect immunofluorescence assays (IFA).
To determine the expression profiles of the OspE/F/Elp paralogs during tick feeding, salivary glands were dissected out of 48 infected ticks that had fed on naive C3H/HeJ mice for 53 to 58 h, and they were placed in PBS (pH 7.4) prior to homogenization by repeated pipetting. The salivary gland suspensions were then spotted onto fluoroslides (Erie Scientific, Portsmouth, N.H.), allowed to air dry, and fixed by immersion in acetone for 10 min. The slides were blocked for 45 min with PBS containing 0.2% bovine serum albumin and then incubated for 1 h at room temperature with mouse polyclonal anti-OspA and anti-OspC (mixture containing a 1:50 dilution of each), rabbit polyclonal anti-FlaB (1:500 dilution), and affinity-purified rat polyclonal anti-OspE, anti-p21, anti-OspF/BbK2.11, anti-BbK2.10, anti-ElpB1, anti-ElpB2, anti-ElpA1, or anti-ElpA2 (1:25 dilution). The slides were subsequently washed, and Alexa 350-labeled goat anti-rabbit IgG, Alexa 488-labeled goat anti-mouse IgG, and Alexa 568-labeled goat anti-rat IgG (all heavy and light chain specific; Molecular Probes, Eugene, Oreg.) were then added at a 1:1,000 dilution. After incubation for 45 min at room temperature, the slides were washed three times in PBS (pH 7.4) and three times in water before they were air dried. One drop of buffered glycerol (Becton Dickinson, Cockeysville, Md.) and a coverslip were then placed over each sample. All samples were examined at a magnification of ×1,000 with a BX-60 fluorescence microscope (Olympus, Melville, N.Y.). For each sample, spirochetes were first identified by anti-FlaB reactivity. Individual spirochetes identified in this manner were then scored for OspA/C expression and for expression of the OspE/F/Elp paralogs from a minimum of three independent salivary gland preparations.
To determine which of the OspE-related, OspF-related, and Elp lipoproteins are surface exposed, spirochetes cultivated following a temperature shift and organisms cultivated in DMCs were probed in solution as previously described (12). Affinity-purified rat polyclonal antiserum directed against OspA, OspC, OspE, p21, OspF/BbK2.11, BbK2.10, ElpA1, ElpA2, ElpB1, or ElpB2 was used at a final dilution of 1:25. Also included in each of the primary antibody incubation mixtures was polyclonal rabbit anti-FlaB (1:500 dilution). Spirochetes were also incubated with prebleed sera diluted appropriately to ensure specific reactivity. After the primary incubation, spirochetes were gently collected by centrifugation for 5 min at 4,000 × g and washed in PBS containing 10 mM magnesium chloride. The spirochetes were subsequently spotted onto fluoroslides (Erie Scientific), air dried, and blocked as described above before Alexa 488-conjugated goat anti-rat IgG and Alexa 568-conjugated goat anti-rabbit IgG (both heavy and light chain specific; Molecular Probes) were added at a dilution of 1:1,000 and the preparations were incubated for 45 min in PBS containing 0.2% bovine serum albumin. To identify all spirochetes in a given microscope field, the permeable DNA-binding fluorophore 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI) (Molecular Probes) also was included in the secondary antibody incubation mixture as previously described (33). The final slide preparations were washed and visualized as described above. The percentages of spirochetes expressing surface-exposed OspE/F/Elp paralogs were quantified by first identifying all spirochetes (i.e., DAPI-positive cells) in each microscopic field, ensuring that the membrane integrity had not been compromised by the absence of FlaB reactivity, and then classifying individual organisms based on their anti-OspE-related, OspF-related, and Elp reactivities. For each OspE/F/Elp paralog analyzed, at least 100 intact organisms from three independent preparations were examined. To determine the total percentages of organisms expressing the OspE/F/Elp parologs, aliquots of spirochetes from the same cultures also were washed, spotted onto slides, allowed to air dry, and fixed by acetone immersion prior to addition of primary antibodies. The acetone fixation step allowed total enumeration of organisms expressing the various lipoproteins, which was not dependent on the lipoprotein being surface localized. Images for all IFA analyses were captured with a Spot II digital camera and associated software (Diagnostic Instruments, Sterling Heights, Mich.). A two-tailed unpaired t test was utilized to determine significant differences between patterns of expression of the OspE/F/Elp paralogs in different environments. The significance of decreases in the amounts of individual lipoproteins that were surface exposed were determined by using a one-tailed unpaired t test. All statistical analyses were performed by using the Prism 3.0 software package (GraphPad, San Diego, Calif.).
Sequence analysis.
Nucleotide sequence analyses were performed and primers were designed by using the MacVector, version 6.5.3, software package (Oxford Molecular Group, Cambell, Calif.).
RESULTS
Expression of the OspE/F/Elp paralogs in tick salivary glands.
Recently, Ohnishi and coworkers determined that stable infection of a mammalian host by B. burgdorferi occurs only after 53 h of tick feeding (30). Interestingly, the majority of spirochetes identified in tick salivary glands during this critical period were found not to express either OspA or OspC, two intensely studied lipoproteins which appear to play important roles in bacterial transmission (30, 32, 40, 41). These findings, in addition to our prior observations which revealed that several of the OspE/F/Elp paralogs are upregulated in the midgut of ticks during feeding (20), led us to ask whether the proteins also are expressed by spirochetes in tick salivary glands during this critical time period of transmission. To examine this possibility, we allowed B. burgdorferi strain 297-infected ticks to feed for 53 to 58 h on naive C3H/HeJ mice. IFA performed on the resulting salivary gland homogenates revealed that OspE, p21, OspF/BbK2.11, ElpB1, and ElpB2 were expressed by spirochetes during this time period. As shown in Table 2, a quantitative analysis revealed that 17.7 to 50.7% of the spirochetes expressed OspE, ElpB1, p21, ElpB2, or OspF/BbK2.11 and that in all cases only a minority expressed OspA or OspC. The percentages of spirochetes expressing OspA or OspC were similar to the percentage reported by Ohnishi et al. (30) for B. burgdorferi strain B31. Although numerous spirochetes in the salivary gland preparations were found to express each of the lipoproteins analyzed, many spirochetes also were identified that did not express either OspE (49.3%), ElpB1 (57.4%), p21 (82.3%), ElpB2 (80.6%), or OspF/BbK2.11 (64.8%), indicating that there is an antigenically diverse population of spirochetes in the salivary gland environment during the transmission process. Of the OspE/F/Elp paralogs, only BbK2.10, ElpA1, and ElpA2 were not expressed by any of the spirochetes examined in the salivary gland preparations. Ear punch biopsy cultures confirmed that all mice used for these experiments were stably infected after the 53- to 58-h feeding period (data not shown).
TABLE 2.
Expression of the OspE-related, OspF-related, and Elp lipoproteins in tick salivary glands after 53 to 58 h of feeding
| Phenotypeb | % of spirochetes expressinga:
|
||
|---|---|---|---|
| OspA+/C+ | OspA−/C− | Totalc | |
| OspE+ | 22.6 | 28.0 | 50.7 |
| OspE− | 9.6 | 39.7 | 49.3 |
| ElpB1+ | 19.6 | 22.9 | 42.6 |
| ElpB1− | 7.4 | 50.1 | 57.4 |
| p21+ | 6.8 | 10.8 | 17.7 |
| p21− | 19.3 | 63.0 | 82.3 |
| ElpB2+ | 8.1 | 11.3 | 19.4 |
| ElpB2− | 12.6 | 68.0 | 80.6 |
| OspF/BbK2.11+ | 13.3 | 22.0 | 35.2 |
| OspF/BbK2.11− | 11.4 | 53.4 | 64.8 |
| BbK2.10+ | 0 | 0 | 0 |
| BbK2.10− | 23.5 | 76.5 | 100 |
| ElpA1+ | 0 | 0 | 0 |
| ElpA1− | 36.0 | 64.0 | 100 |
| ElpA2+ | 0 | 0 | 0 |
| ElpA2− | 18.9 | 81.1 | 100 |
| OspA/C | 26.1 | 73.9 | |
The standard deviations were less than 12.5% for all samples analyzed.
Spirochetes expressed (+) or did not express (−) the lipoprotein.
Percentage of spirochetes that expressed or did not express the individual OspE/F/Elp lipoproteins.
OspE-related, OspF-related, and Elp proteins are not protective antigens.
On the basis of the expression patterns observed for the OspE/F/Elp paralogs during transmission and early infection (20) and on the basis of prior reports that orthologs to these proteins are surface exposed (16, 23), we next sought to determine whether an immune response to these proteins could induce protective immunity. The findings described above, however, which demonstrated that expression of many of the OspE/F/Elp paralogs is restricted to subpopulations of spirochetes suggested that an immunization strategy based on only one of these proteins would probably not confer total protection. Therefore, we actively immunized groups of mice with either (i) all of the OspE/F/Elp paralogs, (ii) members of each paralogous lipoprotein family (i.e., OspE related, OspF related, or Elps), or (iii) individual OspE-related, OspF-related, and Elp lipoproteins. As shown in Table 3, none of the individual OspE-related, OspF-related, and Elp lipoproteins or any of the various combinations protected mice from challenge infections. As expected, all mice immunized with OspA were protected from tick challenge, while GST-immunized mice were not protected.
TABLE 3.
Protection of OspE-related, OspF-related, and Elp protein-vaccinated mice from challenge with infected ticks
| Immunogen | No. of cultures positive/no. testeda |
|---|---|
| OspA | 0/5 |
| GST | 5/5 |
| OspE | 5/5 |
| p21 | 5/5 |
| OspF | 5/5 |
| BbK2.11 | 5/5 |
| BbK2.10 | 5/5 |
| ElpA1 | 5/5 |
| ElpA2 | 5/5 |
| ElpB1 | 5/5 |
| ElpB2 | 5/5 |
| OspE related | 5/5 |
| OspF related | 5/5 |
| Elps | 5/5 |
| OspE related, OspF related, and Elps | 5/5 |
Two weeks after infestation, ear punch biopsies were cultivated in BSK-H medium containing rifampin and amphotericin B. Cultures were examined over a 3-week period and considered positive if spirochetes were observed at any point by dark-field microscopy. For OspA, heart, spleen, and bladder biopsies also were cultured.
Genetic stability of the ospE/F/elp paralogs during infection.
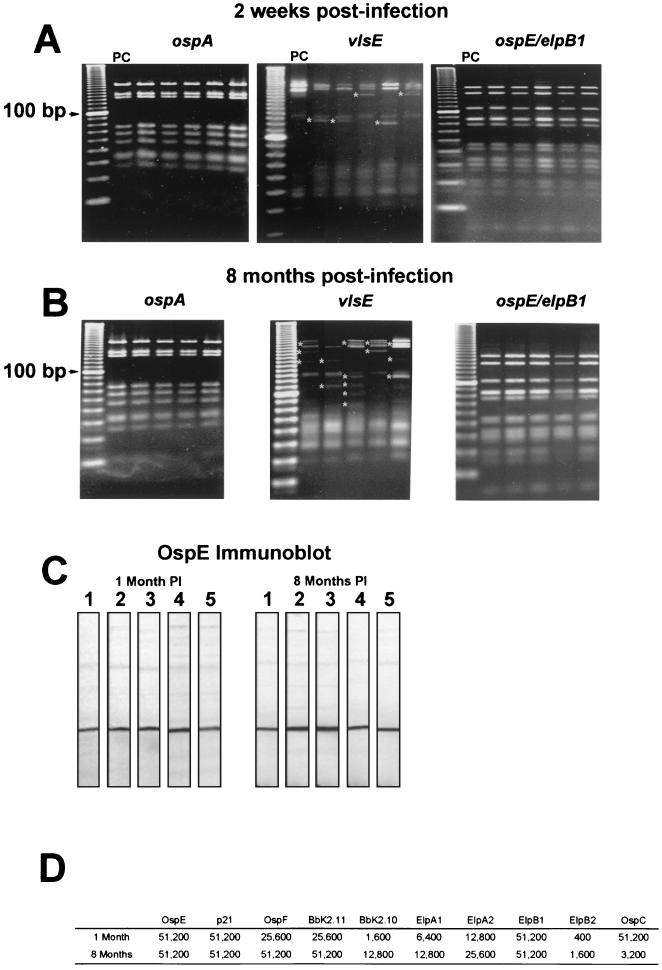
We next explored the possibility that the ospE/F/elp genes undergo recombination or variation during infection, which could help explain the apparent lack of protection described above. Since a prior report indicated that a specific immune response directed against OspE resulted in genetic variation at the ospE locus (49), we hyperimmunized mice with the various OspE/F/Elp paralogs and experimentally infected them to help identify escape mutants. Specific amplicons encompassing more than 95% of the ospE/elpB1 and p21/elpB2 operons, as well as the ospF, bbk2.10, bbk2.11, elpA1, and elpA2 genes, were generated from clonal isolates as outlined in Materials and Methods. Amplicons corresponding to portions of the ospA and vlsE genes also were included as controls for stability and variation, respectively. Because of the high A+T content of B. burgdorferi (>70%) (17), the resulting amplicons were screened by RFLP analysis by using the restriction enzyme Tru9I, which recognizes the sequence TTAA. While variation and recombination in vlsE were apparent in clones isolated from OspE-immunized mice within 2 weeks after infection, no RFLP variants were identified for ospA or the ospE/elpB1 operon in 40 different clones obtained from OspE-immunized animals (Fig. 1A and data not shown). Similarly, no RFLPs were observed for any of the other paralogs when 20 different isogenic clones from each group of immunized mice were examined (data not shown). Sequence analysis of amplicons generated from 10 to 20 different isogenic clones for each of the various groups of immunized mice did not reveal any sequence differences compared with the sequence of the parental strain (data not shown).
FIG. 1.
Genetic stability of the ospE/elpB1 loci and antibody reactivity to the OspE-related, OspF-related, and Elp proteins during infection. (A and B) RFLP analyses of the ospA, vlsE, and ospE/elpB1 loci of isogenic clones generated from mice hyperimmunized with OspE 2 weeks postinfection (A) and from unimmunized mice 8 months postinfection (B) using restriction enzyme Tru9I. The results for five representative clones from the 20 to 40 clones analyzed for each locus are shown. PC, RFLP analysis performed on amplicons generated from parental strain 297. For the vlsE locus, fragments that vary from the fragments in the parental pattern are indicated by asterisks. The left lane in each panel contained a 10-bp DNA ladder. (C) Representative immunoblot of recombinant OspE with serum from mice used to generate clonal isolates 1 and 8 months postinfection (PI). (D) Reciprocal antibody titers for the OspE/F/Elp paralogs and OspC in pooled sera from mice used to generate clonal isolates. Titers were determined 1 and 8 months postinfection.
While no variation was observed after 2 weeks of infection in hyperimmunized mice, we also analyzed isogenic clones generated from mice infected for 8 months. Our underlying assumption was that if antigenic variation occurs in the ospE/F/elp loci, the variation should be detected following 8 months of infection. As observed for the 2-week-postinfection isolates, no RFLPs were identified in the ospE/elpB1 loci for 40 different clones isolated from mice 8 months postinfection (Fig. 1B and data not shown). Sequence analysis of amplicons from 20 of these isogenic clones also failed to reveal any genetic variation in the ospE/elpB1 loci during long-term infection. When the remaining ospE/F/elp paralogs from 20 and 10 different clones were examined by performing RFLP and nucleotide sequence analyses, respectively, no variation was detected (data not shown). Genetic variability was observed 8 months postinfection for the vlsE gene, while ospA was genetically stable, as expected (Fig. 1B). We also analyzed the antibody responses of the five mice used for these analyses to ensure that tick-infected mice generated and maintained a humoral immune response against the various OspE/F/Elp paralogs. ELISA and immunoblot analyses revealed that the antibody titers of mice following 8 months of infection were always equal to or greater than the antibody titers of the same mice after 1 month of infection (Fig. 1C and D). In contrast, the antibody titers against OspC dramatically decreased as the infection progressed during this same time period (Fig. 1D), as has previously been shown (25, 38, 39).
Cellular localization of the OspE-related, OspF-related, and Elp lipoproteins.
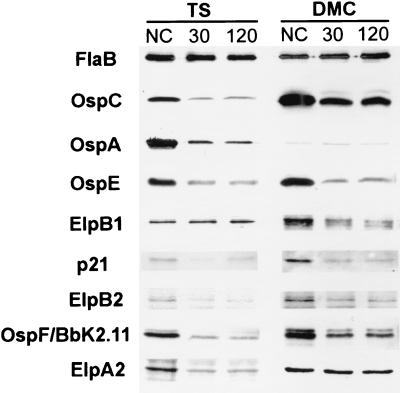
The finding that the OspE/F/Elp paralogs did not protect mice from a natural tick infection, combined with the stability of the genes encoding these lipoproteins, suggested that these proteins may not be located solely on the borrelial surface. To examine the extent of surface localization of the OspE/F/Elp paralogs, we utilized both proteinase K accessibility analysis and IFA with organisms that were temperature shifted in vitro and organisms that were cultivated in DMCs (1). We should note that these localization analyses did not include BbK2.10 or ElpA1 because these proteins are not expressed by temperature-shifted or DMC-cultivated organisms (1, 20). As shown in Fig. 2, even when spirochete suspensions were incubated with as much as 200 μg of proteinase K for 2 h, portions of the intact OspE/F/Elp paralogs could still be detected by immunoblotting. The fact that there did not appear to be a difference in the level of protein degradation between the 30-min and 2-h time points indicated that there was a fraction that was either not accessible to proteinase K or proteinase K resistant. Similarly, OspC and OspA were not completely degraded by proteinase K, even after the 2-h incubation period. Although the overall digestion patterns were similar in the two culture conditions, there were exceptions. For example, the amount of undegraded OspC and ElpA2 appeared to dramatically increase in DMC-cultivated organisms. As expected, OspA was almost undetectable in DMC-cultivated organisms (1, 20). When similar proteinase K accessibility experiments were performed in the presence of 0.05% Triton X-100, a nonionic detergent known to disrupt the borrelial outer membrane (12), complete digestion of all proteins was observed within 30 min (data not shown). The latter finding demonstrates that the undigested protein observed in Fig. 2 was proteinase K inaccessible, not proteinase K resistant.
FIG. 2.
Immunoblot analysis of whole-cell lysates from proteinase K-treated B. burgdorferi. Spirochetes cultivated following a temperature shift (TS) and in DMCs were incubated for 30 or 120 min with 200 μg of proteinase K per ml; control samples (NC) were incubated for 120 min without proteinase K. Proteinase K degradation was assessed by immunoblotting using monoclonal antibodies directed against FlaB (1H6-33) or OspA (14D2-27) or affinity-purified rat polyclonal anti-OspE, anti-p21, anti-OspF/BbK2.11, anti-ElpA2, anti-ElpB1, anti-ElpB2, or anti-OspC using the Enhanced Chemiluminescence Plus Western blotting system as described in Materials and Methods.
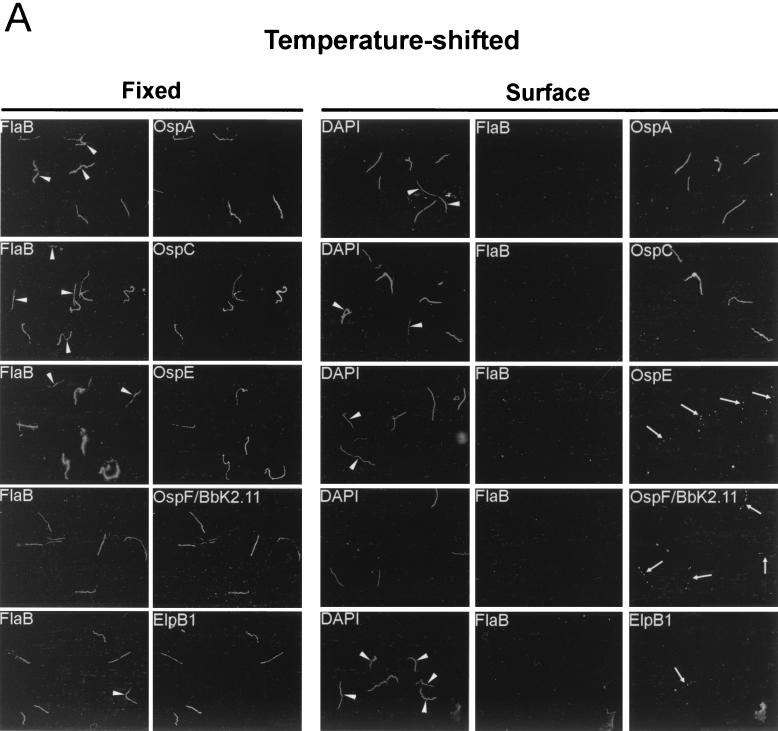
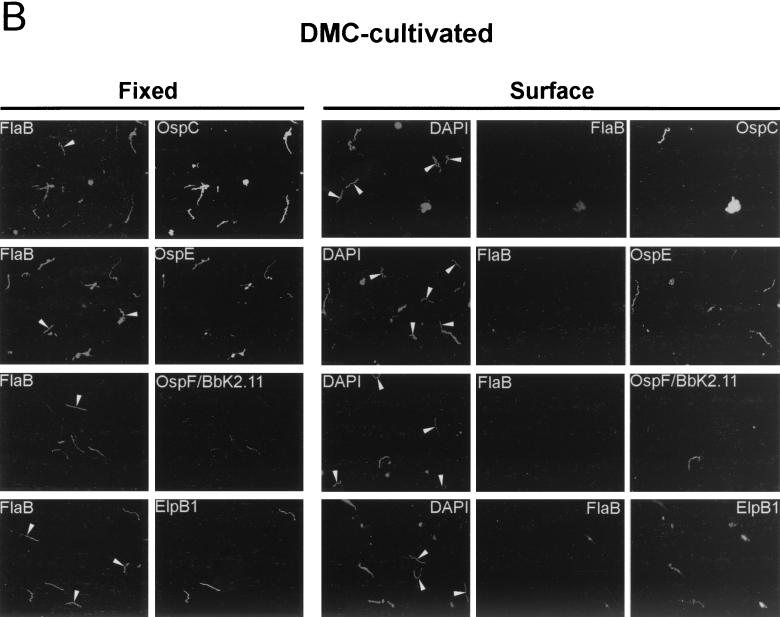
Since we could not rule out the possibility that the undigested fraction of intact lipoproteins was due to constant synthesis of new proteins that had not reached the outer surface, we next used IFA to further examine the cellular location of the OspE/F/Elp paralogs. To analyze the cellular location of these lipoproteins, two different approaches were used. Since B. burgdorferi membranes can be disrupted by acetone fixation, we performed IFA experiments on acetone-fixed organisms to determine the total numbers of spirochetes which expressed the various lipoproteins. We also performed IFA experiments in solution, as previously described (12), to determine which of the lipoproteins were expressed on the surfaces of spirochetes. The combined analyses not only allowed us to determine the total percentage of spirochetes in a population that expressed a given protein (acetone-fixed organisms) but also allowed us to determine the percentage of organisms that localized the proteins to the outer surface (organisms probed in solution). Much to our surprise, as shown in Fig. 3, the total number of organisms expressing OspE, OspF/BbK2.11, and ElpB1 was always greater than the number found to display these proteins on the surface. As would be expected, similar results also were observed for OspC and OspA (12, 36). Additionally, a distinct beaded pattern of expression for OspE, OspF/BbK2.11, and ElpB1 was observed in temperature-shifted organisms that was not evident in organisms cultivated in DMCs (Fig. 3A and B, compare rightmost panels).
FIG. 3.
Expression and cellular localization of OspA, OspC, OspE, OspF/BbK2.11, and ElpB1 within individual organisms. Spirochetes cultivated following a temperature shift (A) or in DMCs (B) were subjected to IFA after acetone fixation (Fixed) or directly in solution prior to fixation (Surface). Samples were probed with rabbit polyclonal serum to FlaB and rat polyclonal serum directed against OspA, OspC, OspE, OspF/BbK2.11, or ElpB1. OspA results are shown only for temperature-shifted organisms because so few organisms were found to express this protein in DMC-cultivated organisms. For surface labeling experiments, DAPI, a permeable DNA-binding dye, was used to identify all spirochetes in a field. The arrows indicate spirochetes labeled with OspE, OspF/BbK2.11, and ElpB1 antibodies. The arrowheads indicate individual spirochetes that either did not express the indicated lipoprotein (Fixed panels) or did not display it on the surface (Surface panels). Organisms were visualized by fluorescence microscopy at a magnification of ×1,000.
As shown in Table 4 , when a more quantitative analysis was performed for all of the OspE/F/Elp paralogs, between 9.7 and 100% were expressed by temperature-shifted organisms, while only 33.3 to 76.7% expressed these proteins during DMC cultivation. Interestingly, significant differences were observed between the total percentages of DMC-cultivated and temperature-shifted organisms expressing the OspE/F/Elp paralogs. Specifically, significantly fewer DMC-cultivated organisms expressed OspE, OspF/BbK2.11, ElpA2, and ElpB1, while the number of organisms expressing p21 and ElpB2 increased significantly during DMC cultivation. As expected, the numbers of DMC-cultivated organisms expressing OspA and OspC also were found to be significantly downregulated and upregulated, respectively (1, 20). We also observed that none of the lipoproteins was located exclusively on the surface of B. burgdorferi in either environment; rather, only 11.7 to 93.3% of the spirochetes expressing an individual lipoprotein also displayed it on the surface. Remarkably, while the percentage of organisms expressing OspC on the surface decreased from 86.9% in temperature-shifted organisms to 22.3% in DMC-cultivated organisms, there was an increase in the overall number of organisms expressing OspC (74.3 versus 93.7%). Like the number of organisms displaying OspC, the number of DMC-cultivated spirochetes displaying OspE, p21, OspF/BbK2.11, and ElpA2 on the surface also was significantly less than the number of temperature-shifted organisms that did this. By contrast, ElpB1 was expressed on the surface of significantly more DMC-cultivated organisms (62.5%) than temperature-shifted organisms (11.7%).
TABLE 4.
Overall expression and cellular localization of the OspE-related, OspF-related, and Elp lipoproteins in temperature-shifted and DMC-cultivated B. burgdorferi
| Antibody | Lipoprotein expression (%)a
|
|||||
|---|---|---|---|---|---|---|
| Temperature shifted
|
DMC cultivated
|
|||||
| Total | Surface | % Surface | Total | Surface | % Surface | |
| OspA | 77.7 | 73.0 | 92.0 | 3.3b | 0c | 0d |
| OspC | 74.3 | 64.3 | 86.9 | 93.7b | 21.0c | 22.3d |
| OspE | 91.3 | 80.0 | 87.8 | 75.7b | 51.3c | 67.8 |
| p21 | 9.7 | 4.3 | 45.5 | 38.3b | 15.3c | 40.7 |
| OspF/BbK2.11 | 100 | 93.3 | 93.3 | 76.7b | 31.7c | 41.2d |
| ElpA2 | 61.3 | 42.7 | 71.1 | 33.3b | 8.3c | 26.2d |
| ElpB1 | 93.3 | 11c | 11.7 | 76.3b | 47.7c | 62.5d |
| ElpB2 | 10.7 | 5.3c | 38.4 | 46b | 19c | 40.5 |
The standard deviations were less than 9% for all samples. The percentage of expression for each lipoprotein was determined from three independent experiments.
The total percentage of DMC-cultivated spirochetes expressing the lipoprotein was significantly different from the percentage of temperature-shifted organisms expressing the lipoprotein (P ≤0.05).
The percentage of the population with surface-exposed lipoprotein was significantly less than the total percentage of organisms expressing the lipoprotein (P ≤0.05).
There was a significant difference in the percentages of DMC-cultivated and temperature-shifted organisms expressing the lipoprotein on the surface.
DISCUSSION
Dramatic changes occur in B. burgdorferi protein expression as the organism adapts to the influx of a blood meal, migrates to the tick salivary glands, and establishes an infection in a mammalian host (13, 30, 41). Prior studies have shown that the OspE/F/Elp paralogs are differentially expressed by temperature- and host-specific factors during transmission and early infection (1, 3, 20, 44, 47). This has led us and other workers to speculate that these lipoproteins are integral to the transmission process and disease establishment. The following observations made in this study not only support these prior contentions but also indicate that the OspE/F/Elp paralogs are important throughout infection: (i) OspE, p21, OspF/BbK2.11, ElpB1, and ElpB2 are expressed in tick salivary glands during the stage of transmission critical for disease establishment, (ii) the majority of spirochetes expressing these lipoproteins in tick salivary glands do not express either OspA or OspC, and (iii) specific antibody titers for the OspE/F/Elp paralogs are maintained or increase as the disease progresses, indicating that the paralogs are continually expressed throughout infection. This conclusion is supported by the finding that all of the OspE/F/Elp lipoproteins previously found to be expressed in tick midguts during a blood meal (20) were expressed in the salivary glands during transmission. In fact, although p21, ElpB2, and ElpA2 are not expressed in the midguts of feeding ticks, all three of these proteins were upregulated and expressed in tick salivary glands during the migration process. While the functions of these proteins are currently unknown, recent reports indicate that OspE binds the host serum component factor H (21, 46), which may help B. burgdorferi evade destruction by the complement system during infection. The fact that OspE can be surface exposed and is upregulated during transmission to a mammalian host is entirely consistent with this recent finding.
The ability of B. burgdorferi to maintain a chronic infection despite the presence of a strong and directed immune response has led many investigators to conclude that this organism utilizes some form of antigenic variation during infection (3, 26, 45, 49). While the plasmid-encoded vlsE system has been shown to undergo genetic rearrangements during mammalian infection (52), it is still unclear whether the ospE/F/elp paralogs also vary during infection and aid in immune evasion (18, 27, 49). The comprehensive analysis which we undertook here indicates that all of the ospE/F/elp loci are genetically stable during infection for at least 8 months. Furthermore, even when mice were hyperimmunized with each of the specific proteins to generate high titers of specific antibodies prior to infection, no escape variants were identified. Therefore, despite the fact that the ospE/F/elp loci and the plasmids that encode them have undergone numerous recombinatorial events in the past (2, 7, 45), we found no evidence that any of these genes mutate during a single infection.
While all of the OspE/F/Elp paralogs analyzed in this study were found to be surface exposed in some organisms, the lack of genetic variation indicates that an alternative system may be utilized by B. burgdorferi to help evade the immune response of the host during infection. The overall heterogeneity in antigen expression and the variable cellular location of the OspE/F/Elp paralogs observed within populations of spirochetes may help explain how B. burgdorferi avoids destruction by the host's immune response. While many previous studies have shown that antigenic diversity can be generated in populations of spirochetes through changes in the temporal patterns of protein regulation (1, 3, 11, 20, 22, 25, 34, 41, 47, 48, 50), our data indicate that there may be another dimension to this phenomenon of differential antigen expression; that is, not only can proteins be regulated by this organism with respect to the stage of the enzootic cycle in which they are expressed (e.g., tick midgut, salivary gland, or mammalian host), but this organism also can alter the cellular location of antigens that are expressed during the various phases of this cycle. The IFA data provide strong evidence for this conjecture. When individual organisms were examined simultaneously for lipoprotein expression and cellular localization, we observed that the OspE/F/Elp paralogs were expressed by many organisms that did not localize them to the outer surface. Consistent with our observations, El-Hage et al. (16) recently reported that only 70% of spirochetes cultivated following a temperature shift localized an OspE ortholog in B. burgdorferi strain B31 to the surface. These observations also may help explain why only subpopulations of spirochetes were killed within tick midguts after the ticks fed on mice immunized with OspE or OspF (29). The combined data lead us to propose a model for generating population heterogeneity and antigenic diversity in B. burgdorferi which relies on both the temporal (i.e., when a protein is expressed) and spatial (i.e., where a protein is localized) regulation of proteins throughout the enzootic cycle. Implicit in this model is the potential for dramatically more antigenic diversity and population heterogeneity during mammalian infection than previously recognized. Furthermore, the generation of antigenic diversity on the borrelial surface through both temporal and spatial changes in protein expression could have major implications on future vaccine development strategies for Lyme disease.
The localization of lipoproteins to the outer surface is energetically unfavorable and does not occur in most organisms with a gram-negative membrane ultrastructure (35). Among gram-negative organisms that localize lipoproteins to the surface, almost all have been found to contain specialized export systems that transport lipoproteins to the cell surface (35). While we cannot rule out the possibility that the lack of surface exposure observed in individual spirochetes is due to a delay in timing of lipoprotein secretion to the outer surface, based on prior studies it seems more plausible that B. burgdorferi regulates the cellular localization of the lipoproteins through a specific export system. In fact, it has previously been speculated that B. burgdorferi contains a lipoprotein export system (12); however, no such system has been identified. This is not surprising given the extensive sequence divergence between the known lipoprotein export systems of other bacteria (35). It is still not known how lipoproteins are exported to the borrelial surface or why they are located in both the outer membrane and periplasmic space of B. burgdorferi, as observed in this study for the OspE/F/Elp paralogs, OspA, and OspC. While this dual localization may seem peculiar, there is substantial evidence that the majority of OspA, OspB, and OspC is located within the periplasmic space (5, 6, 12, 36). Along these lines, it is interesting that the crystal structure of OspC revealed that this protein is structurally homologous to the Salmonella enterica serovar Typhimurium aspartate receptor (15), a protein known to be localized in the periplasm of gram-negative bacteria (10). Given the structural similarities, it is tempting to speculate that OspC may perform a biological function in the periplasm of B. burgdorferi. Obviously, additional biochemical and structural studies are needed to help elucidate why the OspE/F/Elp paralogs also are localized to both the outer surface and the periplasmic compartment in B. burgdorferi. The significance of this dual localization and its relationship to borrelial population heterogeneity, antigenic diversity, and immune evasion represent an important area of research to be addressed in the future.
Acknowledgments
This work was supported in part by grant 0030130N from the American Heart Association and by grant RR-15564 from the National Institutes of Health Center for Research Resources to D.R.A. P.S.H. was supported in part by Molecular Pathogenesis Training Grant AI-07364 from the National Institute of Allergy and Infectious Diseases. M.J.C. was supported in part by grant AI-29735 from the National Institute of Allergy and Infectious Diseases.
We thank Amy Jett for expert technical assistance, Justin Radolf and Chad Brooks for many helpful discussions, and Rodney Tweten for critical review of the manuscript.
Editor: A. D. O'Brien
REFERENCES
- 1.Akins, D. R., K. W. Bourell, M. J. Caimano, M. V. Norgard, and J. D. Radolf. 1998. A new animal model for studying Lyme disease spirochetes in a mammalian host-adapted state. J. Clin. Investig. 101:2240-2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akins, D. R., M. J. Caimano, X. Yang, F. Cerna, M. V. Norgard, and J. Radolf. 1999. Molecular and evolutionary analysis of Borrelia burgdorferi 297 circular plasmid-encoded lipoproteins with OspE- and OspF-like leader peptides. Infect. Immun. 67:1526-1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akins, D. R., S. F. Porcella, T. G. Popova, D. Shevchenko, S. I. Baker, M. Li, M. V. Norgard, and J. D. Radolf. 1995. Evidence for in vivo but not in vitro expression of a Borrelia burgdorferi outer surface protein F (OspF) homolog. Mol. Microbiol. 18:507-520. [DOI] [PubMed] [Google Scholar]
- 4.Benach, J. L., J. L. Coleman, R. A. Skinner, and E. M. Bosler. 1987. Adult Ixodes dammini on rabbits: a hypothesis for the development and transmission of Borrelia burgdorferi. J. Infect. Dis. 155:1300-1306. [DOI] [PubMed] [Google Scholar]
- 5.Bockenstedt, L. K., E. Hodzic, S. Feng, K. W. Bourell, A. de Silva, R. R. Montgomery, E. Fikrig, J. D. Radolf, and S. W. Barthold. 1997. Borrelia burgdorferi strain-specific Osp C-mediated immunity in mice. Infect. Immun. 65:4661-4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brusca, J. S., A. W. McDowall, M. V. Norgard, and J. D. Radolf. 1991. Localization of outer surface proteins A and B in both the outer membrane and intracellular compartments of Borrelia burgdorferi. J. Bacteriol. 173:8004-8008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caimano, M. J., X. Yang, T. G. Popova, M. L. Clawson, D. R. Akins, M. V. Norgard, and J. D. Radolf. 2000. Molecular and evolutionary characterization of the cp32/18 family of supercoiled plasmids in Borrelia burgdorferi 297. Infect. Immun. 68:1574-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Casjens, S., N. Palmer, R. van Vugt, W. M. Huang, B. Stevenson, P. Rosa, R. Lathigra, G. Sutton, J. Peterson, R. J. Dodson, D. Haft, E. Hickey, M. Gwinn, O. White, and C. M. Fraser. 2000. A bacterial genome in flux: the twelve linear and nine circular extrachromosomal DNAs in an infectious isolate of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:490-516. [DOI] [PubMed] [Google Scholar]
- 9.Casjens, S., R. van Vugt, K. Tilly, P. A. Rosa, and B. Stevenson. 1997. Homology throughout the multiple 32-kilobase circular plasmids present in Lyme disease spirochetes. J. Bacteriol. 179:217-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chervitz, S. A., and J. J. Falke. 1996. Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc. Natl. Acad. Sci. USA 93:2545-2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coleman, J. L., J. A. Gebbia, J. Piesman, J. L. Degen, T. H. Bugge, and J. L. Benach. 1997. Plasminogen is required for efficient dissemination of B. burgdorferi in ticks and for enhancement of spirochetemia in mice. Cell 89:1111-1119. [DOI] [PubMed] [Google Scholar]
- 12.Cox, D. L., D. R. Akins, K. W. Bourell, P. Lahdenne, M. V. Norgard, and J. D. Radolf. 1996. Limited surface exposure of Borrelia burgdorferi outer surface lipoproteins. Proc. Natl. Acad. Sci. USA 93:7973-7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Silva, A. M., and E. Fikrig. 1997. Arthropod- and host-specific gene expression by Borrelia burgdorferi. J. Clin. Investig. 99:377-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Silva, A. M., and E. Fikrig. 1995. Growth and migration of Borrelia burgdorferi in Ixodes ticks during blood feeding. Am. J. Trop. Med. Hyg. 53:397-404. [DOI] [PubMed] [Google Scholar]
- 15.Eicken, C., V. Sharma, T. Klabunde, R. T. Owens, D. S. Pikas, M. Hook, and J. C. Sacchettini. 2001. Crystal structure of Lyme disease antigen outer surface protein C from Borrelia burgdorferi. J. Biol. Chem. 276:10010-10015. [DOI] [PubMed] [Google Scholar]
- 16.El-Hage, N., K. Babb, J. A. Carroll, N. Lindstrom, E. R. Fischer, J. C. Miller, J. R. Gilmore, M. L. Mbow, and B. Stevenson. 2001. Surface exposure and protease insensitivity of Borrelia burgdorferi Erp (OspEF-related) lipoproteins. Microbiology 147:821-830. [DOI] [PubMed] [Google Scholar]
- 17.Fraser, C. M., S. Casjens, W. M. Huang, G. G. Sutton, R. Clayton, R. Lathigra, O. White, K. A. Ketchum, R. Dodson, E. K. Hickey, M. Gwinn, B. Dougherty, J.-F. Tomb, R. D. Fleischmann, D. Richardson, J. Peterson, A. R. Kerlavage, J. Quackenbush, S. Salzberg, M. Hanson, R. van Vugt, N. Palmer, M. D. Adams, J. Gocayne, J. Weidman, T. Utterback, L. Watthey, L. McDonald, P. Artiach, C. Bowman, S. Garland, C. Fujii, M. D. Cotton, K. Horst, K. Roberts, B. Hatch, H. O. Smith, and J. C. Venter. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390:580-586. [DOI] [PubMed] [Google Scholar]
- 18.Hage, N. E., L. D. Lieto, and B. Stevenson. 1999. Stability of erp loci during Borrelia burgdorferi infection: recombination is not required for chronic infection of immunocompetent mice. Infect. Immun. 67:3146-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hagman, K. E., P. Lahdenne, T. G. Popova, S. F. Porcella, D. R. Akins, J. D. Radolf, and M. V. Norgard. 1998. Decorin-binding protein of Borrelia burgdorferi is encoded within a two-gene operon and is protective in the murine model of Lyme borreliosis. Infect. Immun. 66:2674-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hefty, P. S., S. E. Jolliff, M. J. Caimano, S. K. Wikel, J. D. Radolf, and D. R. Akins. 2001. Regulation of OspE-related, OspF-related, and Elp lipoproteins of Borrelia burgdorferi strain 297 by mammalian host-specific signals. Infect. Immun. 69:3618-3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellwage, J., T. Meri, T. Heikkila, A. Alitalo, J. Panelius, P. Lahdenne, I. J. Seppala, and S. Meri. 2000. The complement regulator factor H binds to the surface protein OspE of Borrelia burgdorferi. J. Biol. Chem. 276:8427-8435. [DOI] [PubMed] [Google Scholar]
- 22.Lahdenne, P., S. F. Porcella, K. E. Hagman, D. R. Akins, T. G. Popova, D. L. Cox, J. D. Radolf, and M. V. Norgard. 1997. Molecular characterization of a 6.6-kilodalton Borrelia burgdorferi outer membrane-associated lipoprotein (lp6.6) which appears to be downregulated during mammalian infection. Infect. Immun. 65:412-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lam, T. T., T. P. K. Nguyen, R. R. Montgomery, F. S. Kantor, E. Fikrig, and R. A. Flavell. 1994. Outer surface proteins E and F of Borrelia burgdorferi, the agent of Lyme disease. Infect. Immun. 62:290-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane, R. S., J. Piesman, and W. Burgdorfer. 1991. Lyme borreliosis: relation of its causative agent to its vectors and hosts in North America and Europe. Annu. Rev. Entomol. 36:587-609. [DOI] [PubMed] [Google Scholar]
- 25.Liang, F. T., M. B. Jacobs, L. C. Bowers, and M. T. Philipp. 2002. An immune evasion mechanism for spirochetal persistence in Lyme borreliosis. J. Exp. Med. 195:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marconi, R. T., S. Y. Sung, C. A. Norton Hughes, and J. A. Carlyon. 1996. Molecular and evolutionary analyses of a variable series of genes in Borrelia burgdorferi that are related to ospE and ospF, constitute a gene family, and share a common upstream homology box. J. Bacteriol. 178:5615-5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McDowell, J. V., S. Y. Sung, M. Labandeira-Rey, J. T. Skare, and R. T. Marconi. 2001. Analysis of mechanisms associated with loss of infectivity of clonal populations of Borrelia burgdorferi B31MI. Infect. Immun. 69:3670-3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montgomery, R. K., S. E. Malawista, K. J. M. Feen, and L. K. Bockenstedt. 1996. Direct demonstration of antigenic substitution of Borrelia burgdorferi ex vivo: exploration of the paradox of the early immune response to outer surface proteins A and C in Lyme disease. J. Exp. Med. 183:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nguyen, T.-P. K., T. T. Lam, W. W. Barthold, S. R. Telford III, R. A. Flavell, and E. Fikrig. 1994. Partial destruction of Borrelia burgdorferi within ticks that engorged on OspE- or OspF-immunized mice. Infect. Immun. 62:2079-2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohnishi, J., J. Piesman, and A. de Silva. 2001. Antigenic and genetic heterogeneity of Borrelia burgdorferi populations transmitted by ticks. Proc. Natl. Acad. Sci. USA 98:670-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orloski, K. A., E. B. Hayes, G. L. Campbell, and D. T. Dennis. 2000. Surveillance for Lyme disease—United States, 1992-1998. Morbid. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-9. [PubMed] [Google Scholar]
- 32.Pal, U., A. M. de Silva, R. R. Montgomery, D. Fish, J. Anguita, J. F. Anderson, Y. Lobet, and E. Fikrig. 2000. Attachment of Borrelia burgdorferi within Ixodes scapularis mediated by outer surface protein A. J. Clin. Investig. 106:561-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parveen, N., and J. M. Leong. 2000. Identification of a candidate glycosaminoglycan-binding adhesin of the Lyme disease spirochete Borrelia burgdorferi. Mol. Microbiol. 35:1220-1234. [DOI] [PubMed] [Google Scholar]
- 34.Porcella, S. F., T. G. Popova, D. R. Akins, M. Li, J. D. Radolf, and M. V. Norgard. 1996. Borrelia burgdorferi supercoiled plasmids encode multiple tandem open reading frames and a lipoprotein gene family. J. Bacteriol. 178:3293-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pugsley, A. P. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Radolf, J. D., M. S. Goldberg, K. W. Bourell, S. I. Baker, J. D. Jones, and M. V. Norgard. 1995. Characterization of outer membranes isolated from Borrelia burgdorferi, the Lyme disease spirochete. Infect. Immun. 63:2154-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ribeiro, J. M., T. N. Mather, J. Piesman, and A. Spielman. 1987. Dissemination and salivary delivery of Lyme disease spirochetes in vector ticks (Acari: Ixodae). J. Med. Entomol. 24:201-205. [DOI] [PubMed] [Google Scholar]
- 38.Schwan, T. G. 1996. Ticks and Borrelia: model systems for investigating pathogen-arthropod interactions. Infect. Agents Dis. 5:167-181. [PubMed] [Google Scholar]
- 39.Schwan, T. G., K. K. Kime, M. E. Schrumpf, J. E. Coe, and W. J. Simpson. 1989. Antibody response in white-footed mice (Peromyscus leucopus) experimentally infected with the Lyme disease spirochete (Borrelia burgdorferi). Infect. Immun. 57:3445-3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwan, T. G., and J. Piesman. 2000. Temporal changes in outer surface proteins A and C of the Lyme disease-associated spirochete, Borrelia burgdorferi, during the chain of infection in ticks and mice. J. Clin. Microbiol. 38:382-388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steere, A. C. 1989. Lyme disease. N. Engl. J. Med. 321:586-597. [DOI] [PubMed] [Google Scholar]
- 43.Steere, A. C., R. L. Grodzicki, J. E. Craft, M. Shrestha, A. N. Kornblatt, and S. E. Malawista. 1984. Recovery of Lyme disease spirochetes from patients. Yale J. Biol. Med. 57:557-560. [PMC free article] [PubMed] [Google Scholar]
- 44.Stevenson, B., J. L. Bono, T. G. Schwan, and P. A. Rosa. 1998. Borrelia burgdorferi Erp proteins are immunogenic in mammals infected by tick bite, and their synthesis is inducible in cultured bacteria. Infect. Immun. 66:2648-2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stevenson, B., S. Casjens, and P. Rosa. 1998. Evidence of past recombination events among the genes encoding the Erp antigens of Borrelia burgdorferi. Microbiology 144:1869-1879. [DOI] [PubMed] [Google Scholar]
- 46.Stevenson, B., N. El-Hage, M. A. Hines, J. C. Miller, and K. Babb. 2002. Differential binding of host complement inhibitor factor H by Borrelia burgdorferi Erp surface proteins: a possible mechanism underlying the expansive host range of Lyme disease spirochetes. Infect. Immun. 70:491-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suk, K., S. Das, W. Sun, B. Jwang, S. W. Barthold, R. A. Flavell, and E. Fikrig. 1995. Borrelia burgdorferi genes selectively expressed in the infected host. Proc. Natl. Acad. Sci. USA 92:4269-4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sung, S.-Y., J. V. McDowell, J. A. Carlyon, and R. T. Marconi. 2000. Mutation and recombination in the upstream homology box-flanked ospE-related genes of the Lyme disease spirochetes result in the development of new antigenic variants during infection. Infect. Immun. 68:1319-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wallich, R., C. Brenner, M. D. Kramer, and M. M. Simon. 1995. Molecular cloning and immunological characterization of a novel linear-plasmid-encoded gene, pG, of Borrelia burgdorferi expressed only in vivo. Infect. Immun. 63:3327-3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang, X., T. G. Popova, K. E. Hagman, S. K. Wikel, G. B. Schoeler, M. J. Caimano, J. D. Radolf, and M. V. Norgard. 1999. Identification, characterization, and expression of three new members of the Borrelia burgdorferi Mlp (2.9) lipoprotein gene family. Infect. Immun. 67:6008-6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, J. R., J. M. Hardham, A. G. Barbour, and S. J. Norris. 1997. Antigenic variation in Lyme disease borreliae by promiscuous recombination of Vmp-like sequence cassettes. Cell 89:275-285. [DOI] [PubMed] [Google Scholar]