Abstract
Salmonella lipid A is inactive in human macrophages despite being potently active in murine macrophages. We investigated the molecular basis for this species-specific action of Salmonella lipid A. When murine CD14 (mCD14), mTLR4, and mMD-2 were all expressed in human monocytic THP-1 cells, these cells were capable of responding to Salmonella lipid A. Expressing each of these proteins separately did not impart such responsiveness. Expression of mTLR4 plus mMD-2, but not mCD14 plus mTLR4 or mCD14 plus mMD-2, conferred this responsiveness. In THP-1 cells expressing mCD14, mTLR4, and mMD-2, replacing mCD14 with human CD14 had no effect on responsiveness to Salmonella lipid A or synthetic Salmonella-type lipid A (compound 516). When mTLR4 was replaced with human TLR4, the responses to these lipid A preparations were decreased to half, and the replacement of mMD-2 decreased responsiveness to one-third, although the responses to Escherichia coli lipid A or synthetic E. coli-type lipid A (compound 506) were not affected. These results indicate that both TLR4 and MD-2 participate in the species-specific action of Salmonella lipid A.
Bacterial lipopolysaccharide (LPS) is a constituent of the outer membrane of the cell wall of gram-negative bacteria and plays a major role in septic shock in humans (24, 29). The lipid A portion has been identified as the active center responsible for most of the LPS-induced biological effects (17, 24). Lipid A species from Escherichia coli and Salmonella are well characterized in terms of their structures and biological activities (3, 4, 6, 10, 13, 14). Both E. coli-type (compounds called 506) and Salmonella-type (compounds called 516) lipid A molecules have been synthesized chemically (8, 9, 15) and have been demonstrated to be highly active in all test systems examined, although the activity of Salmonella-type lipid A was slightly less than that of E. coli-type lipid A.
CD14 has been recognized as a receptor for LPS (5, 31). However, since CD14 does not possess an intracellular domain, functional receptors transmitting LPS signaling into intracellular components have been sought, and the Toll-like receptor (TLR) 4 (1, 28) was found to confer LPS responsiveness. Toll-like receptors are mammalian homologues of the Drosophila Toll protein (18). TLR4 was initially recognized as a molecule which increases constitutive NF-κB but not LPS-inducible activity (19). However, the finding of a novel accessory molecule, MD-2 (25), which confers LPS responsiveness on TLR4, and analyses of TLR4-deficient mice (7, 20, 22, 26, 30) have provided strong evidence for involvement of the TLR4/MD-2 complex in LPS signaling. Although MD-2 is reported to associate with TLR4 (25), the molecular mechanism by which MD-2 participates in LPS signaling via TLR4 has not been clarified.
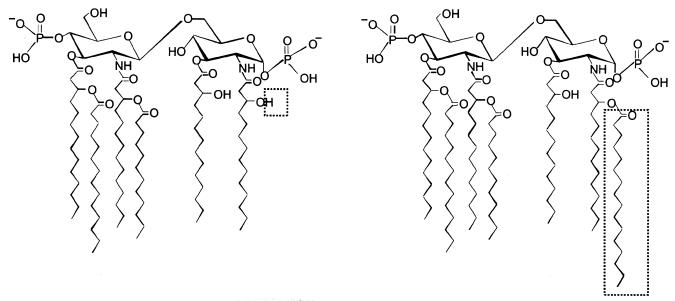
We previously reported that lipid A preparations from various Salmonella strains and synthetic Salmonella-type lipid A (compound 516) possess very little stimulatory activity in human macrophages, despite being potently active in murine macrophages (27). On the other hand, lipid A preparations from E. coli and synthetic E. coli-type lipid A (compound 506) were equally active in human and murine macrophages (27). The only structural difference between the lipid A molecules typically found in these lipid A preparations is the hexadecanoyl acid attached to the hydroxy residue of 3-hydroxy tetradecanoic acid bound to position 2 of the reducing glucosamine (Fig. 1). Thus, human monocytes/macrophages appear capable of discriminating this fine structural difference. To explore the mechanism of this discriminating recognition, we investigated in the present study which molecule of the aforementioned LPS signal transducers participates in this species-specific action of Salmonella lipid A and found that the acquisition of responsiveness is associated with the murine TLR4/MD-2 complex.
FIG. 1.
Chemical structures of lipid A molecules. Structures of E. coli-type (compound 506) (left) and Salmonella-type (compound 516) (right) lipid A molecules are shown. The only difference between the structures of 506 and 516 is indicated by the dotted rectangles.
MATERIALS AND METHODS
Cell culture and reagents.
The human monocyte-like cell line THP-1 (obtained from the Human Science Research Resources Bank, Tokyo, Japan) was grown in RPMI 1640 (Gibco BRL, Rockville, Md.) supplemented with 10% (vol/vol) heat-inactivated fetal calf serum (FCS; Gibco BRL), penicillin (100 U/ml), and streptomycin (100 μg/ml). Diphosphoryl lipid A from E. coli F-583 was obtained from Sigma (St. Louis, Mo.). Salmonella lipid A was prepared from Salmonella enterica serovar Abortus-equi as described previously (27). We confirmed the absence of TLR2-stimulatory activity in both lipid A preparations by checking NF-κB-dependent reporter activity in 293 cells transfected with CD14 and TLR2. Synthetic E. coli-type lipid A (compound 506) and Salmonella-type lipid A (compound 516) were kind gifts of Daiichi Kagaku (Tokyo, Japan).
Expression plasmids.
Plasmids containing human CD14 (hCD14) and mouse CD14 (mCD14) cDNAs were provided by Shunsuke Yamamoto (Medical College of Oita, Oita, Japan). The coding regions of hTLR4, mTLR4, and hMD-2 were amplified by reverse transcription-PCR from total RNA prepared from human spleen (OriGene Technologies, Rockville, Md.), murine fibroblast L929 cells, and THP-1 cells, respectively. The coding region of mMD-2 was amplified from a mouse embryo cDNA library (Clontech, Palo Alto, Calif.). Each PCR product was cloned into a mammalian expression vector, pcDNA3 (Invitrogen, Carlsbad, Calif.). The coding regions of all constructs described above minus their respective signal peptide sequences were subcloned into the downstream region of a modified pcDNA3 vector, in which the preprotrypsin signal peptide sequence precedes the NH2-terminal EIAV-tag epitope (amino acid sequence, ADRRIPGTAEE). A similar LPS response was observed in 293 cells transfected with either tagged or untagged versions of the plasmids described above. The PCR primers used for constructing the plasmids are listed in Table 1. An NF-κB-dependent luciferase reporter plasmid, pELAM-L, was constructed by inserting the PCR fragment (−730 to +52) of the E-selectin (ELAM-1) promoter (23) into the SacI-HindIII site of the pGL3-Basic vector (Promega, Madison, Wis.).
TABLE 1.
Primers used for PCR
| Gene | Orientationa | Sequence (5′ to 3′)b |
|---|---|---|
| hTLR4 | upper-1 | AT GGA TCC ACC ATG ATG TCT GCC TCG CGC CTG |
| upper-2 | AC CGA AGC TTG TCC TGC GTG AGA CCA GAA AGC TG | |
| lower | TAA ATT CTC GAGTCA GAT AGA TGT TGC TTC CTG CCA | |
| mTLR4 | upper-1 | GAA GGA TCCACC ATG ATG CCT CCC TGG CTC CTG |
| upper-2 | GGG GTA CCG TCC TGC CTG ACA CCA GGA AGC TTG | |
| lower | C AGA GTT TGC GGC CGCTCA GGT CCA AGT TGC CGT TTC TTG | |
| hMD-2 | upper-1 | G GAA TTC ACC ATG TTA CCA TTT CTG TTT TTT TCC ACC |
| upper-2 | ACC GAA GCT TTG GAA GCT CAG AAG CAG TAT TGG GTC | |
| lower | GC TCT AGACTA ATT TGA ATT AGG TTG GTG TAG GAT | |
| mMD-2 | upper-1 | G GAA TTC ACC ATG TTG CCA TTT ATT CTC TTT TCG ACG |
| upper-2 | AT TAA AGC TTG GAA TCT GAG AAG CAA CAG TGG | |
| lower | CCG CTC TAG ATT TTT TTT TTT TTT TTT Tc |
The upper-1 and upper-2 primers were used to amplify the entire coding region and the coding region minus the signal peptide sequence, respectively.
Sequences in italics and boldface indicate restriction sites created and the respective coding regions, respectively.
Since the 3′ portion of the mMD-2 sequence had not been determined at the time of creation, an oligo(dT) primer was used.
NF-κB reporter assay.
THP-1 cells (2 × 106/well) were plated in six-well dishes and induced to differentiate by 100 ng of phorbol myristate acetate (Sigma)/ml plus 100 nM 1,25-dihydroxy vitamin D3 (Wako Pure Chemical Industries, Tokyo, Japan) for 3 to 4 days. Following differentiation, the cells were transfected in 1 ml of the culture medium using 3 μl of FuGene 6 transfection reagent (Roche Molecular Laboratories, Burlington, N.C.) according to the manufacturer's instructions with 0.1 μg (each) of the indicated expression plasmids, 1 μg of pELAM-L, and 0.1 μg of pRL-TK (Promega) for normalization. After 24 h, the cells were stimulated in the culture medium containing 10% FCS for 6 h, and reporter gene activity was measured according to the manufacturer's instructions.
RESULTS
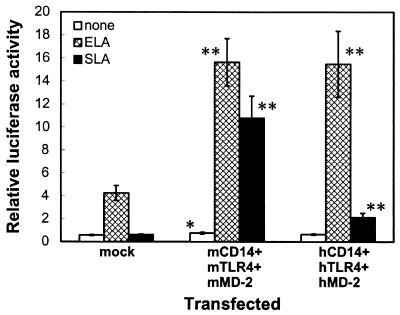
Expression of mCD14, mTLR4, and mMD-2 imparts responsiveness to Salmonella lipid A.
Lipid A preparations from Salmonella induce IκB-α degradation in mouse macrophages but not in human macrophages (27). To explore the molecular basis for this species-specific action of Salmonella lipid A, we first transfected human macrophage THP-1 cells with an NF-κB-dependent luciferase reporter plasmid and then stimulated them with either an E. coli or a Salmonella lipid A preparation. Stimulation with E. coli lipid A caused an approximately sevenfold increase in reporter activity, whereas stimulation with Salmonella lipid A did not increase this activity (Fig. 2). When mCD14, mTLR4, and mMD-2 were all expressed in THP-1 cells, not only was the response to E. coli lipid A increased, but the cells also became responsive to Salmonella lipid A (Fig. 2). Expression of these human proteins also increased the response to E. coli lipid A, whereas the response to Salmonella lipid A was only slightly increased (Fig. 2).
FIG. 2.
Expression of mCD14, mTLR4, and mMD-2 imparts responsiveness to Salmonella lipid A. THP-1 cells were transiently transfected with either a control vector alone (0.3 μg) (mock) or plasmids for either mCD14, mTLR4, and mMD-2 or hCD14, hTLR4, and hMD-2 (0.1 μg each) together with the pELAM-L luciferase reporter plasmid. After 24 h, the cells were stimulated for 6 h without (none) or with 10 ng of E. coli lipid A/ml (ELA) or 100 ng of Salmonella lipid A/ml (SLA) in the presence of 10% (vol/vol) FCS, and the luciferase activities were measured. The values are means ± standard errors of the means from at least 17 independent experiments. ∗, P < 0.05; ∗∗, P < 0.01 (compared with the respective responses in mock-transfected cells by two-tailed Student's t test).
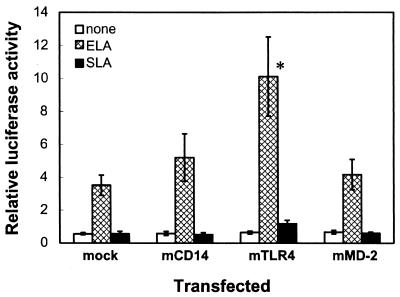
Expression of mCD14, mTLR4, or mMD-2 alone does not impart responsiveness to Salmonella lipid A.
To determine which molecule of mCD14, mTLR4, and mMD-2 is involved in the acquisition of the responsiveness to Salmonella lipid A, we expressed each of these molecules in THP-1 cells and then stimulated them with an E. coli or a Salmonella lipid A preparation (Fig. 3). Expression of mCD14 or mMD-2 did not significantly affect responses to the two lipid A preparations. Expression of mTLR4 clearly increased the response to E. coli lipid A, whereas the response to Salmonella lipid A was not significantly increased. This indicates that the acquisition of responsiveness to Salmonella lipid A cannot be explained by a single molecule.
FIG. 3.
Expression of mCD14, mTLR4, or mMD-2 alone does not impart responsiveness to Salmonella lipid A. THP-1 cells were transiently transfected with either a control vector (mock) or a plasmid for mCD14, mTLR4, or mMD-2 (all 0.1 μg) together with the pELAM-L luciferase reporter plasmid. After 24 h, the cells were stimulated for 6 h without (none) or with 10 ng of E. coli lipid A/ml (ELA) or 100 ng of Salmonella lipid A/ml (SLA) in the presence of 10% (vol/vol) FCS, and the luciferase activities were measured. The values are means ± standard errors of the means from seven independent experiments. ∗, P < 0.05 compared with the respective responses in mock-transfected cells by two-tailed Student's t test.
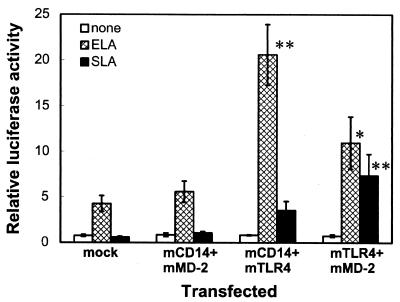
Expression of both mTLR4 and mMD-2 imparts responsiveness to Salmonella lipid A.
Since mCD14, mTLR4, or mMD-2 alone did not impart responsiveness to Salmonella lipid A, we expressed two of these proteins in THP-1 cells, and the reporter activity in response to each lipid A preparation was examined (Fig. 4). Expression of mCD14 plus mMD-2 did not affect responses to either of the lipid A preparations. Expression of mCD14 plus mTLR4 markedly enhanced the response to E. coli lipid A, and these cells were capable of responding to Salmonella lipid A to some extent. In contrast, expression of mTLR4 plus mMD-2 greatly increased the response to Salmonella lipid A, whereas only a twofold increase was observed in the response to E. coli lipid A. This indicates that both mTLR4 and mMD-2 are required to impart responsiveness to Salmonella lipid A.
FIG. 4.
Expression of both mTLR4 and mMD-2 imparts responsiveness to Salmonella lipid A. THP-1 cells were transiently transfected with either a control vector alone (mock) or plasmids for mCD14 and mMD-2, mCD14 and mTLR4, or mTLR4 and mMD-2 (0.1 μg each) together with the pELAM-L luciferase reporter plasmid. After 24 h, the cells were stimulated for 6 h without (none) or with 10 ng of E. coli lipid A/ml (ELA) or 100 ng of Salmonella lipid A/ml (SLA) in the presence of 10% (vol/vol) FCS, and the luciferase activities were measured. The values are means ± standard errors of the means from at least four independent experiments. ∗, P < 0.05; ∗∗, P < 0.01 (compared with the respective responses in mock-transfected cells by two-tailed Student's t test).
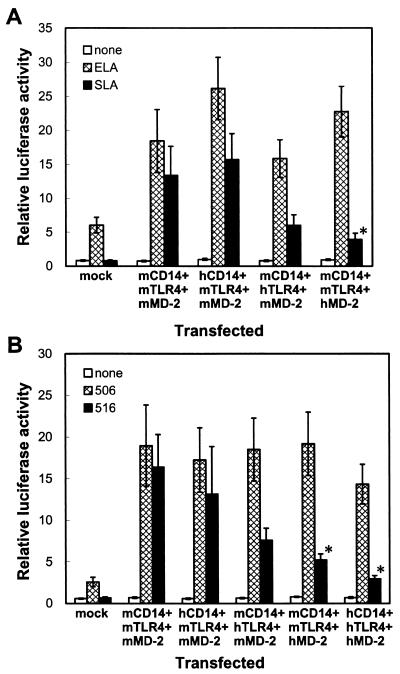
Replacement of mMD-2 with hMD-2 severely impaired responsiveness to Salmonella lipid A.
To further explore whether mTLR4 or mMD-2 is essential for responsiveness to Salmonella lipid A, mCD14, mTLR4, or mMD-2 was replaced with the respective human protein instead of expressing all three mouse proteins, and reporter activities in response to these lipid A preparations, as well as synthetic E. coli-type lipid A (compound 506) and Salmonella-type lipid A (compound 516), were examined in THP-1 cells (Fig. 5). Upon expression of mCD14, mTLR4, and mMD-2 in THP-1 cells, responses to both the E. coli and the Salmonella lipid A preparations were greatly increased, as shown in Fig. 2. Expression of hCD14 instead of mCD14 in THP-1 cells transfected with mTLR4 and mMD-2 did not affect the response to either lipid A preparation. When mTLR4 was replaced with hTLR4, the response to Salmonella lipid A was decreased to some extent, and the response to E. coli lipid A was unaffected. The replacement of mMD-2 with hMD-2 greatly reduced the response to Salmonella lipid A, although the response to E. coli lipid A was not affected (Fig. 5A). Similar results were obtained by using synthetic lipid A compounds instead of lipid A preparations of bacterial origin (Fig. 5B). Taken together, these results indicate that both TLR4 and MD-2 participate in the species-specific action of Salmonella lipid A.
FIG. 5.
Replacement of mMD-2 with hMD-2 severely impaired responsiveness to Salmonella lipid A. THP-1 cells were transiently transfected with either a control vector alone (mock) or plasmids for mCD14, mTLR4, and mMD-2; hCD14, mTLR4, and mMD-2; mCD14, hTLR4, and mMD-2; mCD14, mTLR4, and hMD-2; or hCD14, hTLR4, and hMD-2 (0.1 μg each) together with the pELAM-L luciferase reporter plasmid. After 24 h, the cells were stimulated for 6 h without (none) or with 10 ng of E. coli lipid A/ml (ELA) or 100 ng of Salmonella lipid A/ml (SLA) (A) or 3 ng of synthetic E. coli-type lipid A/ml (compound 506) or 30 ng of synthetic Salmonella-type lipid A/ml (compound 516) (B) in the presence of 10% (vol/vol) FCS, and the luciferase activities were measured. The values are means ± standard errors of the means from at least five independent experiments. ∗, P < 0.05 compared with the respective responses in cells transfected with plasmids for mCD14, mTLR4, and mMD-2 by two-tailed Student's t test (comparison of responses between mock-transfected cells and cells transfected with plasmids for mCD14, mTLR4, and mMD-2 was not performed).
DISCUSSION
The structures of lipid A molecules found in E. coli and Salmonella lipid A preparations have been characterized chemically (27). The only structural difference between the lipid A molecules typically found in these lipid A preparations is the hexadecanoyl acid attached to the hydroxy residue of 3-hydroxy tetradecanoic acid bound to position 2 of the reducing glucosamine (Fig. 1). Typical E. coli-type and Salmonella-type lipid A molecules possess six and seven fatty acids, respectively (27). In contrast to this slight structural difference, the activities of these lipid A molecules are clearly different in human macrophages. We previously demonstrated that lipid A preparations from various Salmonella strains and synthetic Salmonella-type lipid A (compound 516) possess very little stimulatory activity in human macrophages, while they are potently active in murine macrophages. Furthermore, lipid A preparations from E. coli and synthetic E. coli-type lipid A (compound 506) were equally active in both human and murine macrophages (27). Similar species-specific actions were also reported with a tetraacylated lipid A precursor (also called compound 406 or lipid IVa) and Rhodobacter sphaeroides lipid A (2, 16, 21). In these studies, Poltorak et al. (21), and Lien et al. (16) investigated species specificity by transfecting TLR4 proteins from different species. They found that expression of only mTLR4 in immortalized C3H/HeJ macrophages or THP-1 cells was enough to confer responsiveness to 406 and concluded that TLR4 was responsible for species specificity, although they did not examine the involvement of MD-2. In our present study, expression of only mTLR4 in THP-1 was not enough to confer responsiveness to Salmonella lipid A, and mMD-2 with the aid of mTLR4 was required for the acquisition of responsiveness. Therefore, the species-specific action of Salmonella lipid A is clearly different from that of 406.
Taxol has also been reported to exhibit species-specific actions. Taxol has been shown to possess LPS-mimetic activities in mouse macrophages but not in human macrophages (11). Kawasaki et al. (12) showed Gln22 of mMD-2 to be essential for taxol signaling but not for LPS signaling. We also examined the effects of E. coli and Salmonella lipid A preparations on THP-1 cells expressing an mMD-2 mutant, in which Gln22 was replaced with Tyr, together with mCD14 and mTLR4 and found that these cells still responded to Salmonella lipid A (data not shown). In addition, it has been reported (11) that CD14, which is required for lipid A responsiveness, was not required for taxol responsiveness. Therefore, the site of action of Salmonella lipid A is clearly different from that of taxol. We are attempting to narrow the region in MD-2 responsible for the species-specific recognition by using human-mouse chimeric MD-2.
CD14 is known to recognize LPS and lipid A molecules on the macrophage surface, and this is widely believed to be the first step in the LPS signal transduction pathway. However, the species dependency of responsiveness to lipid A molecules is not explained by interspecies differences in CD14 structure because our data and those of others (2) indicate that CD14 is not involved in species dependency. Therefore, species-dependent discrimination of lipid A structures apparently takes place after the binding of lipid A to CD14 has occurred. Poltorak et al. (21) and Lien et al. (16) found that TLR4 is involved in species-specific actions of 406 and R. sphaeroides lipid A and explained this species-dependent discrimination in two ways: TLR4 is capable of discriminating between lipid A structures, or TLR4 discriminates between CD14 molecule conformations induced by the binding of these lipid A molecules. In the present study, we showed MD-2 to be involved in species-dependent discrimination between E. coli and Salmonella lipid A molecules. This suggests that MD-2 is also capable of discriminating among lipid A structures or CD14 molecule conformations induced by these lipid A molecules. Salmonella lipid A may serve as a useful tool for understanding the signaling mechanism which operates between the lipid A/CD14 complex and the TLR4/MD-2 complex.
Acknowledgments
This work was supported in part by grants from the Japan Health Sciences Foundation and from the Ministry of Education of Japan.
We thank Shunsuke Yamamoto (Medical College of Oita, Oita, Japan) for providing plasmids.
Editor: A. D. O'Brien
REFERENCES
- 1.Chow, J. C., D. W. Young, D. T. Golenbock, W. J. Christ, and F. Gusovsky. 1999. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J. Biol. Chem. 274:10689-10692. [DOI] [PubMed] [Google Scholar]
- 2.Delude, R. L., R. J. Savedra, H. Zhao, R. Thieringer, S. Yamamoto, M. J. Fenton, and D. T. Golenbock. 1995. CD14 enhances cellular responses to endotoxin without imparting ligand-specific recognition. Proc. Natl. Acad. Sci. USA 92:9288-9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galanos, C., O. Lüderitz, M. Freudenberg, L. Brade, U. Schade, E. T. Rietschel, S. Kusumoto, and T. Shiba. 1986. Biological activity of synthetic heptaacyl lipid A representing a component of Salmonella minnesota R595 lipid A. Eur. J. Biochem. 160:55-59. [DOI] [PubMed] [Google Scholar]
- 4.Galanos, C., O. Lüderitz, E. T. Rietschel, O. Westphal, H. Brade, L. Brade, M. Freudenberg, U. Schade, M. Imoto, H. Yoshimura, et al. 1985. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 148:1-5. [DOI] [PubMed] [Google Scholar]
- 5.Hailman, E., H. S. Lichenstein, M. M. Wurfel, D. S. Miller, D. A. Johnson, M. Kelley, L. A. Busse, M. M. Zukowski, and S. D. Wright. 1994. Lipopolysaccharide (LPS)-binding protein accelerates the binding of LPS to CD14. J. Exp. Med. 179:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Homma, J. Y., M. Matsuura, S. Kanegasaki, Y. Kawakubo, Y. Kojima, N. Shibukawa, Y. Kumazawa, A. Yamamoto, K. Tanamoto, and T. Yasuda. 1985. Structural requirements of lipid A responsible for the functions: a study with chemically synthesized lipid A and its analogues. J. Biochem. (Tokyo) 98:395-406. [DOI] [PubMed] [Google Scholar]
- 7.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 8.Imoto, M., H. Yoshimura, S. Sakaguchi, S. Kusumoto, and T. Shiba. 1984. Total synthesis of Escherichia coli lipid A. Tetrahedron Lett. 26:1545-1548. [Google Scholar]
- 9.Imoto, M., H. Yoshimura, M. Yamamoto, T. Shimamoto, S. Kusumoto, and T. Shiba. 1984. Chemical synthesis of phosphorylated tetraacyl disaccharide corresponding to a biosynthetic precursor of lipid A. Tetrahedron Lett. 25:2667-2670. [Google Scholar]
- 10.Kanegasaki, S., K. Tanamoto, T. Yasuda, J. Y. Homma, M. Matsuura, M. Nakatsuka, Y. Kumazawa, A. Yamamoto, T. Shiba, and S. Kusumoto. 1986. Structure-activity relationship of lipid A: comparison of biological activities of natural and synthetic lipid A's with different fatty acid compositions. J. Biochem. (Tokyo) 99:1203-1210. [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki, K., S. Akashi, R. Shimazu, T. Yoshida, K. Miyake, and M. Nishijima. 2000. Mouse toll-like receptor 4·MD-2 complex mediates lipopolysaccharide-mimetic signal transduction by Taxol. J. Biol. Chem. 275:2251-2254. [DOI] [PubMed] [Google Scholar]
- 12.Kawasaki, K., K. Gomi, and M. Nishijima. 2001. Gln22 of mouse MD-2 is essential for species-specific lipopolysaccharide mimetic action of taxol. J. Immunol. 166:11-14. [DOI] [PubMed] [Google Scholar]
- 13.Kotani, S., H. Takada, I. Takahashi, M. Tsujimoto, T. Ogawa, T. Ikeda, K. Harada, H. Okamura, T. Tamura, and S. Tanaka. 1986. Low endotoxic activities of synthetic Salmonella-type lipid A with an additional acyloxyacyl group on the 2-amino group of beta (1-6) glucosamine disaccharide 1,4′-bisphosphate. Infect. Immun. 52:872-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kotani, S., H. Takada, M. Tsujimoto, T. Ogawa, I. Takahashi, T. Ikeda, K. Otsuka, H. Shimauchi, N. Kasai, and J. Mashimo. 1985. Synthetic lipid A with endotoxic and related biological activities comparable to those of a natural lipid A from an Escherichia coli Re-mutant. Infect. Immun. 49:225-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kusumoto, S., H. Yoshimura, M. Imoto, T. Shimamoto, and T. Shiba. 1985. Chemical synthesis of 1-dephospho derivative Escherichia coli lipid A. Tetrahedron Lett. 26:909-912. [Google Scholar]
- 16.Lien, E., T. K. Means, H. Heine, A. Yoshimura, S. Kusumoto, K. Fukase, M. J. Fenton, M. Oikawa, N. Qureshi, B. Monks, R. W. Finberg, R. R. Ingalls, and D. T. Golenbock. 2000. Toll-like receptor 4 imparts ligand-specific recognition of bacterial lipopolysaccharide. J. Clin. Investig. 105:497-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lüderitz, O., M. Freudenberg, C. Galanos, E. T. Lehmann, E. T. Rietschel, and D. H. Shaw. 1982. Lipopolysaccharides of gram-negative bacteria. Curr. Top. Membr. Transplant. 17:79-151. [Google Scholar]
- 18.Medzhitov, R., P. Preston-Hurlburt, and C. A. J. Janeway. 1997. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature 388:394-397. [DOI] [PubMed] [Google Scholar]
- 19.Muzio, M., G. Natoli, S. Saccani, M. Levrero, and A. Mantovani. 1998. The human Toll signaling pathway: divergence of nuclear factor κB and JNK/SAPK activation upstream of tumor necrosis factor receptor-associated factor 6 (TRAF6). J. Exp. Med. 187:2097-2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poltorak, A., X. He, I. Smirnova, M. Y. Liu, C. V. Huffel, X. Du, D. Birdwell, E. Alejos, M. Silva, C. Galanos, M. Freudenberg, P. Ricciardi-Castagnoli, B. Layton, and B. Beutler. 1998. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science 282:2085-2088. [DOI] [PubMed] [Google Scholar]
- 21.Poltorak, A., P. Ricciardi-Castagnoli, S. Citterio, and B. Beutler. 2000. Physical contact between lipopolysaccharide and Toll-like receptor 4 revealed by genetic complementation. Proc. Natl. Acad. Sci. USA 97:2163-2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qureshi, S. T., L. Larivire, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp. Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schindler, U., and V. R. Baichwal. 1994. Three NF-kappa B binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol. Cell. Biol. 14:5820-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schletter, J., H. Heine, A. J. Ulmer, and E. T. Rietschel. 1995. Molecular mechanisms of endotoxin activity. Arch. Microbiol. 164:383-389. [DOI] [PubMed] [Google Scholar]
- 25.Shimazu, R., S. Akashi, H. Ogata, Y. Nagai, K. Fukudome, K. Miyake, and M. Kimoto. 1999. MD-2, a molecule that confers lipopolysaccharide responsiveness on Toll-like receptor 4. J. Exp. Med. 189:1777-1782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takeuchi, O., K. Hoshino, T. Kawai, H. Sanjo, H. Takada, T. Ogawa, K. Takeda, and S. Akira. 1999. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity 11:443-451. [DOI] [PubMed] [Google Scholar]
- 27.Tanamoto, K., and S. Azumi. 2000. Salmonella-type heptaacylated lipid A is inactive and acts as an antagonist of lipopolysaccharide action on human line cells. J. Immunol. 164:3149-3156. [DOI] [PubMed] [Google Scholar]
- 28.Tapping, R. I., S. Akashi, K. Miyake, P. J. Godowski, and P. S. Tobias. 2000. Toll-like receptor 4, but not Toll-like receptor 2, is a signaling receptor for Escherichia and Salmonella lipopolysaccharides. J. Immunol. 165:5780-5787. [DOI] [PubMed] [Google Scholar]
- 29.Ulevitch, R. J., and P. S. Tobias. 1995. Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu. Rev. Immunol. 13:437-457. [DOI] [PubMed] [Google Scholar]
- 30.Vogel, S. N., D. Johnson, P. Y. Perera, A. Medvedev, L. Larivire, S. T. Qureshi, and D. Malo. 1999. Functional characterization of the effect of the C3H/HeJ defect in mice that lack an Lpsn gene: in vivo evidence for a dominant negative mutation. J. Immunol. 162:5666-5670. [PubMed] [Google Scholar]
- 31.Wright, S. D., R. A. Ramos, P. S. Tobias, R. J. Ulevitch, and J. C. Mathison. 1990. CD14, a receptor for complexes of lipopolysaccharide (LPS) and LPS binding protein. Science 249:1431-1433. [DOI] [PubMed] [Google Scholar]