Abstract
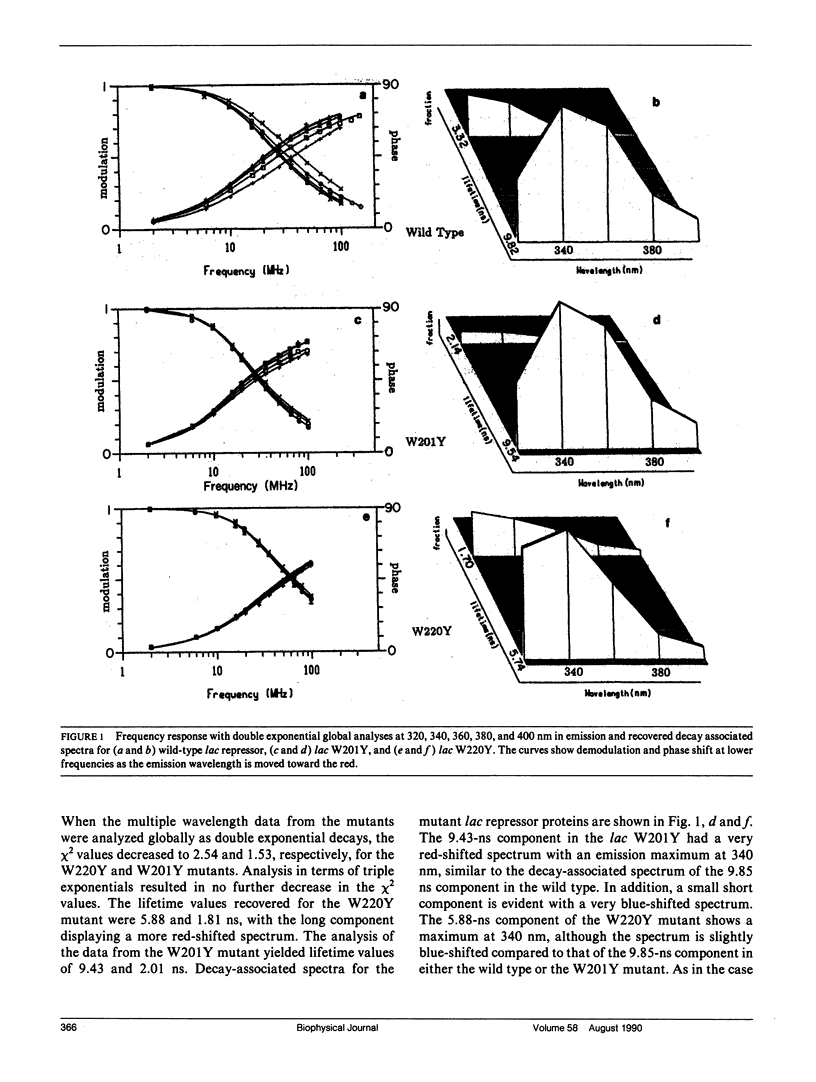
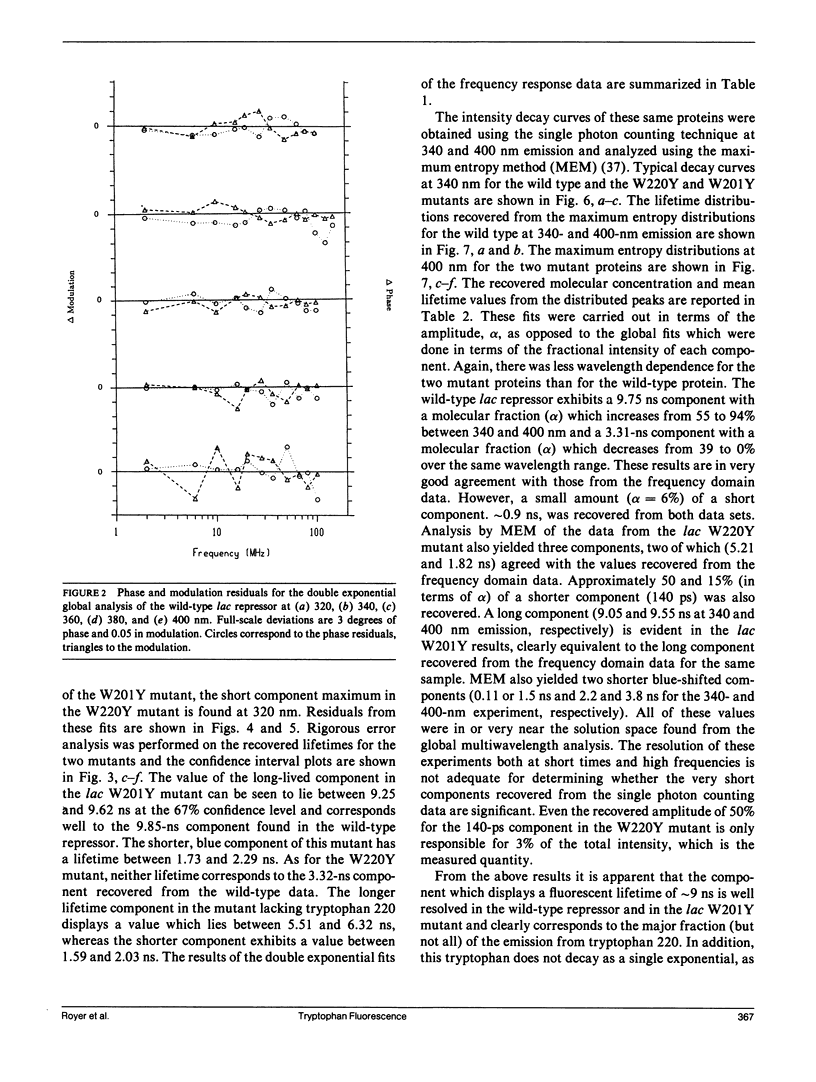
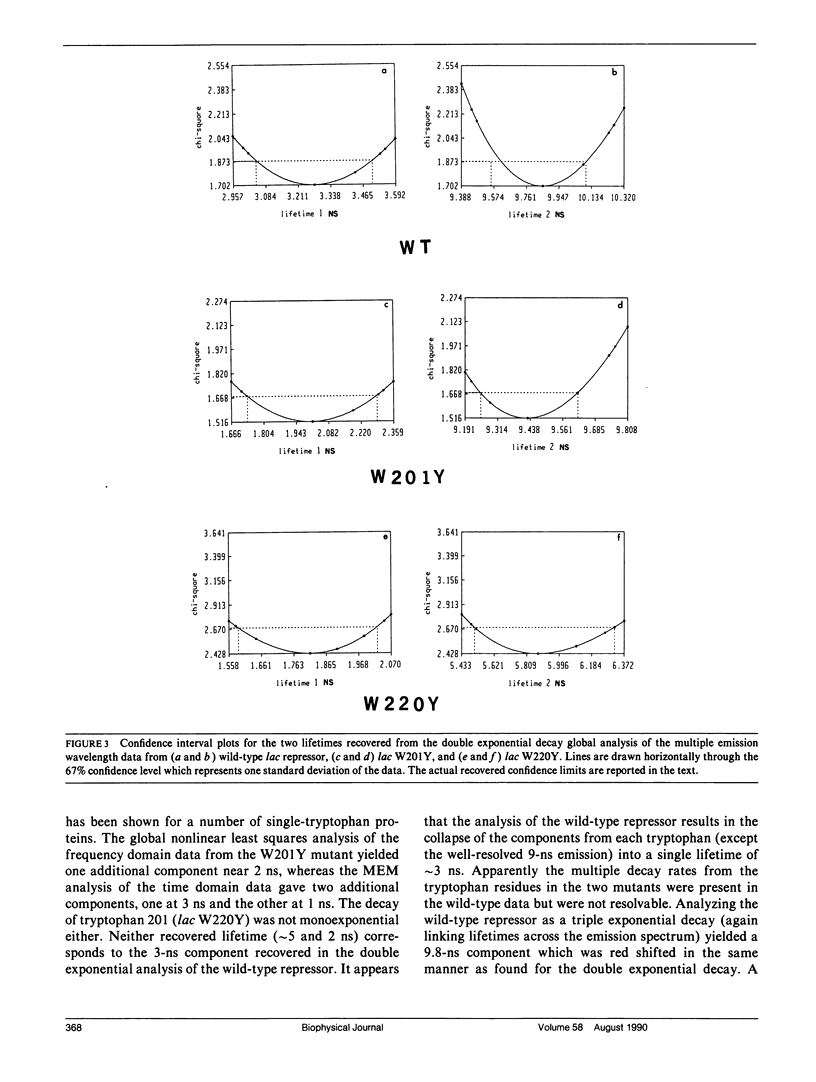
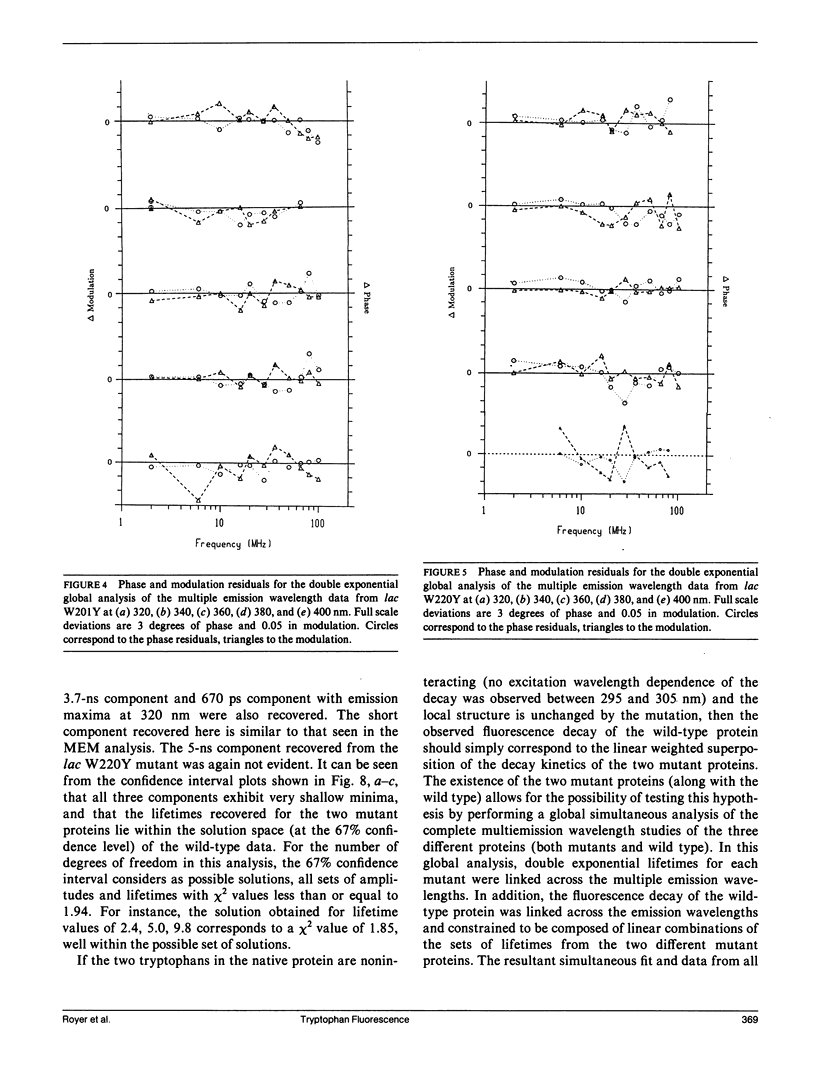
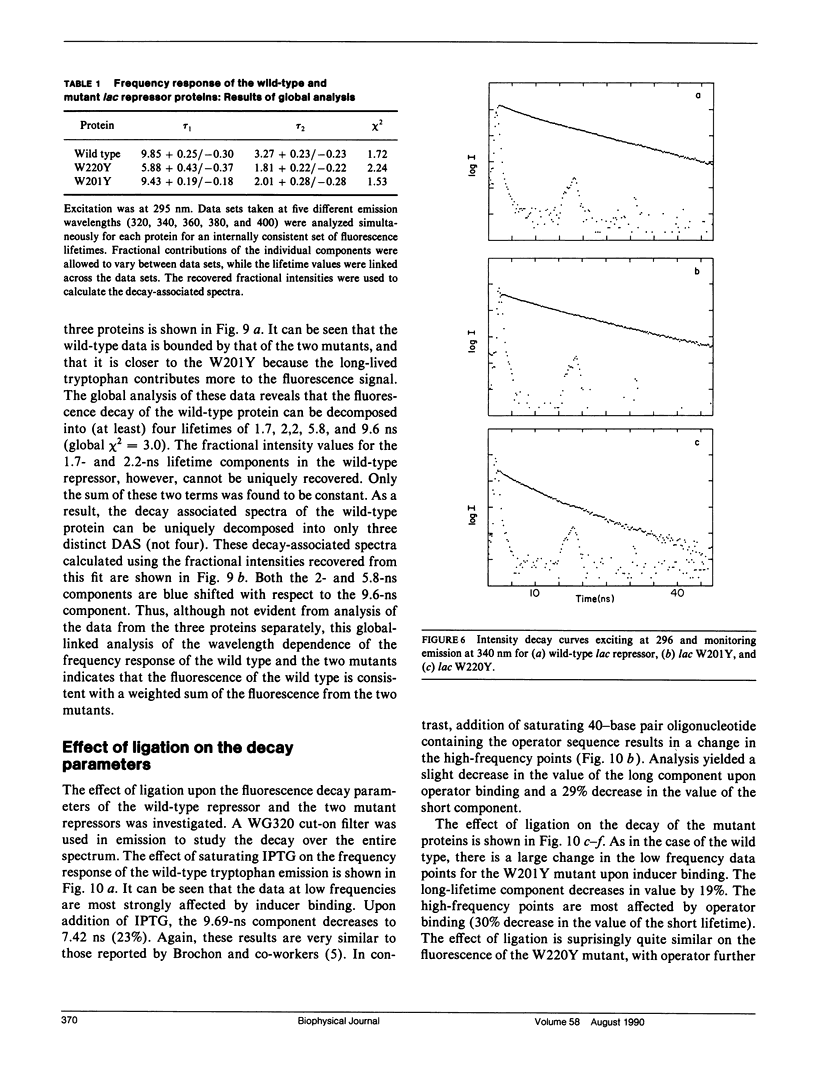
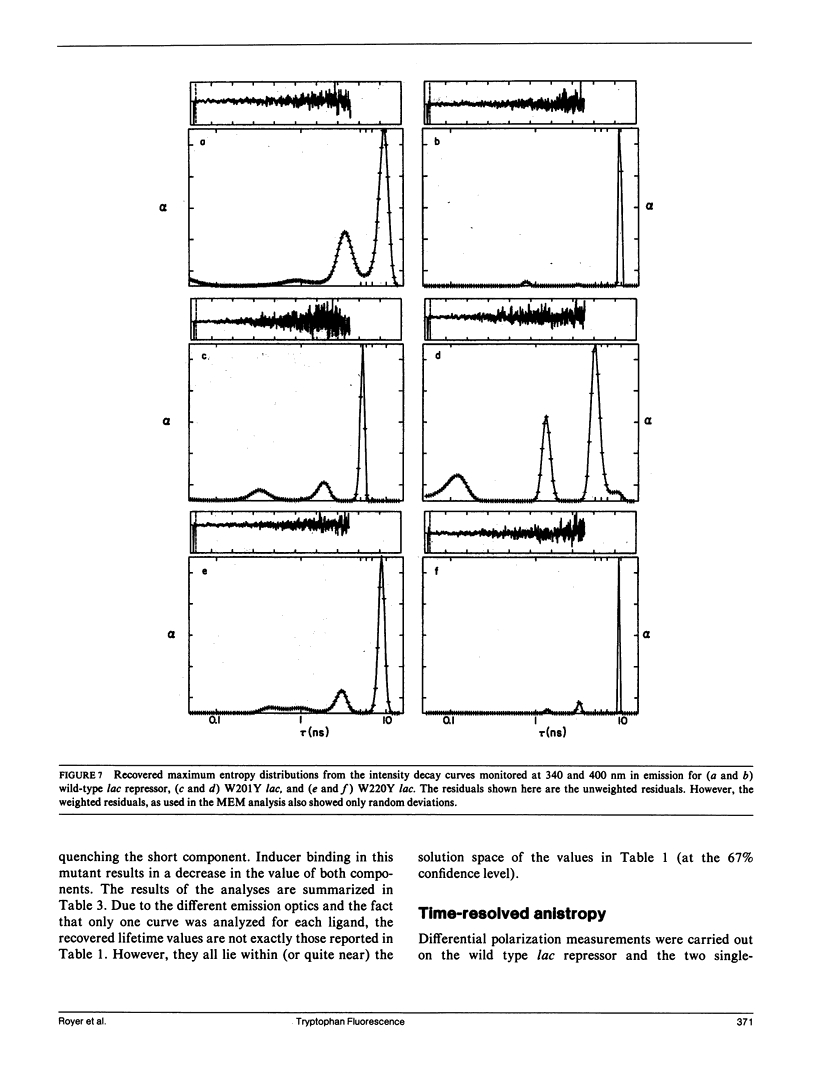
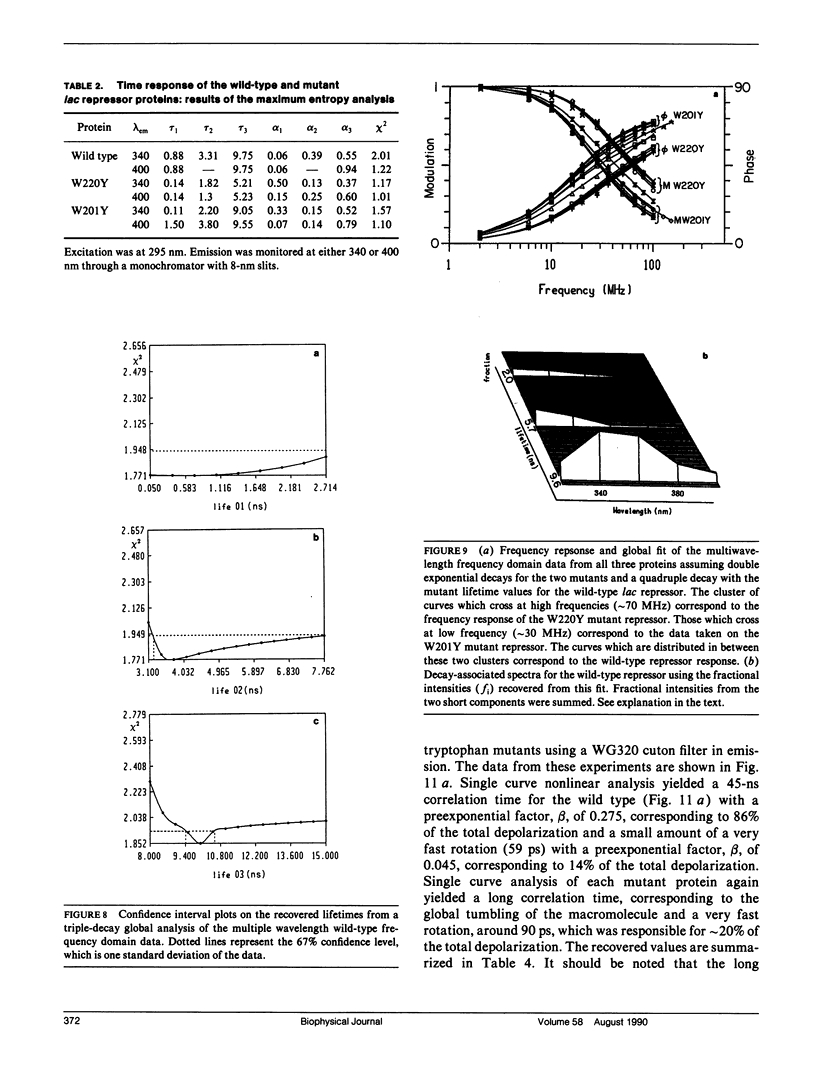
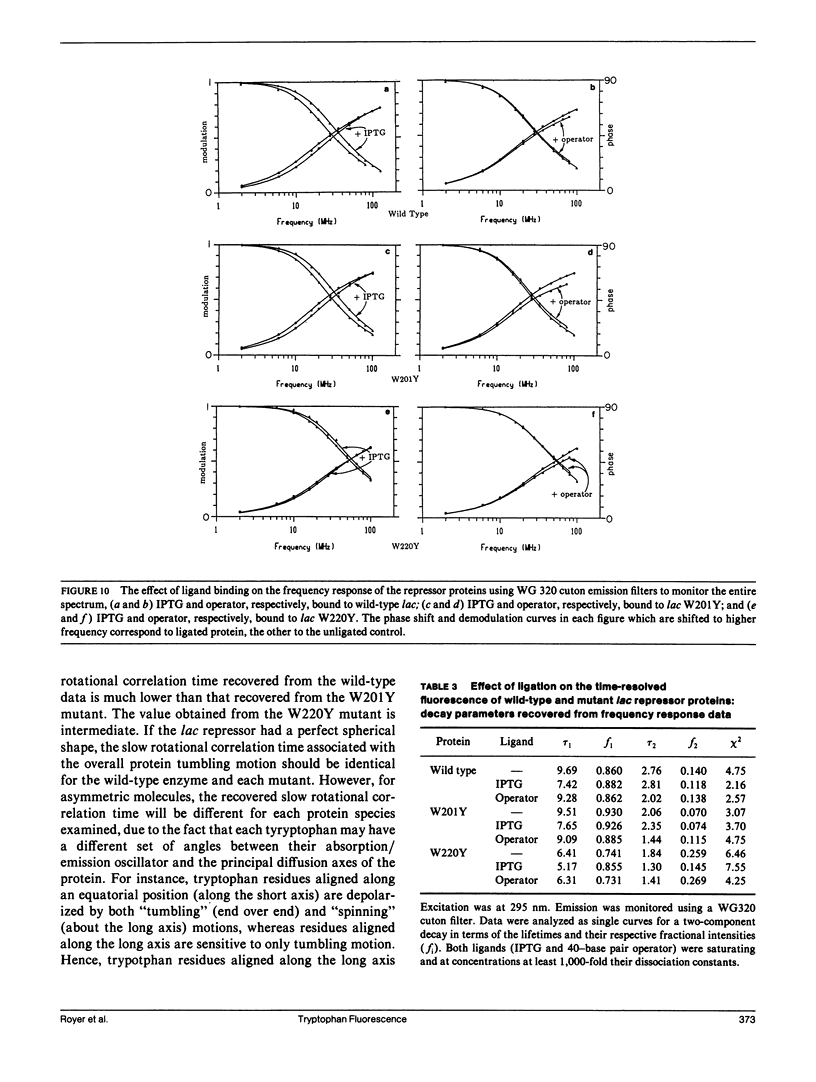
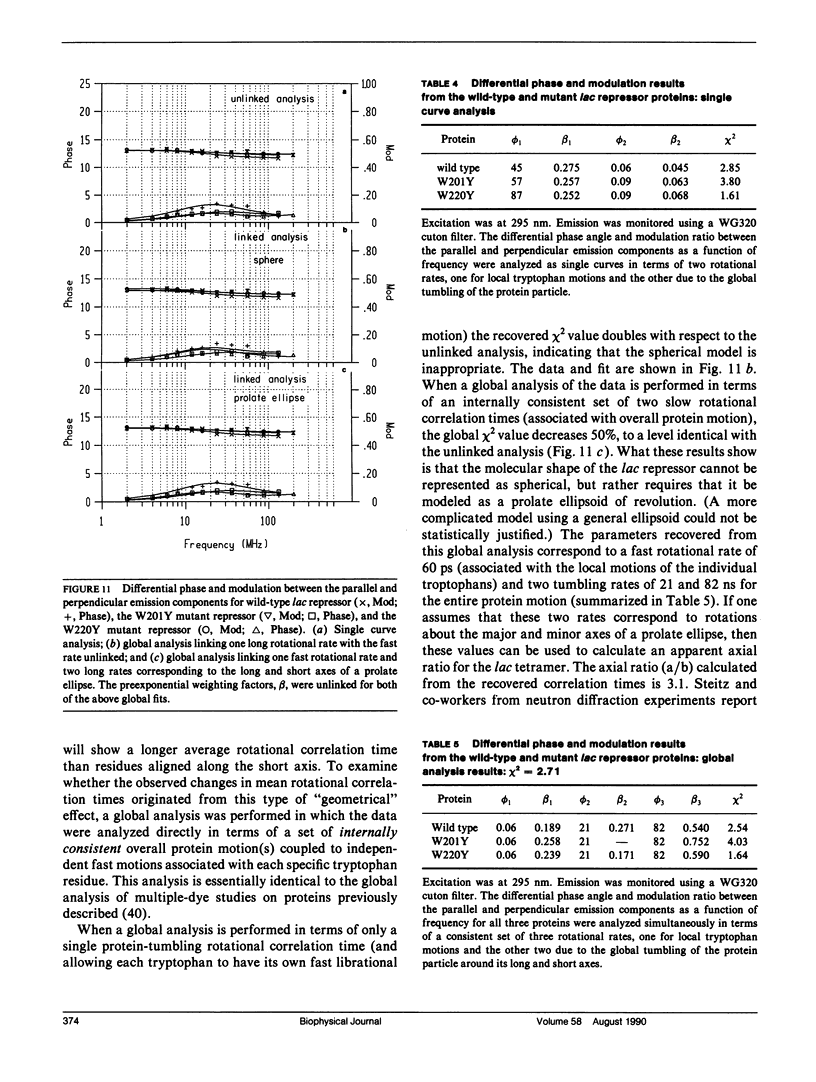
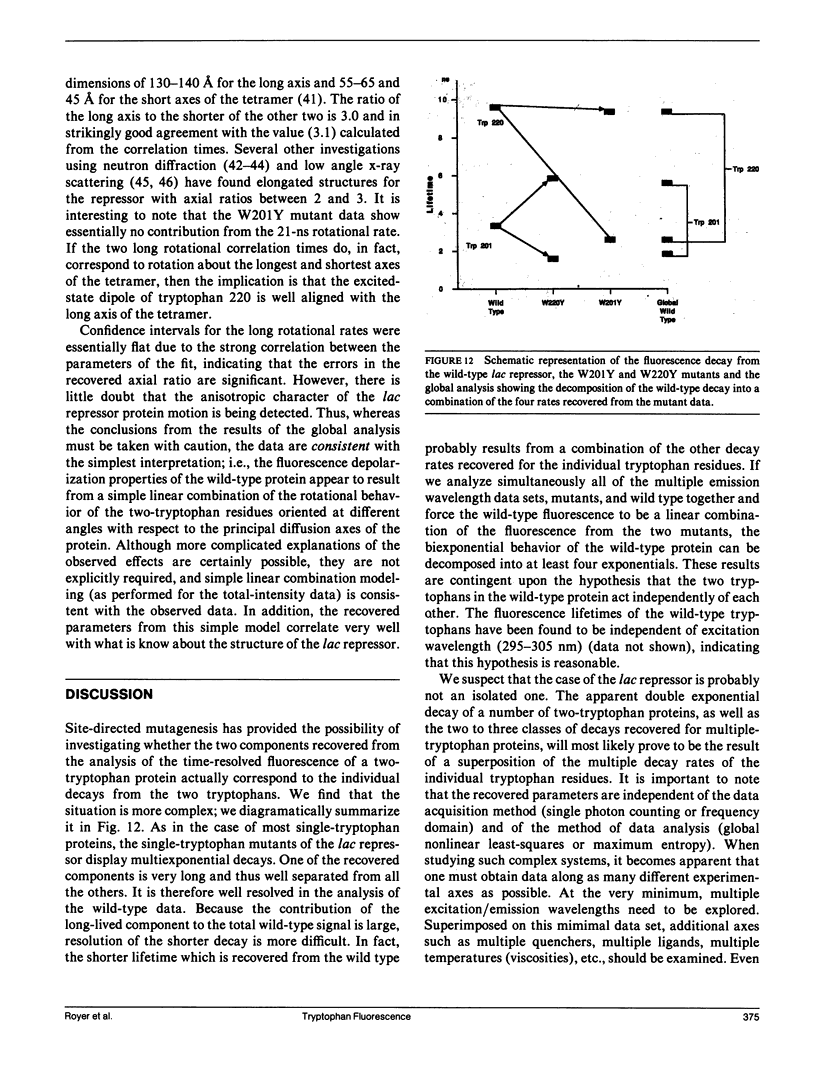
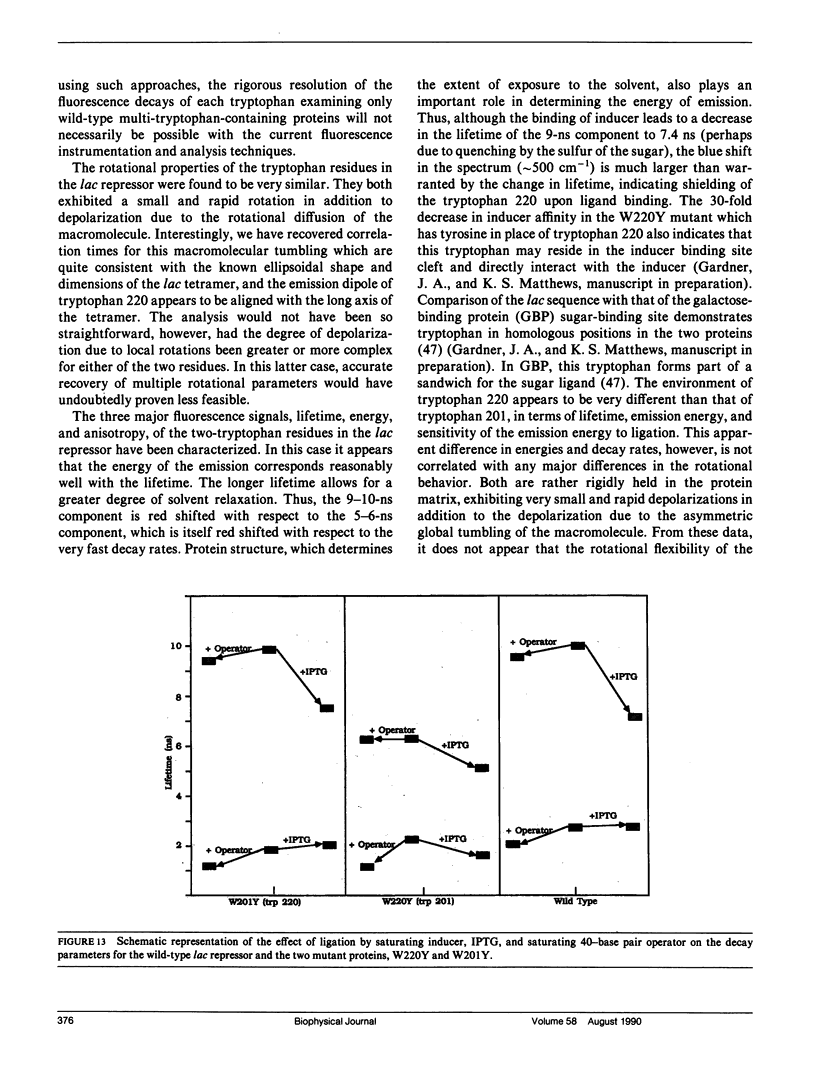
We have studied the time-resolved intrinsic tryptophan fluorescence of the lac repressor (a symmetric tetramer containing two tryptophan residues per monomer) and two single-tryptophan mutant repressors obtained by site-directed mutagenesis, lac W201Y and lac W220Y. These mutant repressor proteins have tyrosine substituted for tryptophan at positions 201 and 220, respectively, leaving a single tryptophan residue per monomeric subunit at position 220 for the W201Y mutant and at position 201 in the W220Y mutant. It was found that the two decay rates recovered from the analysis of the wild type data do not correspond to the rates recovered from the analysis of the decays of the mutant proteins. Each of these residues in the mutant repressors displays at least two decay rates. Global analysis of the multiwavelength data from all three proteins, however, yielded results consistent with the fluorescence decay of the wild type lac repressor corresponding simply to the weighted linear combination of the decays from the mutant proteins. The effect of ligation by the antagonistic ligands, inducer and operator DNA, was similar for all three proteins. The binding of the inducer sugar resulted in a quenching of the long-lived species, while binding by the operator decreased the lifetime of the short components. Investigation of the time-resolved anisotropy of the intrinsic tryptophan fluorescence in these three proteins revealed that the depolarization of fluorescence resulted from a fast motion and the global tumbling of the macromolecule. Results from the simultaneous global analysis of the frequency domain data sets from the three proteins revealed anisotropic rotations for the macromolecule, consistent with the known elongated shape of the repressor tetramer. In addition, it appears that the excited-state dipole of tryptophan 220 is alighed with the long axis of the repressor.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Beechem J. M., Knutson J. R., Brand L. Global analysis of multiple dye fluorescence anisotropy experiments on proteins. Biochem Soc Trans. 1986 Oct;14(5):832–835. doi: 10.1042/bst0140832. [DOI] [PubMed] [Google Scholar]
- Brochon J. C., Wahl P., Charlier M., Maurizot J. C., Hélène C. Time resolved spectroscopy of the tryptophyl fluorescence of the E. coli LAC repressor. Biochem Biophys Res Commun. 1977 Dec 21;79(4):1261–1271. doi: 10.1016/0006-291x(77)91142-1. [DOI] [PubMed] [Google Scholar]
- Charlier M., Maurizot J. C., Zaccai G. Neutron scattering studies of lac repressor. Nature. 1980 Jul 24;286(5771):423–425. doi: 10.1038/286423a0. [DOI] [PubMed] [Google Scholar]
- Charlier M., Maurizot J. C., Zaccai G. Neutron-scattering studies of lac repressor: a low-resolution model. J Mol Biol. 1981 Nov 25;153(1):177–182. doi: 10.1016/0022-2836(81)90533-7. [DOI] [PubMed] [Google Scholar]
- Cockle S. A., Szabo A. G. Time-resolved fluorescence spectra of tryptophan in monomeric glucagon. Photochem Photobiol. 1981 Jul;34(1):23–27. doi: 10.1111/j.1751-1097.1981.tb08955.x. [DOI] [PubMed] [Google Scholar]
- Dente L., Cesareni G., Cortese R. pEMBL: a new family of single stranded plasmids. Nucleic Acids Res. 1983 Mar 25;11(6):1645–1655. doi: 10.1093/nar/11.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafni A., Werber M. M. Ferredoxin from Halobacterium of the Dead Sea. Structural properties revealed by fluorescence techniques. Arch Biochem Biophys. 1979 Sep;196(2):363–370. doi: 10.1016/0003-9861(79)90588-5. [DOI] [PubMed] [Google Scholar]
- Grinvald A., Steinberg I. Z. The fluorescence decay of tryptophan residues in native and denatured proteins. Biochim Biophys Acta. 1976 Apr 14;427(2):663–678. doi: 10.1016/0005-2795(76)90210-5. [DOI] [PubMed] [Google Scholar]
- Hochstrasser R. M., Negus D. K. Picosecond fluorescence decay of tryptophans in myoglobin. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4399–4403. doi: 10.1073/pnas.81.14.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonas A., Privat J. P., Wahl P., Osborne J. C., Jr Nanosecond rotational motions of apolipoprotein C-I in solution and in complexes with dimyristoylphosphatidylcholine. Biochemistry. 1982 Nov 23;21(24):6205–6211. doi: 10.1021/bi00267a028. [DOI] [PubMed] [Google Scholar]
- Knutson J. R., Walbridge D. G., Brand L. Decay-associated fluorescence spectra and the heterogeneous emission of alcohol dehydrogenase. Biochemistry. 1982 Sep 14;21(19):4671–4679. doi: 10.1021/bi00262a024. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., O'Kane D. J., Visser A. J. Spectral properties and function of two lumazine proteins from Photobacterium. Biochemistry. 1985 Mar 12;24(6):1476–1483. doi: 10.1021/bi00327a028. [DOI] [PubMed] [Google Scholar]
- Livesey A. K., Brochon J. C. Analyzing the distribution of decay constants in pulse-fluorimetry using the maximum entropy method. Biophys J. 1987 Nov;52(5):693–706. doi: 10.1016/S0006-3495(87)83264-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay D. B., Pickover C. A., Steitz T. A. Escherichia coli lac repressor is elongated with its operator DNA binding domains located at both ends. J Mol Biol. 1982 Mar 25;156(1):175–183. doi: 10.1016/0022-2836(82)90465-x. [DOI] [PubMed] [Google Scholar]
- Munro I., Pecht I., Stryer L. Subnanosecond motions of tryptophan residues in proteins. Proc Natl Acad Sci U S A. 1979 Jan;76(1):56–60. doi: 10.1073/pnas.76.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mérola F., Rigler R., Holmgren A., Brochon J. C. Picosecond tryptophan fluorescence of thioredoxin: evidence for discrete species in slow exchange. Biochemistry. 1989 Apr 18;28(8):3383–3398. doi: 10.1021/bi00434a038. [DOI] [PubMed] [Google Scholar]
- O'Gorman R. B., Dunaway M., Matthews K. S. DNA binding characteristics of lactose repressor and the trypsin-resistant core repressor. J Biol Chem. 1980 Nov 10;255(21):10100–10106. [PubMed] [Google Scholar]
- Pilz I., Goral K., Kratky O., Bray R. P., Wade-Jardetzky N. G., Jardetzky O. Small-angle X-ray studies of the quaternary structure of the lac repressor from Escherichia coli. Biochemistry. 1980 Aug 19;19(17):4087–4090. doi: 10.1021/bi00558a028. [DOI] [PubMed] [Google Scholar]
- Privat J. P., Wahl P., Auchet J. C., Pain R. H. Time resolved spectroscopy of tryptophyl fluorescence of yeast 3-phosphoglycerate kinase. Biophys Chem. 1980 Apr;11(2):239–248. doi: 10.1016/0301-4622(80)80026-3. [DOI] [PubMed] [Google Scholar]
- Ross J. B., Schmidt C. J., Brand L. Time-resolved fluorescence of the two tryptophans in horse liver alcohol dehydrogenase. Biochemistry. 1981 Jul 21;20(15):4369–4377. doi: 10.1021/bi00518a021. [DOI] [PubMed] [Google Scholar]
- Royer C. A., Tauc P., Hervé G., Brochon J. C. Ligand binding and protein dynamics: a fluorescence depolarization study of aspartate transcarbamylase from Escherichia coli. Biochemistry. 1987 Oct 6;26(20):6472–6478. doi: 10.1021/bi00394a027. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Barrell B. G., Smith A. J., Roe B. A. Cloning in single-stranded bacteriophage as an aid to rapid DNA sequencing. J Mol Biol. 1980 Oct 25;143(2):161–178. doi: 10.1016/0022-2836(80)90196-5. [DOI] [PubMed] [Google Scholar]
- Sommer H., Lu P. Lac Repressor. Fluorescence of the two tryptophans. J Biol Chem. 1976 Jun 25;251(12):3774–3779. [PubMed] [Google Scholar]
- Steitz T. A., Richmond T. J., Wise D., Engelman D. The lac repressor protein: molecular shape, subunit structure, and proposed model for operator interaction based on structural studies of microcrystals. Proc Natl Acad Sci U S A. 1974 Mar;71(3):593–597. doi: 10.1073/pnas.71.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabo A. G., Stepanik T. M., Wayner D. M., Young N. M. Conformational heterogeneity of the copper binding site in azurin. A time-resolved fluorescence study. Biophys J. 1983 Mar;41(3):233–244. doi: 10.1016/S0006-3495(83)84433-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Brochon J. C., Merola F., Jordi W., Gallay J. Nanosecond dynamics of horse heart apocytochrome c in aqueous solution as studied by time-resolved fluorescence of the single tryptophan residue (Trp-59). Biochemistry. 1988 Nov 29;27(24):8752–8761. doi: 10.1021/bi00424a010. [DOI] [PubMed] [Google Scholar]
- Yguerabide J. Nanosecond fluorescence spectroscopy of macromolecules. Methods Enzymol. 1972;26:498–578. doi: 10.1016/s0076-6879(72)26026-8. [DOI] [PubMed] [Google Scholar]
- de Lauder W. B., Wahl P. Fluorescence studies on human serum albumin. Biochem Biophys Res Commun. 1971 Feb 5;42(3):398–404. doi: 10.1016/0006-291x(71)90384-6. [DOI] [PubMed] [Google Scholar]


