Abstract
Cryptosporidium sp. is a significant cause of diarrheal disease, particularly in human immunodeficiency virus (HIV)-infected patients in developing countries. We recently cloned and sequenced several alleles of the highly polymorphic single-copy Cryptosporidium parvum gene Cpgp40/15. This gene encodes a precursor protein that is proteolytically cleaved to yield mature cell surface glycoproteins gp40 and gp15, which are implicated in zoite attachment to and invasion of enterocytes. The most-striking feature of the Cpgp40/15 alleles and proteins is their unprecedented degree of sequence polymorphism, which is far greater than that observed for any other gene or protein studied in C. parvum to date. In this study we analyzed nucleic acid and amino acid sequence polymorphism at the Cpgp40/15 locus of 20 C. parvum isolates from HIV-infected South African children. Fifteen isolates exhibited one of four previously identified genotype I alleles at the Cpgp40/15 locus (Ia, Ib, Ic, and Id), while five displayed a novel set of polymorphisms that defined a new Cpgp40/15 genotype I allele, designated genotype Ie. Surprisingly, only 15 of these isolates exhibited concordant type I alleles at the thrombospondin-related adhesive protein of Cryptosporidium and Cryptosporidium oocyst wall protein loci, while five isolates (all of which displayed Cpgp40/15 genotype Ic alleles) displayed genotype II alleles at these loci. Furthermore, the last five isolates also manifested chimeric genotype Ic/Ib or Ic/II alleles at the Cpgp40/15 locus, raising the possibility of sexual recombination within and between prototypal parasite genotypes. Lastly, children infected with isolates having genotype Ic alleles were significantly older than those infected with isolates displaying other genotype I alleles.
Cryptosporidium sp., a protozoan parasite of the phylum Apicomplexa, is a globally important enteric pathogen (15, 17). Cryptosporidiosis in the immunocompetent host is most often a self-limited diarrheal illness, but in immunosuppressed individuals, such as those with human immunodeficiency virus (HIV) infection, the disease can be severe and life-threatening (12). Because of the magnitude of the AIDS epidemic in developing countries, such as those in sub-Saharan Africa (43), the consequences of cryptosporidial infection in these countries are more devastating than in other parts of the world. Most of these patients do not have access to antiretroviral therapy and are therefore at higher risk for many of the complications of HIV infection, including cryptosporidiosis. In developing countries, diarrhea caused by Cryptosporidium parvum early in childhood may be associated with subsequent impaired physical and cognitive development (10, 16, 21).
Very little is known about the epidemiology of cryptosporidiosis, particularly in developing countries. Studies using restriction fragment length polymorphism (RFLP) analysis and sequence analysis of several different genes have identified genetic differences among Cryptosporidium isolates and have facilitated the development of molecular approaches to study the epidemiology of this infection. These investigations have identified two major genotypes of C. parvum, the most-common species causing human cryptosporidiosis. The anthroponotic genotype (human or genotype I) is found exclusively in human (and some primate) infections, and the zoonotic genotype (calf, bovine, or genotype II) is able to infect both humans and animals (30, 46). The majority of human infections appear to be caused by genotype I parasites (30, 48). Recent studies examining nucleic acid sequence polymorphisms at diverse C. parvum genetic loci have identified allelic subgroups within both major genotypes [3, 4, 13, 39; A. E. Aiello et al., abstract from the 52nd Annual Meeting of the Society of Protozoologists 1999, J. Eukaryot. Microbiol. 46(Suppl. 5):46S-47S, 1999].
We recently cloned and sequenced several alleles of a highly polymorphic single-copy gene, named Cpgp40/15 (9) or gp60/45/15 (39). This gene encodes a precursor protein that is proteolytically cleaved to yield mature cell surface glycoproteins gp40 (also named gp45) and gp15, both of which are implicated in zoite attachment to and invasion of enterocytes (8, 9). The deduced amino acid sequences are all characterized by an N-terminal signal sequence, a polyserine tract, a C-terminal glycophosphatidylinositol (GPI) anchor site, and numerous predicted sites of mucin-type O glycosylation. The most-striking features of the Cpgp40/15 alleles and proteins are their unprecedented degree of sequence polymorphism, particularly among genotype I isolates, which is far greater than that of any other gene or protein studied in C. parvum to date. In a previous study, numerous single-nucleotide polymorphisms (SNPs) and single-amino-acid polymorphisms (SAAPs) were detected among 29 different isolates of C. parvum from different geographic regions (39). Based on these polymorphisms, isolates were classified into five allelic subgroups, one of which included all genotype II isolates and four, designated Ia, Ib, Ic, and Id, which discretely subdivided genotype I isolates. This study suggested that further analysis of Cpgp40/15 SNPs and SAAPs, particularly among geographically and clinically well-defined Cryptosporidium isolates, might prove useful to study the molecular epidemiology of cryptosporidiosis.
The aims of the present study were to determine the species and genotypes of Cryptosporidium isolated from HIV-infected children admitted to hospital with diarrhea in Durban, South Africa; to analyze polymorphisms at the Cpgp40/15 locus from these isolates; and to correlate the results with clinical and epidemiological findings.
MATERIALS AND METHODS
Study population.
HIV-infected South African children with diarrhea were recruited as part of an ongoing study to determine the effect of enhanced nutritional support on clinical outcome (N. C. Rollins et al., Abstr. Pediatr. Acad. Soc. Annu. Meet. 2000, abstr. 750931, 2000). One hundred and one children, 6 to 36 months of age, who had had diarrhea for 3 or more days on admission to the hospital (the time when stool samples were obtained) and whose diarrhea continued for at least 7 days in total were eligible for entry into the nutrition study. All patients were admitted to the King Edward VIII Hospital in Durban, South Africa, from January 1999 to October 2000. Informed consent was obtained from the parents or guardians of the patients according to the guidelines of the institutional review boards of New England Medical Center and the Nelson R. Mandela School of Medicine. Blood and stool samples were obtained, and stool samples were processed for detection of enteric (bacterial, viral, and protozoan) pathogens. Modified acid-fast staining and immunofluorescence were used to determine the presence of Cryptosporidium in stool samples. Stool samples were available for genotyping from 24 of the 25 children identified as having Cryptosporidium in the stool upon initial evaluation in Durban. These 24 children were included in the present study.
DNA isolation.
Stool samples were frozen at −80°C and shipped to Boston on dry ice from the study site. DNA was isolated by modification of a previously described method (23). Briefly, samples were disrupted with glass beads in lysis buffer containing 100 mM NaCl, 25 mM EDTA, 10 mM Tris (pH 8), and 2% sodium dodecyl sulfate. Samples were vortexed, and this was followed by centrifugation at 16,000 × g for 5 min at 4°C. The aqueous phase was extracted with phenol-chloroform-isoamyl alcohol (25:24:1). Following extraction, DNA was further purified using a Geneclean kit (Bio 101, Vista, Calif.) to remove any potential inhibitors of PCR.
PCR-RFLP analysis for species and genotype determination.
PCR-RFLP analysis at the small-subunit (SSU) rRNA locus was used to determine the species of Cryptosporidium (50). A nested PCR using primers and conditions described previously was performed (50). The final PCR product was digested with SspI at 37°C for 1 h. For genotype determination, PCR-RFLP analysis of the thrombospondin-related adhesive protein of Cryptosporidium (TRAP C1) and Cryptosporidium oocyst wall protein (COWP) was performed using previously described primers and conditions (36, 38). PCR products were digested with RsaI at 37°C for 1 h. Fragments were resolved on 1% agarose gels and visualized by ethidium bromide staining.
Cpgp40/15 sequence analysis.
The Cpgp40/15 locus was amplified by PCR using previously published primers 5′TTACTCTCCGTTATAGTCTCCGCT-3′ and 5′-CGAATAAGGCTGCAAAGATTGC3′, which span most of the open reading frame (9). The conditions used were 95°C for 2 min; 72°C for 30 s (hot start); and 30 cycles of 95°C for 40 s, 55°C for 50 s, and 72°C for 1 min; this was followed by a 10-min 72°C extension. PCR products were purified using a QIAquick kit (QIAGEN, Inc., Valencia, Calif.) and sequenced by the dye-terminator method at the Tufts University Core Facility using a Perkin-Elmer ABI 377 sequencer. For samples in which PCR products were not obtained using the first set of primers, and for samples from the Ic allelic subgroup, a second PCR was performed using primers 5′-ATGAGATTGTCGCTCATT-3′ and 5′-TTACAACACGAATAAGGC-3′, which span the entire Cpgp40/15 open reading frame. The PCR conditions were the same except for an annealing temperature of 49°C. PCR products were directly sequenced or gel isolated, purified using a QIAquick kit (QIAGEN), and cloned into pCRII-TOPO (Invitrogen Corp., Carlsbad, Calif.), and inserts were sequenced. The nucleotide and deduced amino acid sequences were compared using the Clustal W alignment algorithm of the Align X program of Vector NTI, Suite 6 (Informax, North Bethesda, Md.), in a pairwise analysis. Putative N-glycosylation sites and predicted O-glycosylation sites were identified with the PROSCAN program utilizing the PROSITE database (2) and NetOGlyc 2.0 (19) programs, respectively.
Statistical analysis.
Results for continuous variables are expressed as the median and 25th and 75th centiles. The Mann-Whitney U test was used to test the significance of differences in continuous variables, and the Pearson chi-square test was used to test the significance of differences in categorical variables. P values of <0.05 were considered significant.
Nucleotide sequence accession numbers.
Cpgp40/15 sequences obtained in this study have been deposited in GenBank under accession numbers AF440621 to AF440640.
RESULTS
Clinical findings.
The results of the nutrition study obtained to date have been described previously (Rollins et al., Abstr. Pediatr. Acad. Soc. Annu. Meet. 2000). Cryptosporidium was the pathogen most commonly identified and was isolated in samples from 25 of 101 (24.8%) children enrolled in the study (Rollins et al., Abstr. Pediatr. Acad. Soc. Annu. Meet. 2000). The clinical details of the 24 children with Cryptosporidium from whom stool samples were available for genotyping are shown in Table 1. Briefly, the median age of the 24 patients was 12 months (25th and 75th percentiles, 8 and 18 months, respectively). Thirteen of the children were males and 11 were females. The median duration of diarrhea before admission was 10 days (25th and 75th percentiles, 7 and 25 days, respectively). The median CD4 count was 424 × 103/ml (25th and 75th percentiles, 235 × 103 and 1,014 × 103/ml, respectively). The median HIV RNA level was 1.59 × 106 copies/ml (25th and 75th percentiles, 1.02 × 106 and 1.93 × 106 copies/ml, respectively). Eight weeks after admission to the hospital 15 patients were still living, 7 patients had died, and 2 were lost to follow-up.
TABLE 1.
Clinical data on Cryptosporidium-infected children and genetic analysis of Cryptosporidium isolates obtained from them
| Patient no. | Age (mo) | Sexc | Diarrhea (days)a | CD4 count (103/ml) | HIV RNA (106 copies/ml) | Clinical outcomeb | Species | Genotype
|
C. parvum gp40/15 allelic subgroup | |
|---|---|---|---|---|---|---|---|---|---|---|
| TRAP C1 | COWP | |||||||||
| 1 | 15 | M | 12 | 1,904 | 0.63 | Alive | C. parvum | IId | II | Ic |
| 2 | 22 | M | 8 | 298 | 1.96 | NKe | C. parvum | If | I | Id |
| 3 | 26 | F | 14 | 464 | 1.36 | Alive | C. parvum | II | II | Ic |
| 4 | 8 | F | 7 | 855 | 11.6 | Dead | C. parvum | I | I | Id |
| 5 | 9 | M | 8 | 146 | 1.6 | Dead | C. parvum | —g | — | — |
| 6 | 13 | F | 7 | 424 | 0.02 | Dead | C. parvum | I | I | Id |
| 7 | 6 | M | 7 | 172 | 2.28 | Alive | C. parvum | I | I | Ib |
| 8 | 8 | F | 3 | 1,240 | 1.68 | Alive | C. parvum | II | II | Ic |
| 9 | 12 | F | 14 | 418 | 1.68 | Alive | C. parvum | I | I | Ib |
| 10 | 6 | M | 60 | 24 | 0.88 | Dead | C. parvum | I | I | Ie |
| 11 | 10 | M | 30 | 1,134 | 1.92 | Alive | C. parvum | I | I | Ie |
| 12 | 21 | M | 7 | 344 | 5.8 | Alive | C. parvum | II | II | Ic |
| 13 | 14 | M | 14 | 410 | 1.93 | Alive | C. parvum | I | I | Id |
| 14 | 12 | M | 7 | 628 | 1.22 | Alive | C. parvum | I | I | — |
| 15 | 7 | F | 7 | 1,932 | 0.19 | Alive | C. parvum | I | I | Ie |
| 16 | 25 | F | 5 | 352 | 0.62 | Alive | C. parvum | I | I | Ia |
| 17 | 12 | F | 8 | 848 | 1.00 | Alive | C. parvum | I | I | Id |
| 18 | 34 | M | 21 | 60 | 1.58 | Dead | C. parvum | II | II | Ic |
| 19 | 6 | F | 29 | 598 | 1.55 | NK | C. parvum | I | I | Ib |
| 20 | 17 | M | 14 | 138 | 2.41 | Alive | — | — | — | — |
| 21 | 7 | M | 30 | 1,776 | 1.03 | Alive | — | — | — | — |
| 22 | 8 | M | 7 | 39 | 2.38 | Dead | C. parvum | I | I | Ie |
| 23 | 19 | F | 14 | 1070 | 1.88 | Alive | C. parvum | I | I | Ie |
| 24 | 11 | F | 15 | NDh | 1.08 | Dead | C. parvum | I | I | Ib |
Duration of diarrhea in days prior to admission.
Clinical outcome was recorded as alive or dead by 8 weeks postadmission or postrecruitment.
Abbreviations: M, male; F, female.
Genotype II (calf).
NK, not known, as lost to followup.
Genotype I (human).
—, no result.
ND, not done.
All isolates were C. parvum.
To determine the species of Cryptosporidium present in these 24 stool samples, we used a nested PCR for the SSU rRNA locus followed by RFLP analysis as previously described (50). PCR products could be amplified in 22 of the 24 (92%) samples (Table 1). All the isolates in which PCR products could be amplified displayed the previously published RFLP pattern and the expected sizes of restriction fragments for C. parvum, indicating that all 22 isolates were of this species (Table 1).
All isolates displayed Cpgp40/15 genotype I alleles containing both highly polymorphic and strongly conserved sequence regions.
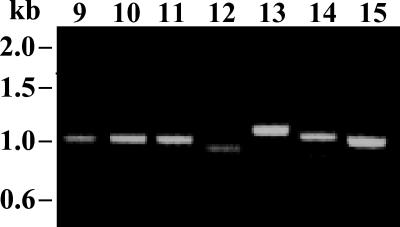
Cpgp40/15 sequences were obtained from 20 of the 24 (83.3%) isolates. PCR fragments ranging from 731 to 1,018 bp were amplified from 18 samples using the first set of primers (representative examples are shown in Fig. 1) and were directly sequenced. In samples failing to yield PCR products with these primers, the PCR was repeated using the second set of primers. PCR products were amplified from two additional isolates using this set of primers. PCR products could not be amplified from 4 of the 24 samples using either set of primers.
FIG. 1.
PCR products at the Cpgp40/15 locus from representative isolates. PCR was performed on DNA extracted from stool samples using the first set of primers. PCR products for samples 9 to 15 are shown. Note the differences in size of PCR products among isolates.
All of the Cpgp40/15 nucleotide and deduced amino acid sequences were compared with each other and with sequences from each of the allelic subgroups originally described and deposited in GenBank by Strong et al. (39). Fifteen of the 20 sequences could be placed into one of the four previously described genotype I allelic subgroups. However, 5 of the 20 sequences were significantly different from those of the other type I and type II Cpgp40/15 alleles and were, therefore, classified within a fifth and new allelic subgroup, which we have designated Ie. None of the isolates displayed genotype II alleles at the Cpgp40/15 locus.
The Cpgp40/15 nucleotide and deduced amino acid sequences from all 20 isolates revealed extensive polymorphisms (Fig. 2). Comparison of these sequences revealed several specific polymorphic as well as conserved regions. The N-terminal 5 amino acids, DVSVE, following the putative cleavage site for the signal peptide (9, 39) were conserved among all isolates (Fig. 2). The polyserine tract was conserved but varied greatly in length (from 9 to 24 residues) among isolates. Downstream of the polyserine tract, all isolates contained a hypervariable region characterized by numerous SAAPs. The potential N-glycosylation sites, ANSS and NGST, identified previously (39) in isolates of the Ia and Ic allelic subgroups were also present in isolates of the same subgroups in the present study. A potential N-glycosylation site, NVSF, was identified in isolates of the Id group. No N-glycosylation sites were predicted for isolates of the Ib and Ie subgroups. The presence (or absence) of a particular potential N-glycosylation site was conserved within subgroups. Sites of predicted O-glycosylation also varied among isolates, but were conserved within allelic subgroups. The putative proteolytic cleavage site, EE, between gp40 and gp15 (9, 39) was conserved among all isolates. The previously determined (32) putative attachment site for the GPI anchor in the C-terminal region was also mostly conserved among isolates.
FIG. 2.
Polymorphisms in the deduced amino acid sequences of the Cpgp40/15 gene. The deduced amino acid sequences of 20 isolates were aligned using the Clustal W algorithm of the Align X program of Vector NTI. The five genotype I allelic subgroups—Ia, Ib, Ic, Id, and Ie—are indicated on the left-hand side. Numbers in parentheses indicate the amino acid position within that particular sequence. Hyphens indicate gaps, and conserved residues are shaded. Putative N-glycosylation sites are indicated in boldface and italic type, and predicted O-glycosylation sites are indicated in boldface and underlined type (note that although several S and T residues are conserved among the same allelic subgroup, not all of them are predicted by the NetOGlyc 2.0 program to be sites of O glycosylation). The putative cleavage sites of gp40 and gp15 (EE) are boxed. The predicted site for addition of a GPI anchor is underlined.
Isolates manifesting the Cpgp40/15 Ic genotype displayed discordant genotype II alleles at the TRAP C1 and COWP loci.
The vast majority of C. parvum isolates that have been genotyped at more than one locus have exhibited concordant haplotypes; i.e., all loci examined displayed exclusively genotype I or genotype II alleles. To determine whether the South African isolates examined in this study, which all displayed Cpgp40/15 genotype I alleles, displayed concordant alleles at other loci, we used PCR-RFLP analysis at the TRAP C1 and COWP loci to further genotype the isolates. Surprisingly, only 15 of the 20 (75%) isolates displayed genotype I alleles at the TRAP C1 and COWP loci (Table 1), and interestingly, each of the five isolates that displayed genotype II alleles at these loci exhibited genotype Ic alleles at the Cpgp40/15 locus. Thus, all isolates examined in this investigation that manifested type Ia, Ib, Id, or Ie alleles at the Cpgp40/15 locus also displayed genotype I alleles at both the TRAP C1 and COWP loci, while all isolates manifesting Ic alleles at the former locus displayed genotype II alleles at both of the latter loci. At this point we do not know the generality of these observations.
The Cpgp40/15 genotype Ic isolates comprised two subgroups exhibiting hybrid genotype Ic/Ib or Ic/II sequences.
To further investigate the genotypically discordant Cpgp40/15 type Ic isolates, we made pairwise sequence comparisons of the Cpgp40/15 nucleic acid and deduced amino acid sequences of these isolates. In contrast to similar comparisons made among the sequences of other Cpgp40/15 allelic subgroups, which yielded pairwise nucleotide and predicted amino acid sequence identities approaching or equal to 100%, these allele Ic sequences displayed nucleotide and amino acid sequence identities as low as 94.6 and 89.7%, respectively (Table 2). Indeed, the isolates displaying type 1c alleles could be further divided into two distinct groups based on 27 polymorphic amino acid positions where the deduced SAAPs differed from one another. At these sites the proteins encoded by the various Ic alleles displayed amino acid polymorphisms that were either more similar to those of the Ib allelic class or the genotype II allele. For example, the Cpgp40/15 Ic alleles from isolates 3 and 8 shared identity of 17 of these 27 SAAPs with the Ib allele, while the Ic alleles from isolates 1, 12, and 18 shared identity of 17 of the 27 SAAPs with the genotype II allele. Most of the 27 polymorphic sites at which the deduced Cpgp40/15 protein sequences of isolates 3 and 8 differed from those of isolates 1, 12, and 18 were present in the C-terminal half of the protein (examples of these from the gp15 portion of the deduced amino acid sequence of Cpgp40/15 are shown in Fig. 3), whereas the predicted N-terminal sequences of all allele Ic Cpgp40/15 proteins were essentially identical.
TABLE 2.
Sequence identitiesa define five Cpgp40/15 genotype I alleles: Ia, Ib, Ic, Id, and Ie
| Allele (isolate no.) | % Identity with isolate:
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ia | Ib
|
Ic
|
Id
|
Ie
|
||||||||||||||||
| 16 | 7 | 9 | 19 | 24 | 1 | 3 | 8 | 12 | 18 | 2 | 4 | 6 | 13 | 17 | 10 | 11 | 15 | 22 | 23 | |
| Ia (16) | 100 | 74.8 | 74.8 | 74.8 | 74.8 | 71.5 | 70.9 | 70.7 | 71.5 | 71.5 | 76.7 | 75.7 | 75.6 | 76.3 | 76.4 | 73.3 | 73.3 | 73.3 | 75 | 73.9 |
| Ib (7) | 63.7 | 100 | 100 | 100 | 100 | 77.4 | 79.7 | 79.4 | 77.4 | 77.4 | 73.9 | 74.6 | 74.8 | 71.9 | 74.2 | 75.5 | 75.5 | 75.5 | 74.3 | 75 |
| Ib (9) | 63.7 | 100 | 100 | 100 | 100 | 77.4 | 79.7 | 79.4 | 77.4 | 77.4 | 73.9 | 74.6 | 74.8 | 71.9 | 74.2 | 75.5 | 75.5 | 75.5 | 74.3 | 75 |
| Ib (19) | 64 | 100 | 100 | 100 | 100 | 77.4 | 79.7 | 79.4 | 77.4 | 77.4 | 73.9 | 74.6 | 74.8 | 72.1 | 74.2 | 75.5 | 75.5 | 75.5 | 74.3 | 75 |
| Ib (24) | 64 | 100 | 100 | 100 | 100 | 77.4 | 79.7 | 79.4 | 77.4 | 77.4 | 73.9 | 74.6 | 74.8 | 72.1 | 74.2 | 75.5 | 75.5 | 75.5 | 74.3 | 75 |
| Ic (1) | 63.8 | 66.8 | 66.8 | 66.8 | 66.8 | 100 | 94.9 | 94.6 | 100 | 100 | 63.4 | 70 | 70.2 | 62.5 | 63.6 | 75.3 | 75.3 | 75.3 | 73.9 | 74.8 |
| Ic (3) | 61.3 | 69 | 69 | 69 | 69 | 90.5 | 100 | 99.8 | 94.9 | 94.9 | 62.9 | 68.7 | 68.9 | 62 | 63.2 | 75.8 | 75.8 | 75.8 | 73 | 75.3 |
| Ic (8) | 60.7 | 68.4 | 68.4 | 68.4 | 68.4 | 89.7 | 99.3 | 100 | 94.6 | 94.6 | 62.7 | 68.4 | 68.8 | 61.8 | 63 | 75.6 | 75.6 | 75.6 | 73.4 | 75.1 |
| Ic (12) | 63.8 | 66.8 | 66.8 | 66.8 | 66.8 | 100 | 90.5 | 89.7 | 100 | 100 | 63.4 | 69.9 | 70.3 | 62.5 | 63.6 | 75.3 | 75.3 | 75.3 | 73 | 74.8 |
| Ic (18) | 63.8 | 66.8 | 66.8 | 66.8 | 66.8 | 100 | 90.5 | 89.7 | 100 | 100 | 63.4 | 69.9 | 70.3 | 62.5 | 63.6 | 75.3 | 75.3 | 75.3 | 73 | 74.8 |
| Id (2) | 63.7 | 60.1 | 60.1 | 60.1 | 60.1 | 58.4 | 58.1 | 57.5 | 58.4 | 58.4 | 100 | 98.9 | 98.5 | 98.5 | 99.4 | 71.4 | 71.4 | 71.4 | 73 | 72.1 |
| Id (4) | 63.4 | 61 | 61 | 61 | 61 | 58.9 | 58.9 | 58 | 59.2 | 59.2 | 98.8 | 100 | 99.7 | 97.7 | 99.3 | 72.2 | 71.4 | 72.2 | 72.4 | 72.9 |
| Id (6) | 62.8 | 61.2 | 61.2 | 61.2 | 61.2 | 59.1 | 59.1 | 58.5 | 59.1 | 59.1 | 98.4 | 99.7 | 100 | 97.4 | 99 | 72.6 | 72.6 | 72.6 | 72.3 | 72.7 |
| Id (13) | 63.4 | 59.4 | 59.4 | 59.4 | 59.4 | 57.7 | 57.7 | 57.1 | 57.7 | 58 | 98.2 | 97.5 | 97.2 | 100 | 98.3 | 70.6 | 70.6 | 70.7 | 72.2 | 71.3 |
| Id (17) | 64 | 60.6 | 60.6 | 60.9 | 60.9 | 58.6 | 58.6 | 57.9 | 58.6 | 58.6 | 99.1 | 99.1 | 98.8 | 97.8 | 100 | 71.9 | 71.9 | 71.9 | 73 | 72.5 |
| Ie (10) | 62.2 | 64.1 | 64.1 | 64.1 | 64.1 | 66.7 | 67.7 | 67 | 66.7 | 66.7 | 60.4 | 61.9 | 62.1 | 60.3 | 61.6 | 100 | 100 | 100 | 98.3 | 99.3 |
| Ie (11) | 62.2 | 64.1 | 64.1 | 64.1 | 64.1 | 66.7 | 67.7 | 67 | 66.7 | 66.7 | 60.4 | 61.9 | 62.1 | 60.3 | 61.6 | 100 | 100 | 100 | 98.3 | 99.3 |
| Ie (15) | 62.2 | 64.1 | 64.1 | 64.1 | 64.1 | 66.7 | 67.7 | 67.7 | 67 | 66.7 | 60.4 | 61.9 | 62.1 | 60.3 | 61.6 | 100 | 100 | 100 | 98.3 | 99.3 |
| Ie (22) | 63.8 | 63 | 63 | 63 | 63 | 65.6 | 66.6 | 65.9 | 65.6 | 65.6 | 61.7 | 62 | 61.7 | 61.8 | 62.3 | 98.3 | 98.3 | 98.3 | 100 | 99 |
| Ie (23) | 62.8 | 63.6 | 63.6 | 63.6 | 63.6 | 66.2 | 99 | 66.6 | 66.2 | 66.2 | 61.1 | 62.3 | 61.9 | 60.9 | 62.2 | 99.3 | 99.3 | 99.3 | 99 | 100 |
Nucleotide (above diagonal) and amino acid (below diagonal) identity are shown. Intra-allelic sequence comparisons are shown in boldface type.
FIG. 3.
Comparison of SAAPs in the Cpgp40/15 allelic Ic/Ib and Ic/II subclasses with those of their hypothesized allele Ib and genotype II parents, respectively. The gp15 portion of the Cpgp40/15 deduced amino acid sequences of the Ic and Ib isolates from this study and of genotype II isolates Peru (accession number AAF78348), Brazil (accession number AAF78349), Iowa (accession number AAF78345), and KSU-1 (accession number AAF78351) and genotype Ic isolates 0676I (accession number AAF78357) and SFGH23 (accession number AAF78347), previously deposited in GenBank, were aligned using the Clustal W algorithm of the Align X program of Vector NTI. Hyphens indicate gaps. All SAAPs that distinguish the various Ic alleles are indicated in boldface type, allele Ic/II SAAPs that are shared with hypothesized parent allele II are shaded in yellow, and allele Ic amino acid residues that are not shared with their hypothesized parent are shaded in gray.
As described above, three of these isolates (isolates 1, 12, and 18) shared most of the deduced Cpgp40/15 C-terminal SAAPs with genotype II isolates (Fig. 3). This raised the unlikely possibility that these Cpgp40/15 Ic sequences might somehow have arisen from artifactual PCR fragments derived from genotypically heterogeneous isolates generated by mixed infection with genotype I and genotype II parasites. To exclude this possibility, we PCR amplified Cpgp40/15 sequences from these three Ic isolates using the second set of primers and directly sequenced these PCR fragments or cloned them into pCRII-TOPO and sequenced the clones. In all cases there was 100% identity among sequences obtained from each isolate using different primer sets or sequencing templates (not shown), making the possibility of a PCR artifact highly unlikely.
Correlation of clinical parameters and Cpgp40/15 genotype I alleles.
There were no statistically significant associations between a particular C. parvum Cpgp40/15 allelic subgroup and duration of diarrhea, CD4+ T-cell count, or HIV RNA levels using the Mann-Whitney test or between subgroup and sex of the patient or clinical outcome using the Pearson chi-square test. However, there was a significant association between age of the patient and allelic subgroup. Children infected with isolates displaying genotype Ic alleles at the Cpgp40/15 locus were significantly (P = 0.025) older (median age, 21 months) than those infected with isolates displaying other genotype I alleles (median age, 11.5 months).
DISCUSSION
Although the burden of disease from cryptosporidiosis is in developing countries such as those in sub-Saharan Africa (1, 7, 11, 44), most genetic studies of Cryptosporidium have focused on infections in Europe and the United States (18, 22-24, 29, 31, 34, 42). Other than a recent genotypic analysis of isolates obtained from six HIV-infected individuals from Kenya (24), no genetic studies of Cryptosporidium isolates from sub-Saharan Africa have been reported. To initiate such studies in well-defined cohorts of Cryptosporidium-infected individuals in this region where the prevalence of HIV infection is among the highest in the world (43), we analyzed isolates obtained from HIV-infected children admitted to hospital with diarrhea in Durban, South Africa.
All isolates examined in this investigation were found to be C. parvum based on the RFLP pattern observed in PCR fragments amplified from the SSU rRNA loci. While this species has been most commonly identified in human infections, recent studies have documented the presence of non-C. parvum species such as C. felis and C. meleagridis in immunocompetent as well as HIV-infected individuals (24, 28, 29, 31, 49). Oocysts suggestive of those of C. muris were also reported to be isolated from two healthy Indonesian children (20). In addition to describing the human and bovine genotypes of C. parvum, recent studies have described the presence of unusual genotypes such as the “dog” genotype in immunocompetent and HIV-infected patients (28, 31, 49). In our study 21 of 22 isolates identified as C. parvum by PCR-RFLP at the SSU rRNA locus manifested either the human or bovine genotype at both the TRAP C1 and COWP loci. However, we were unable to amplify PCR fragments from one of the C. parvum isolates (isolate 5) using either the TRAP C1 or COWP primers; it is possible that this isolate may have an unusual C. parvum genotype. It is also possible that analysis of a larger number of isolates from this study population may reveal the occurrence of non-C. parvum species or unusual C. parvum genotypes, but this study suggests that their prevalence is likely to be low in South Africa.
Previous investigations have examined the genetic diversity at many C. parvum loci, including the 18S SSU rRNA (27, 49-51), the ITS1 region of rRNA (5, 24), and the genes encoding HSP70 (41), dihydrofolate reductase-thymidylate synthase (27, 45), acetyl coenzyme A synthetase (25), polythreonine (6), β-tubulin (40, 47), COWP (38), and TRAP C1 and C2 (36, 37, 42). In each case these studies have identified two predominant C. parvum genotypes, one exclusively associated with isolates derived from human infections (genotype I) and one associated with isolates derived from both human and animal infections (genotype II). In the cohort of HIV-infected children described in this study, we found that the majority of infections were caused by genotype I parasites as defined by sequence analysis at the Cpgp40/15 locus and PCR-RFLP analysis at the TRAP C1 and COWP loci. This is consistent with epidemiologic studies in which C. parvum genotype I isolates have been shown to be more frequently identified in immunocompetent (49) as well as HIV-infected individuals (48). However, two studies of sporadic cases and small outbreaks in the United Kingdom and one survey of immunocompetent and immunosuppressed French patients have shown that genotype II isolates were predominant (18, 22, 23). Recent studies have shown that in human infections, genotype I was associated with increased quantity and duration of oocyst shedding (23, 49). The clinical significance and prognostic significance of these genotypic distinctions are not clear at the present time.
In spite of the documented genetic diversity that has served to define the two major C. parvum genotypes, most previous studies have not identified substantial sequence variation at protein-coding loci within either of the two prototypal genotypes. The majority of the genes previously studied encode cytoplasmic proteins; however, even genes that encode cell surface-associated proteins such as TRAP C1 (37) have shown remarkably little sequence variation within, or even between, the two canonical genotypes. Strong et al. recently documented extensive nucleic acid and amino acid sequence polymorphism at the gp60/45/15 (same as Cpgp40/15) locus among isolates from different geographic areas (39). This gene encodes the glycoproteins gp40 (also called gp45) and gp15, which are expressed on the apical and surface regions of C. parvum invasive stages, and are implicated in attachment to and invasion of host cells (8, 39). The finding of extensive polymorphism at this locus is therefore consistent with the hypothesis that its gene products are surface-associated virulence determinants that may be under host immune selection.
The present study also demonstrates a remarkable degree of genetic heterogeneity at the Cpgp40/15 locus among C. parvum isolates in this cohort of children in South Africa. These findings confirm and extend the observations of Strong et al. (39). Comparing sequences of the Cpgp40/15 locus from this study with those described by Strong et al., we found that 15 of the South African sequences fell into one of the previously described Ia, Ib, Ic, and Id allelic subgroups; the remaining five South African sequences defined a new subgroup, which we have designated Cpgp40/15 allele Ie. The Ie subgroup was not identified in any of the previously described isolates from different geographic locations—including the United States, Brazil, Peru, Guatemala, and Zaire (39)—and may represent a geographically restricted allele specifically found in South Africa. Surprisingly, none of the isolates in our study displayed the genotype II allele at the Cpgp40/15 locus (as opposed to the TRAP-C1 and COWP loci); it is possible that this genotype is rare in HIV-infected South African children.
In this study, five isolates which displayed genotype Ic alleles at the Cpgp40/15 locus, exhibited genotype II alleles at the TRAP C1 and COWP loci. These isolates could be further divided into two subgroups based on the particular SAAPs present in the C-terminal half of their deduced Cpgp40/15 protein sequences. Three isolates displayed SAAPs that were most nearly identical to those present in the deduced genotype II Cpgp40/15 protein sequence (isolates 1, 12, and 18), while two isolates (numbers 3 and 8) displayed C-terminal SAAPs most similar to those present in allele Ib Cpgp40/15 sequences (Fig. 3). Two additional, previously characterized isolates displaying the Cpgp40/15 Ic allele, viz., 0676I (GenBank accession number AAF78357) and SFGH23 (GenBank accession number AAF78347), were also found to have similar hybrid Cpgp40/15 allele Ic/II or Ic/Ib SAAP patterns, respectively (39). Interestingly, like the five South African isolates described here, isolate 0676I was also genotyped as a type II isolate at other genetic loci (48), but unlike the other type II isolates examined in that study or others, it was not infective for neonatal mice and, therefore, appeared genotypically anomalous. Furthermore, isolate 0676I was also atypical in that it displayed an unusual β-tubulin intron allele genotype, viz., a hybrid genotype I/II SNP pattern (47) that was not seen in a large number of other isolates examined (35, 40).
The finding that the five South African isolates that were genotyped as Ic at the Cpgp40/15 locus displayed discordant genotype II alleles at the TRAP C1 and COWP loci suggests that these mixed genotype lineages were derived by sexual recombination between genotype I and genotype II parasites, as hypothesized previously by Strong et al. (39). The recombinant haplotypes appear to have arisen through both of the usual mechanisms that create genetic diversity during sexual reproduction, i.e., independent assortment of chromosomes and crossing over, as both the TRAP C1 gene on chromosome I (33) and the COWP gene on chromosome VI (33) segregated independently of the Cpgp40/15 gene also located on chromosome VI (W. Strong and R. Nelson, unpublished results). The independent segregation of type I and type II alleles at the latter two loci would require at least one crossover on chromosome VI. In addition, the chimeric nature of the Cpgp40/15 Ic genes, which contain conserved allele Ic sequence at their 5′ ends joined to either allele Ib- or allele II-like sequence at their 3′ ends, suggests that the progenitors of these hybrid genes were derived by recombination between distinct genotype I Cpgp40/15 alleles and between genotype I and genotype II Cpgp40/15 alleles, respectively. Although meiotic recombination between genotype II parasites has recently been experimentally demonstrated in an animal host (14) no similar data are available regarding sexual recombination between the genotype I and genotype II parasite lineages. This is thought to be an exceedingly rare or nonexistent event in natural populations, as isolates manifesting recombinant haplotypes have not been previously detected (23, 38; however, see reference 41), and in fact, their absence has been used to argue that the prototypal C. parvum genotypes may actually be reproductively isolated and represent discrete species (26). The present investigation indicates, however, that while the C. parvum genetic population structure is almost certainly highly clonal and dominated by these two widespread clonal lineages, occasional interlineage recombination does occur naturally and produces mixed genotype progeny lineages that are viable and infectious. Although such events are thought to be very rare based on previous data, 5 of 20 isolates genotyped in this investigation revealed two such recombinant multilocus genotypes (Cpgp40/15 Ic subtypes Ic/Ib or Ic/II, TRAP C1 type II, and COWP type II). The reason for the high frequency of recombinant genotypes observed in this study is unclear, but is probably not related to geographic restriction or the patient population per se, as both of the Cpgp40/15 allele Ic subtypes have also been observed in human isolates from the United States (39).
In summary, the results of this study indicate that genotype I isolates are predominant in this population of HIV-infected children with diarrhea in South Africa. Further, this study confirms the presence of extensive nucleic acid and amino acid sequence polymorphism, which define at least six allelic subgroups (one genotype II and five genotype I subgroups), at the Cpgp40/15 locus. Other than age of the patient, there was no significant association between allelic subgroup and clinical parameters. However, the number of isolates was small. It is possible that examination of a larger number of isolates will identify additional correlations between allelic subgroups and clinical or epidemiological parameters. Further, the genetic fingerprinting of isolates based on Cpgp40/15 SNPs may serve to identify the source and mode of transmission (waterborne, food-borne, zoonotic, or anthroponotic) of Cryptosporidium infection in defined cohorts in various geographic locations. Moreover, additional studies on the putative recombinant Cpgp40/15 allelic Ic subgroup may facilitate analysis of the population structure of Cryptosporidium.
Acknowledgments
This work was supported by grants AI46299 and ICIDR opportunity pool funds (to H.D.W.); grants AI40319 and AI42565 (to R.G.N.) and grant AI45508 (to M.L.B.) from the NIAID, NIH; grant P30 DK 34928 from the NIDDK, NIH, to the GRASP Digestive Diseases Center at Tufts—New England Medical Center; and grant PGR-50991 (to M.L.B.) from the Pediatric AIDS Foundation. B. A. Leav was supported by training grant AI07438 from the NIAID, NIH. M. R. Mackay was supported by training grant TW05571 from the Fogarty International Center, NIH. A. Anyanwu was supported in part by the Fellowship Program in Academic Medicine for Minority Students sponsored by Bristol-Myers Squibb Company. R. M. O'Connor was supported by training grant AI07389 from the NIAID, NIH. M. L. Bennish was supported in part by Mid-Career Award AI01671 from the NIAID, NIH, and by a grant from the Wellcome Trust.
We thank Cheleste Thorpe and Shiv Pillai for critical reading of the manuscript and helpful suggestions.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Amadi, B., P. Kelly, M. Mwiya, E. Mulwazi, S. Sianongo, F. Changwe, M. Thomson, J. Hachungula, A. Watuka, J. Walker-Smith, and C. Chintu. 2001. Intestinal and systemic infection, HIV, and mortality in Zambian children with persistent diarrhea and malnutrition. J. Pediatr. Gastroenterol. Nutr. 32:550-554. [DOI] [PubMed] [Google Scholar]
- 2.Bairoch, A., P. Bucher, and K. Hofmann. 1997. The PROSITE database, its status in 1997. Nucleic Acids Res. 25:217-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caccio, S., W. Homan, R. Camilli, G. Traldi, T. Kortbeek, and E. Pozio. 2000. A microsatellite marker reveals population heterogeneity within human and animal genotypes of Cryptosporidium parvum. Parasitology 120:237-244. [DOI] [PubMed] [Google Scholar]
- 4.Caccio, S., F. Spano, and E. Pozio. 2001. Large sequence variation at two microsatellite loci among zoonotic (genotype C) isolates of Cryptosporidium parvum. Int. J. Parasitol. 31:1082-1086. [DOI] [PubMed] [Google Scholar]
- 5.Carraway, M., S. Tzipori, and G. Widmer. 1996. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl. Environ. Microbiol. 62:712-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carraway, M., S. Tzipori, and G. Widmer. 1997. A new restriction fragment length polymorphism from Cryptosporidium parvum identifies genetically heterogeneous parasite populations and genotypic changes following transmission from bovine to human hosts. Infect. Immun. 65:3958-3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cegielski, J. P., Y. R. Ortega, S. McKee, J. F. Madden, L. Gaido, D. A. Schwartz, K. Manji, A. F. Jorgensen, S. E. Miller, U. P. Pulipaka, A. E. Msengi, D. H. Mwakyusa, C. R. Sterling, and L. B. Reller. 1999. Cryptosporidium, Enterocytozoon, and Cyclospora infections in pediatric and adult patients with diarrhea in Tanzania. Clin. Infect. Dis. 28:314-321. [DOI] [PubMed] [Google Scholar]
- 8.Cevallos, A. M., N. Bhat, R. Verdon, D. H. Hamer, B. Stein, S. Tzipori, M. E. Pereira, G. T. Keusch, and H. D. Ward. 2000. Mediation of Cryptosporidium parvum infection in vitro by mucin-like glycoproteins defined by a neutralizing monoclonal antibody. Infect. Immun. 68:5167-5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cevallos, A. M., X. Zhang, M. K. Waldor, S. Jaison, X. Zhou, S. Tzipori, M. R. Neutra, and H. D. Ward. 2000. Molecular cloning and expression of a gene encoding Cryptosporidium parvum glycoproteins gp40 and gp15. Infect. Immun. 68:4108-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Checkley, W., L. D. Epstein, R. H. Gilman, R. E. Black, L. Cabrera, and C. R. Sterling. 1998. Effects of Cryptosporidium parvum infection in Peruvian children: growth faltering and subsequent catch-up growth. Am. J. Epidemiol. 148:497-506. [DOI] [PubMed] [Google Scholar]
- 11.Chintu, C., C. Luo, S. Baboo, B. Khumalo-Ngwenya, J. Mathewson, H. L. DuPont, and A. Zumla. 1995. Intestinal parasites in HIV-seropositive Zambian children with diarrhoea. J. Trop. Pediatr. 41:149-152. [DOI] [PubMed] [Google Scholar]
- 12.Colford, J. M., Jr., I. B. Tager, A. M. Hirozawa, G. F. Lemp, T. Aragon, and C. Petersen. 1996. Cryptosporidiosis among patients infected with human immunodeficiency virus. Factors related to symptomatic infection and survival. Am. J. Epidemiol. 144:807-816. [DOI] [PubMed] [Google Scholar]
- 13.Feng, X., S. M. Rich, D. Akiyoshi, J. K. Tumwine, A. Kekitiinwa, N. Nabukeera, S. Tzipori, and G. Widmer. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, X., S. M. Rich, S. Tzipori, and G. Widmer. 2002. Experimental evidence for recombination in the opportunistic pathogen Cryptosporidium parvum. Mol. Biochem. Parasitol. 119:55-62. [DOI] [PubMed] [Google Scholar]
- 15.Griffiths, J. K. 1998. Human cryptosporidiosis: epidemiology, transmission, clinical disease, treatment, and diagnosis. Adv. Parasitol. 40:37-85. [DOI] [PubMed] [Google Scholar]
- 16.Guerrant, D. I., S. R. Moore, A. A. Lima, P. D. Patrick, J. B. Schorling, and R. L. Guerrant. 1999. Association of early childhood diarrhea and cryptosporidiosis with impaired physical fitness and cognitive function four-seven years later in a poor urban community in northeast Brazil. Am. J. Trop. Med. Hyg. 61:707-713. [DOI] [PubMed] [Google Scholar]
- 17.Guerrant, R. L. 1997. Cryptosporidiosis: an emerging, highly infectious threat. Emerg. Infect. Dis. 3:51-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guyot, K., A. Follet-Dumoulin, E. Lelievre, C. Sarfati, M. Rabodonirina, G. Nevez, J. C. Cailliez, D. Camus, and E. Dei-Cas. 2001. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J. Clin. Microbiol. 39:3472-3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hansen, J. E., O. Lund, N. Tolstrup, A. A. Gooley, K. L. Williams, and S. Brunak. 1998. NetOglyc: prediction of mucin type O-glycosylation sites based on sequence context and surface accessibility. Glycoconj. J. 15:115-130. [DOI] [PubMed] [Google Scholar]
- 20.Katsumata, T., D. Hosea, I. G. Ranuh, S. Uga, T. Yanagi, and S. Kohno. 2000. Short report: possible Cryptosporidium muris infection in humans. Am. J. Trop. Med. Hyg. 62:70-72. [DOI] [PubMed] [Google Scholar]
- 21.Lima, A. A., S. R. Moore, M. S. Barboza, A. M. Soares, M. A. Schleupner, R. D. Newman, C. L. Sears, J. P. Nataro, D. P. Fedorko, T. Wuhib, J. B. Schorling, and R. L. Guerrant. 2000. Persistent diarrhea signals a critical period of increased diarrhea burdens and nutritional shortfalls: a prospective cohort study among children in northeastern Brazil. J. Infect. Dis. 181:1643-1651. [DOI] [PubMed] [Google Scholar]
- 22.McLauchlin, J., C. Amar, S. Pedraza-Diaz, and G. L. Nichols. 2000. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J. Clin. Microbiol. 38:3984-3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McLauchlin, J., S. Pedraza-Diaz, C. Amar-Hoetzeneder, and G. L. Nichols. 1999. Genetic characterization of Cryptosporidium strains from 218 patients with diarrhea diagnosed as having sporadic cryptosporidiosis. J. Clin. Microbiol. 37:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morgan, U., R. Weber, L. Xiao, I. Sulaiman, R. C. Thompson, W. Ndiritu, A. Lal, A. Moore, and P. Deplazes. 2000. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J. Clin. Microbiol. 38:1180-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morgan, U. M., K. D. Sargent, P. Deplazes, D. A. Forbes, F. Spano, H. Hertzberg, A. Elliot, and R. C. Thompson. 1998. Molecular characterization of Cryptosporidium from various hosts. Parasitology 117:31-37. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. Thompson. 1999. Variation in Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 27.Ong, C. S., D. L. Eisler, S. H. Goh, J. Tomblin, F. M. Awad-El-Kariem, C. B. Beard, L. Xiao, I. Sulaiman, A. Lal, M. Fyfe, A. King, W. R. Bowie, and J. L. Isaac-Renton. 1999. Molecular epidemiology of cryptosporidiosis outbreaks and transmission in British Columbia, Canada. Am. J. Trop. Med. Hyg. 61:63-69. [DOI] [PubMed] [Google Scholar]
- 28.Pedraza-Diaz, S., C. Amar, A. M. Iversen, P. J. Stanley, and J. McLauchlin. 2001. Unusual Cryptosporidium species recovered from human faeces: first description of Cryptosporidium felis and Cryptosporidium "dog type' from patients in England. J. Med. Microbiol. 50:293-296. [DOI] [PubMed] [Google Scholar]
- 29.Pedraza-Diaz, S., C. Amar, and J. McLauchlin. 2000. The identification and characterisation of an unusual genotype of Cryptosporidium from human faeces as Cryptosporidium meleagridis. FEMS Microbiol. Lett. 189:189-194. [DOI] [PubMed] [Google Scholar]
- 30.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, A. C. Weltman, C. S. Ong, W. R. Mac Kenzie, A. A. Lal, and C. B. Beard. 1997. Genetic polymorphism among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pieniazek, N. J., F. J. Bornay-Llinares, S. B. Slemenda, A. J. da Silva, I. N. Moura, M. J. Arrowood, O. Ditrich, and D. G. Addiss. 1999. New Cryptosporidium genotypes in HIV-infected persons. Emerg. Infect. Dis. 5:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Priest, J. W., L. T. Xie, M. J. Arrowood, and P. J. Lammie. 2001. The immunodominant 17-kDa antigen from Cryptosporidium parvum is glycosylphosphatidylinositol-anchored. Mol. Biochem. Parasitol. 113:117-126. [DOI] [PubMed] [Google Scholar]
- 33.Putignani, L., P. Sallicandro, P. Alano, M. S. Abrahamsen, A. Crisanti, and F. Spano. 1999. Chromosome mapping in Cryptosporidium parvum and establishment of a long-range restriction map for chromosome VI. FEMS Microbiol. Lett. 175:231-238. [DOI] [PubMed] [Google Scholar]
- 34.Quiroz, E. S., C. Bern, J. R. MacArthur, L. Xiao, M. Fletcher, M. J. Arrowood, D. K. Shay, M. E. Levy, R. I. Glass, and A. Lal. 2000. An outbreak of cryptosporidiosis linked to a foodhandler. J. Infect. Dis. 181:695-700. [DOI] [PubMed] [Google Scholar]
- 35.Rochelle, P. A., E. M. Jutras, E. R. Atwill, R. De Leon, and M. H. Stewart. 1999. Polymorphisms in the beta-tubulin gene of Cryptosporidium parvum differentiate between isolates based on animal host but not geographic origin. J. Parasitol. 85:986-989. [PubMed] [Google Scholar]
- 36.Spano, F., L. Putignani, A. Crisanti, P. Sallicandro, U. M. Morgan, S. M. Le Blancq, L. Tchack, S. Tzipori, and G. Widmer. 1998. Multilocus genotypic analysis of Cryptosporidium parvum isolates from different hosts and geographical origins. J. Clin. Microbiol. 36:3255-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spano, F., L. Putignani, S. Guida, and A. Crisanti. 1998. Cryptosporidium parvum: PCR-RFLP analysis of the TRAP-C1 (thrombospondin-related adhesive protein of Cryptosporidium-1) gene discriminates between two alleles differentially associated with parasite isolates of animal and human origin. Exp. Parasitol. 90:195-198. [DOI] [PubMed] [Google Scholar]
- 38.Spano, F., L. Putignani, J. McLauchlin, D. P. Casemore, and A. Crisanti. 1997. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiol. Lett. 150:209-217. [DOI] [PubMed] [Google Scholar]
- 39.Strong, W. B., J. Gut, and R. G. Nelson. 2000. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infect. Immun. 68:4117-4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sulaiman, I. M., A. A. Lal, M. J. Arrowood, and L. Xiao. 1999. Biallelic polymorphism in the intron region of beta-tubulin gene of Cryptosporidium parasites. J. Parasitol. 85:154-157. [PubMed] [Google Scholar]
- 41.Sulaiman, I. M., U. M. Morgan, R. C. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sulaiman, I. M., L. Xiao, C. Yang, L. Escalante, A. Moore, C. B. Beard, M. J. Arrowood, and A. A. Lal. 1998. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 4:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.UNAIDS. 2000. AIDS epidemic update: December 2000. World Health Organization, Geneva, Switzerland.
- 44.van den Ende, G. M. 1986. Cryptosporidiosis among black children in hospital in South Africa. J. Infect. 13:25-30. [DOI] [PubMed] [Google Scholar]
- 45.Vasquez, J. R., L. Gooze, K. Kim, J. Gut, C. Petersen, and R. G. Nelson. 1996. Potential antifolate resistance determinants and genotypic variation in the bifunctional dihydrofolate reductase-thymidylate synthase gene from human and bovine isolates of Cryptosporidium parvum. Mol. Biochem. Parasitol. 79:153-165. [DOI] [PubMed] [Google Scholar]
- 46.Widmer, G. 1998. Genetic heterogeneity and PCR detection of Cryptosporidium parvum. Adv. Parasitol. 40:223-239. [DOI] [PubMed] [Google Scholar]
- 47.Widmer, G., L. Tchack, C. L. Chappell, and S. Tzipori. 1998. Sequence polymorphism in the beta-tubulin gene reveals heterogeneous and variable population structures in Cryptosporidium parvum. Appl. Environ. Microbiol. 64:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Widmer, G., S. Tzipori, C. J. Fichtenbaum, and J. K. Griffiths. 1998. Genotypic and phenotypic characterization of Cryptosporidium parvum isolates from people with AIDS. J. Infect. Dis. 178:834-840. [DOI] [PubMed] [Google Scholar]
- 49.Xiao, L., C. Bern, J. Limor, I. Sulaiman, J. Roberts, W. Checkley, L. Cabrera, R. H. Gilman, and A. A. Lal. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis. 183:492-497. [DOI] [PubMed] [Google Scholar]
- 50.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao, L., U. M. Morgan, J. Limor, A. Escalante, M. Arrowood, W. Shulaw, R. C. Thompson, R. Fayer, and A. A. Lal. 1999. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl. Environ. Microbiol. 65:3386-3391. [DOI] [PMC free article] [PubMed] [Google Scholar]