Abstract
Enterohemorrhagic Escherichia coli (EHEC) strains adhere to the intestinal mucosa and produce an attaching and effacing (A/E) lesion. Most of the genes required to produce A/E lesions are thought to be encoded by the 36-kb pathogenicity island termed the locus for enterocyte effacement (LEE). Although the mechanisms underlying the bacterial adherence, including the genes involved, are still poorly understood, the preferential adherence phenotype of EHEC is thought to depend on the nature of the genes and/or the response of these genes to changes in environmental conditions. To explore the environmental factors affecting EHEC adherence, we used an O157:H7 strain and investigated the optimal growth conditions for its adherence to Caco-2 cells. We observed that EHEC grown in Dulbecco's modified Eagle's medium (DMEM) adhered more efficiently to Caco-2 cells than EHEC grown in Luria-Bertani (LB) broth. Among the components of DMEM, only NaHCO3 was found to remarkably stimulate bacterial adherence. When bacteria were grown in LB broth containing NaHCO3, the production of intimin, Tir, EspA, and EspB was greatly enhanced compared with the production in LB broth. Indeed, the transcription of ler required for LEE-encoded gene expression was promoted in response to the concentration of NaHCO3 in LB broth. Since the concentration of NaHCO3 in the lower intestinal tract has been shown to be relatively high compared with that in the upper small intestine, our results may imply that NaHCO3 is an important signaling factor for promoting colonization of EHEC in the lower intestinal tract in humans.
Enterohemorrhagic Escherichia coli (EHEC) is a leading cause of hemorrhagic colitis, bloody diarrhea, and hemolytic uremic syndrome (31, 42). Early in infection leading to illness, the ability of the bacteria to colonize the intestinal epithelial surface and cause histopathological alterations at so-called attaching and effacing lesions (A/E lesions) is thought to be vital. The formation of A/E lesions is characterized by localized destruction of the brush border microvilli, intimate attachment to the host cell, and reconstitution of cytoskeletal components beneath the attached bacteria (11). Thus, EHEC shares pathogenic features with enteropathogenic E. coli (EPEC), rabbit enteropathogenic E. coli, and Citrobacter rodentium, although the extent to which A/E lesion formation, including the target host tissue, varies among the pathogens is not clear.
Most of the genes necessary to form A/E lesions are located in a pathogenicity island termed the locus for enterocyte effacement (LEE) (7, 27, 32). This locus contains (i) sep and esc genes encoding a type III secretion system (17), (ii) eae encoding an adhesin called intimin that is necessary for intimate attachment to epithelial cells (18), (iii) espA, espB, espD, and tir genes encoding proteins secreted by the type III secretion system, including EspA, EspB, EspD, and Tir (3, 20, 22, 23, 25), and (iv) ler encoding a positive regulator of LEE (LEE-encoded regulator) (6, 30). Esp proteins are required for translocation of Tir into the host cell membrane (23). Translocated Tir serves as a receptor for intimin, and intimin-Tir interaction leads to intimate attachment of bacteria to the surface of the host cells (13). Recently, studies have shown that the EHEC type III secretion system is involved in the initial diffuse adherence of the bacteria to epithelial cells (40). Type III secretion systems are highly conserved among EHEC and EPEC strains, and espA, espB, espD, tir, and eae are much less highly conserved. However, the esp protein-dependent translocation mechanisms of Tir are thought to be identical in EPEC and EHEC (11).
Meanwhile, the regulatory systems involved in the expression of LEE genes in the two pathogens are rather different. For example, two distinctive quorum-sensing systems have recently been shown to be involved in the expression of LEE-encoded genes in EHEC. The expression of the LEE in both EHEC and EPEC was shown to be activated by the autoinducer of quorum-sensing system 2 (termed AI-2) during the transition from the late exponential phase to the stationary phase (39). In the stationary phase, however, the autoinducer of quorum-sensing system 1 (termed AI-1) was shown to be downregulated by the expression of EHEC LEE-encoded genes but not by the expression of EPEC LEE-encoded genes through the involvement of SdiA, an E. coli homologue of quorum-sensing regulators (19; K. Kanamaru, unpublished results). Although EHEC and EPEC share coordinate regulation of the LEE operons by Ler, EPEC has the perABC genes on pEAF (12, 41), and the Per proteins act as a positive regulator for the expression of bfpA, which encodes bundle forming pili required for EPEC to adhere to epithelial cells (4). The Per proteins also regulate the expression of LEE genes through the transcription of ler (30). Transfer of the LEE from EPEC into a nonpathogenic E. coli K-12 strain was shown to be sufficient by itself to confer on the recipient the capacity to cause A/E lesions (28), while introduction of the LEE from EHEC into the E. coli K-12 strain was unable to confer on the recipient the ability to cause A/E lesions or the ability to secrete Esp proteins into the medium (8). Thus, studies have indicated that the regulatory systems required for LEE-encoded gene expression differ in EHEC and EPEC. Although the system for regulation of LEE gene expression in EHEC remains to be elucidated, the difference in the regulatory systems between EHEC and EPEC may affect the target tissue colonized in the intestine. In fact, several studies have suggested that EPEC strains preferentially colonize small intestinal epithelial cells, while EHEC strains tend to colonize large intestinal mucosa (10, 30, 43, 44), although some disparity has been reported (34, 35).
Studies have indicated that production and secretion of the Esp proteins required for attachment of EHEC and EPEC to host epithelial cells are affected by various factors, including the phase of bacterial growth, temperature, cations, pH, osmolarity (2, 21), and quorum-sensing systems. In addition, it is accepted that both pathogens adhere more efficiently to epithelial cells when they are grown in a tissue culture medium, such as Dulbecco's modified Eagle's medium (DMEM), than when they are grown in Luria-Bertani (LB) broth (5). In the present study, therefore, we investigated the conditions that stimulate adherence of EHEC to epithelial cells, such as Caco-2 cells. We found that this adherence is strongly affected by the NaHCO3 present in DMEM; NaHCO3 stimulates LEE-encoded gene expression, thus promoting the initial adherence of EHEC to Caco-2 cells in vitro. Importantly, the effect of NaHCO3 was very profound when EHEC was grown in LB broth containing a physiological concentration of NaHCO3. The significance of EHEC adherence capacity that is affected by the presence of NaHCO3 in the growth medium for infection of the human intestine is discussed below.
MATERIALS AND METHODS
Bacterial strains and plasmids.
EHEC strain O157Sakai (RIMD 0509952) has been described previously (40). EPEC strains E2348/69 and B171-8 have also been described previously (7, 41). Clinically isolated EHEC strains O157Okayama01, O157V425, O157Oku133, O157Oku702, and O157Niimi14 were supplied by T. Honda (Research Institute for Microbial Diseases, University of Osaka). E. coli DH5α and HB101 were used as transformant recipients. pBR322 was used for cloning of PCR products.
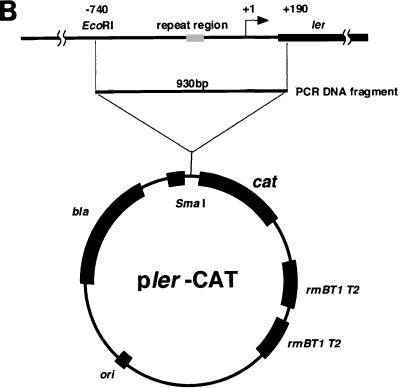
By using a PCR technique, a DNA fragment encompassing the ler gene was generated with primers ler-1 (5′-GCCTTATCAAAGAATTCTC-3′) and ler-4 (5′-TTTTTGGATCCAGTTTGATAATATCATCAG-3′). Plasmid pBRler is a derivative of pBR322 containing 989 bp of this PCR fragment digested with Sau3AI encompassing the ler gene. Plasmid plerCAT is a derivative of pKK232-8 containing 930 bp of the PCR fragment digested with EcoRI encompassing the ler regulatory region, generated with primers ler-1 and ler-2 (5′-GTGAATTCTTTTCCATATTC-3′).
Media.
DMEM without fetal calf serum was obtained from Gibco Life Technologies Inc. The DMEM which we used (catalogue no. 11965) was supplemented with 25 mM HEPES and 1 mM sodium pyruvate. LB broth has been described previously (39).
Tissue culture and adherence assay.
Caco-2 cells were maintained in DMEM supplemented with gentamicin (100 μg/ml), kanamycin (50 μg/ml), l-glutamine (2 mM), 100 μM minimum essential medium nonessential amino acid solution (Gibco BRL), and 10% fetal calf serum in a humidified atmosphere containing 5% CO2 at 37°C. EHEC strain O157Sakai was grown in LB broth overnight at 37°C, and then the culture was diluted 1:20 with fresh medium. Bacteria were preincubated in test tubes containing several media at 37°C with gentle shaking (80 rpm). After preincubation, the bacterial cultures were removed to measure the optical density at 600 nm and then allowed to adhere to monolayers of Caco-2 cells. The infected monolayers were incubated for 120 min in a humidified atmosphere containing 5% CO2 at 37°C and washed five times with phosphate-buffered saline. After an additional 150 min of incubation, the monolayers were washed five times with phosphate-buffered saline, fixed with methanol, and stained with Giemsa solution to visualize the adherent bacterial colonies. The adherence efficiency was determined by counting the number of bacteria adhering to Caco-2 cells. Clusters containing at least eight bacteria were counted as microcolonies. The adherence data obtained in this study were collected blind. The numbers of microcolonies that developed on Caco-2 cell monolayers were adjusted by using the CFU measured by plating the inoculated bacteria on agar plates.
Immunoblot analysis.
Bacterial strains were grown at 37°C in several media. The bacterial cultures were spun down, and each cell pellet was dissolved in 2× sodium dodecyl sulfate (SDS) sample buffer (62.5 mM Tris-HCl [pH 6.8], 2% SDS, 10% glycerol, 5% β-mercaptethanol, 0.1% bromophenol blue). The supernatant fluid was passed through a 0.45-μm-pore-size filter and precipitated by addition of 24% (vol/vol) trichloroacetic acid and 5% (vol/vol) deoxycholic acid, incubation for 20 min at 4°C, and subsequent centrifugation at 15,000 × g for 10 min at 4°C. The pellet was resuspended in both 1.5 M Tris base and 2× SDS sample buffer. The samples were analyzed by SDS-12% polyacrylamide gel electrophoresis (PAGE). Immunoblotting with intimin-, EspA-, EspB-, or Tir-specific antiserum was performed as described previously (39).
Northern blot analysis.
Total RNAs were prepared by the hot-phenol extraction method. The total RNAs (10 μg) were resolved by 1.0% agarose gel electrophoresis in the presence of formaldehyde and blotted onto a Hybond-N+ membrane (Amersham) as described previously (1). The membrane was hybridized to a DNA probe labeled with biotin by using a Psoralen biotin kit (Ambion) and washed. The signals were visualized with a BrightStar Biodetect kit (Ambion) by following the manufacturer's protocol. The 989-bp DNA fragment digested with Sau3AI containing the ler gene, prepared from the DNA fragment obtained by PCR with primers ler-1 and ler-4, was used as a probe for ler. The 526-bp DNA fragment containing ihfα amplified by PCR with primers ihfα-1 (5′-CCATAAGCCGGATCCTGCAAGATACCAGCCG-3′) and ihfα (5′-GCATGGGATCCGTTCTGCTGAAGTG-3′) was used as a probe for ihfα.
Promoter activity analysis.
A DNA fragment containing the regulatory region of ler was isolated by PCR from chromosomal DNA of EHEC strain O157Sakai. This fragment contains 740 bp of the upstream region and a 190-bp downstream region transcription start site for ler. After digestion with appropriate restriction enzymes, the DNA fragment was inserted upstream of the cat gene in pKK232-8 (Pharmacia). plerCAT and control plasmid pKK232-8 were introduced into EHEC strain O157Sakai and used for experiments. Bacteria were grown in LB broth with or without 44 mM NaHCO3 with gentle shaking after dilution (50-fold) of an overnight culture in LB broth. At each sampling time, an aliquot of culture was removed, the optical density at 600 nm was measured, and the bacteria were collected by centrifugation. The chloramphenicol acetyltransferase (CAT) activity of the protein extract of bacteria, which was prepared by sonication following centrifugation to remove insoluble material and debris, was measured as described by Shaw (38).
RESULTS
The capacity to adhere to Caco-2 cells is greater in EHEC grown in DMEM than in EHEC grown in LB broth.
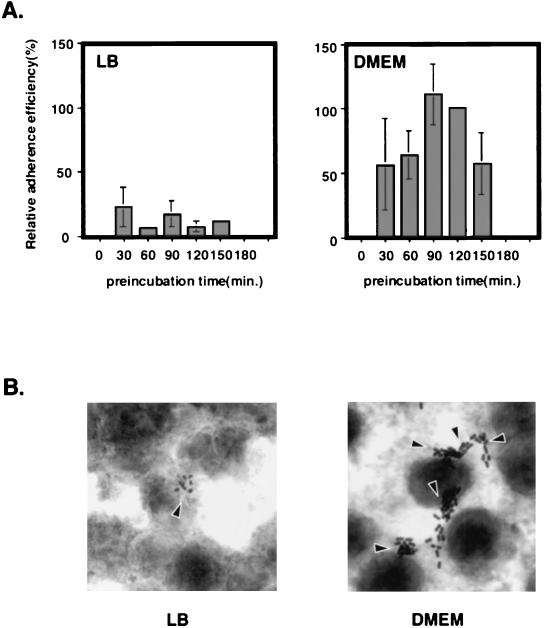
To assess the stimulatory effects of DMEM on the adherence to epithelial cells, EHEC strain O157Sakai cultures grown in DMEM-glucose (referred to as DMEM below unless indicated otherwise) or in LB broth to various growth phases were investigated for the capacity to adhere to Caco-2 cells at 4.5 h postinfection (see Materials and Methods). O157Sakai grown to the late log phase in DMEM for 90 to 120 min showed a marked increase in adherence compared to a culture grown in LB broth (Fig. 1A). The increased number of adherent bacteria in DMEM cultures at 4.5 h postinfection resulted from an increase in the number of microcolonies, as well as an increase in the number of bacteria, suggesting that the increase in adherence was due to intimate attachment to the epithelial cells (Fig. 1B).
FIG. 1.
Effect of preincubation conditions on EHEC adherence to Caco-2 cells. (A) EHEC grown in DMEM shows a greater capacity to adhere to Caco-2 cells than EHEC grown in LB broth. After 30, 60, 90, 120, and 150 min, bacterial cultures were removed to measure optical density at 600 nm, and the bacteria were allowed to adhere to Caco-2 cells. The adherence assay was performed in triplicate, and values are expressed relative to the value for EHEC preincubated in DMEM for 120 min. The data are the means and standard errors of the means for microscopic fields. (B) Caco-2 cells were infected with EHEC strain O157Sakai preincubated in LB broth or DMEM for 120 min. Microcolony formation was examined by Giemsa staining and phase-contrast microscopy (40).
A component of DMEM can stimulate EHEC adherence.
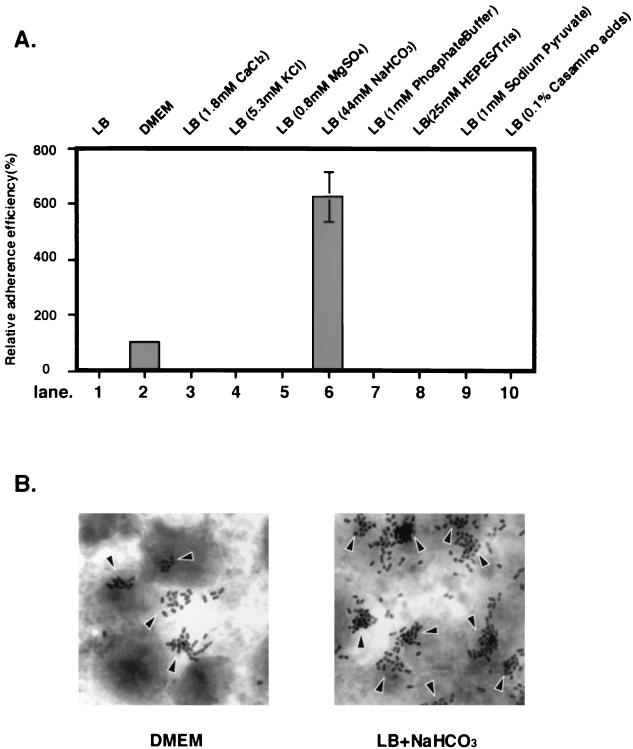
To examine the possibility that some component(s) present in DMEM improved the adherence capacity, components of DMEM were added to LB broth. First, the components of DMEM were categorized into four groups: inorganic salts, amino acids, vitamins, and other compounds (glucose, phenol red, HEPES, and sodium pyruvate). Previous reports showed that the presence of several salts and buffer in minimum medium stimulates the expression of LEE-encoded genes in EHEC and EPEC (2, 21). Furthermore, our preliminary data showed that addition of an amino acid mixture or a vitamin mixture to LB broth did not affect the expression of LEE-encoded genes in EHEC (data not shown). Thus, eight components, CaCl2, KCl, MgSO4, NaHCO3, NaH2PO4, HEPES, sodium pyruvate, and Casamino Acids, were each added to LB broth. In each of the LB broth preparations, the pH and the concentration of the component added were adjusted to the pH and the concentration in DMEM. Addition of CaCl2, KCl, MgSO4, NaH2PO4, HEPES, sodium pyruvate, or Casamino Acids had no significant effect on the adherence capacity (Fig. 2A). However, NaHCO3 markedly increased the adherence capacity; the adherence of bacteria grown in LB broth containing 44 mM NaHCO3 was five times more efficient than the adherence of bacteria grown in DMEM (Fig. 2A). Under the conditions used, neither hyperosmotic LB broth nor LB broth containing a high concentration of HEPES, both of which have been reported to promote expression of the esp operon (2), had a significant effect on the adherence of O157Sakai to Caco-2 cells (data not shown). Although the adherent bacteria grown under both conditions produced microcolonies on Caco-2 cells, the number of microcolonies and the number of bacteria in each colony were greater for O157Sakai grown in LB broth with NaHCO3 (44 mM) than for O157Sakai grown in DMEM (Fig. 2B). Thus, these results suggested that NaHCO3 is a component of DMEM that stimulates EHEC adherence.
FIG. 2.
Effect of DMEM components on EHEC adherence to Caco-2 cells. Bacteria were preincubated in test tubes containing LB broth supplemented with 1.8 mM CaCl2, with 5.3 mM KCl, with 0.8 mM MgSO4, with 44 mM NaHCO3, with 1 mM phosphate buffer (pH 7.4), with 25 mM HEPES-Tris (pH 7.4), with 1 mM sodium pyruvate, or with 0.1% Casamino Acids. (A) EHEC grown in LB broth supplemented with NaHCO3 shows a greater capacity to adhere to Caco-2 cells than EHEC grown in LB broth and DMEM. After 120 min, bacterial cultures were allowed to adhere to Caco-2 cells. The adherence assay was performed in triplicate, and values are expressed relative to the value for EHEC preincubated in DMEM for 120 min. The data are the means and standard errors of the means for microscopic fields. (B) Caco-2 cells were infected with EHEC O157Sakai preincubated in LB broth supplemented with NaHCO3 or in DMEM for 120 min. Microcolony formation was examined by Giemsa staining and phase-contrast microscopy.
NaHCO3 can stimulate LEE gene expression.
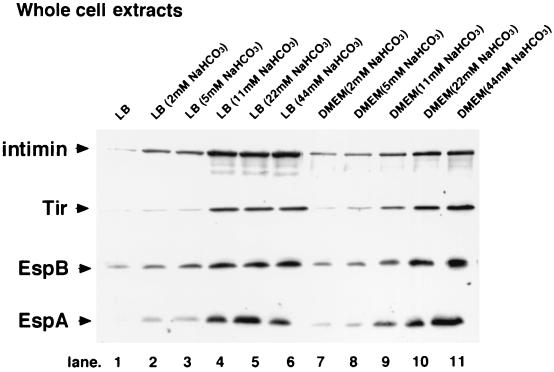
To investigate whether the stimulatory effect of NaHCO3 on O157Sakai could result from increased secretion of EspA, EspB, or Tir, the proteins secreted into the medium were precipitated with trichloroacetic acid and analyzed by immunoblotting with anti-EspA, anti-EspB, and anti-Tir antibodies (see Materials and Methods). The results showed that protein secretion was significantly enhanced for bacteria grown in LB broth with NaHCO3 compared to bacteria grown in LB broth (data not shown). Bacteria grown in LB broth with various concentrations of NaHCO3 were subsequently investigated for the production of EspA, EspB, Tir, and intimin. As shown in Fig. 3, the production of these proteins in O157Sakai was stimulated upon addition of NaHCO3 to LB broth, and the levels of production increased as the concentration of NaHCO3 increased. The same was true for DMEM (Fig. 3). In this study, we performed a Western blot experiment to test the effects of NaHCO3 on expression of the LEE genes together with visualization of bacterial products by Coomassie blue and silver staining of SDS-PAGE gels. However, there was no significant difference in the profiles of proteins visualized in the SDS-PAGE gels with and without NaHCO3 in LB broth and DMEM (data not shown). In a medium, NaHCO3 at concentrations of 11 to 44 mM resulted in high levels of production of EspA, EspB, Tir, and intimin, suggesting that expression of the LEE-encoded genes could be coordinately activated by the presence of NaHCO3.
FIG. 3.
Effect of NaHCO3 on the expression of LEE-encoded genes. Whole-cell extracts of EHEC strain O157Sakai grown in LB broth supplemented with 0, 2, 5, 11, 22, and 44 mM NaHCO3 (lanes 1, 2, 3, 4, 5, and 6, respectively) and in DMEM supplemented with 2, 5, 11, 22, and 44 mM NaHCO3 (lanes 7, 8, 9, 10, and 11, respectively) were analyzed. The expression levels of intimin, Tir, EspB, and EspA were determined by Western blotting with protein-specific antiserum (40).
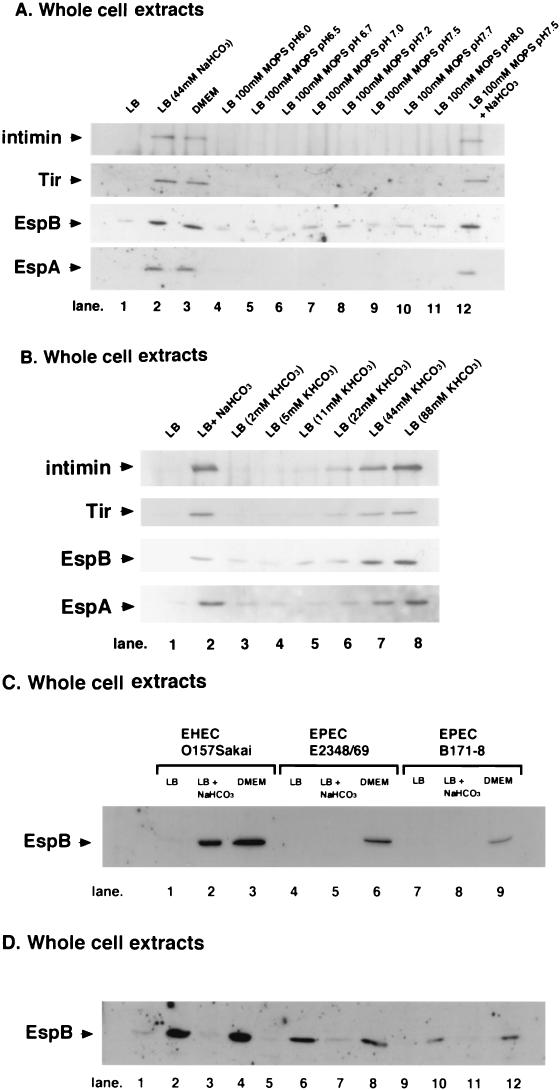
NaHCO3 is slightly alkaline in solution, in which it acts as a pH buffer through equilibrium with CO2. Since addition of NaHCO3 to LB broth at a final concentration of 44 mM resulted in a slightly alkaline medium (pH 7.5 to 7.7) (Abe, unpublished results), morpholinepropanesulfonic acid (MOPS)-containing LB broth (MOPS-LB broth) preparations with pH values ranging from 6.0 to 8.0 (pH 6.0, 6.5, 6.7, 7.0, 7.2, 7.5, 7.7, and 8.0) were investigated to determine whether there was a stimulatory effect on the production of EspA, EspB, and Tir by immunoblotting whole bacterial lysates with anti-EspA, anti-EspB, anti-Tir, and anti-intimin antibodies. MOPS-LB broth was prepared by adding MOPS (final concentration, 100 mM) to LB broth and adjusting the pH with NaOH or HCl. As shown in Fig. 4A, a change in the pH of the medium from pH 6.0 to 8.0 had no significant effect on the production of protein, suggesting that the HCO3− ion itself is the factor that stimulates LEE-encoded gene expression in O157Sakai. To test this hypothesis, O157Sakai grown in MOPS-LB broth (pH 7.5) with NaHCO3 was investigated for the capacity to stimulate the production of EspA, EspB, and Tir by immunoblotting whole bacterial lysates with anti-EspA, anti-EspB, anti-Tir, and anti-intimin antibodies. The results showed that the levels of protein production were significantly increased in bacteria grown in MOPS-LB broth with NaHCO3 compared to the levels of protein production in bacteria grown in LB broth (Fig. 4A, lane 12). In addition, O157Sakai grown in LB broth with and without KHCO3 was investigated by the same procedure. As shown in Fig. 4B, production of all the proteins was enhanced when the bacteria were grown in LB broth with KHCO3 at a concentration of 22 to 88 mM, strongly indicating that HCO3− ions are responsible for the induction of LEE-encoded gene expression.
FIG.4.
Effect of the HCO3− ion on the expression of LEE-encoded genes. EHEC strain O157Sakai was grown in LB broth overnight and then diluted 1:20 with fresh medium. Bacteria were grown for 120 min at 37°C, and 1 ml of the culture was removed to measure optical density and to prepare whole-cell extracts. (A) The pH of LB broth did not affect the expression of LEE-encoded genes. Whole-cell extracts of EHEC strain O157Sakai grown in LB broth supplemented with 0 and 44 mM NaHCO3 (lanes 1 and 2) and in DMEM (lane 3) were analyzed by using the experimental procedure. Whole-cell extracts of EHEC strain O157Sakai grown in MOPS-LB broth at pH 6.0, 6.5, 6.7, 7.0, 7.2, 7.5, 7.7, and 8.0 (lanes 4, 5, 6, 7, 8, 9, 10, and 11, respectively) and at pH 7.5 supplemented with 44 mM NaHCO3 (lane 13) were analyzed. (B) KHCO3 also stimulated the expression of intimin, Tir, EspB, and EspA. Whole-cell extracts of EHEC strain O157Sakai grown in LB broth supplemented with 0, 2, 5, 11, 22, 44, and 88 mM KHCO3 (lanes 1, 3, 4, 5, 6, 7, and 8, respectively) and with 44 mM NaHCO3 (lane 2) were analyzed by using the experimental procedure described above. (C) Whole-cell extracts of EHEC strain O157Sakai (lanes 1 to 3), EPEC strain E2348/69 (lanes 4 to 6), and EPEC strain B171-8 (lanes 7 to 9) grown in LB broth (lanes 1, 4, and 7), in LB broth supplemented with 44 mM NaHCO3 (lanes 2, 5, and 8), and in DMEM (lanes 3, 6, and 9) were analyzed by using experimental procedure described above. The expression levels of EspB were determined by Western blotting with EspB-specific antiserum. (D) Whole-cell extracts of EHEC strains O157Sakai (lanes 1 and 2), O157Okayama01 (lanes 3 and 4), O157V425 (lanes 5 and 6), O157Oku133 (lanes 7 and 8), O157Oku702 (lanes 9 and 10), and O157Niimi14 (lanes 11 and 12) grown in LB broth (lanes 1, 3, 5, 7, 9, and 11) and in LB broth supplemented with 44 mM NaHCO3 (lanes 2, 4, 6, 8, 10, and 12) were analyzed by using the experimental procedure described above.
A previous report of Kenny et al. showed that the NaHCO3 concentration in DMEM can affect the secretion of LEE-encoded proteins (21). We also observed that the NaHCO3 concentration in DMEM could affect the expression of LEE-encoded proteins in EHEC (Fig. 3). Thus, the effects of NaHCO3 in LB broth on the expression of EspB were examined by using two additional EPEC strains. Although NaHCO3 in LB broth affected the expression of EspB in EHEC, no effect was observed for EPEC strains (Fig. 4C). Furthermore, stimulation of the expression of EspB was demonstrated with several clinically isolated O157:H7 strains (Fig. 4D). These data clearly indicate that the effect in EHEC of NaHCO3, which triggers expression of LEE-encoded genes in EHEC, was different from the effect in EPEC under the growth conditions used.
Activation of the ler gene by NaHCO3.
Since LEE-encoded gene expression can be positively regulated by Ler encoded by the ler gene (30, 6), we hypothesized that the enhanced production of EspA, EspB, Tir, and intimin in O157Sakai grown in LB broth with NaHCO3 resulted from activation of the ler gene, which led to production of LEE-encoded genes, including espA, espB, tir, and eae. Analysis of the mRNA extracted from O157Sakai grown in LB broth with NaHCO3 by Northern blotting with a DNA probe specific for eae indicated that the level of eae mRNA was greater in bacteria grown in LB broth containing 11 to 44 mM NaHCO3 than in bacteria grown in LB broth (data not shown).
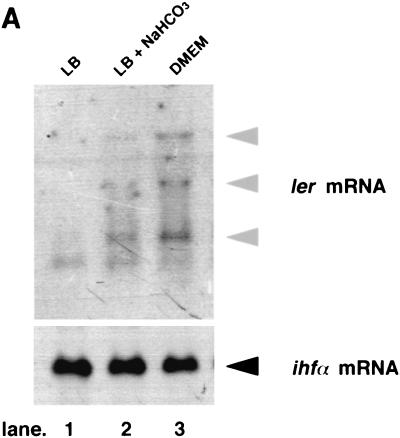
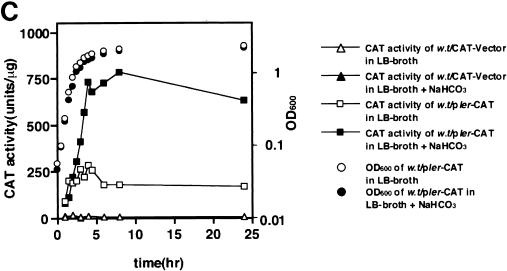
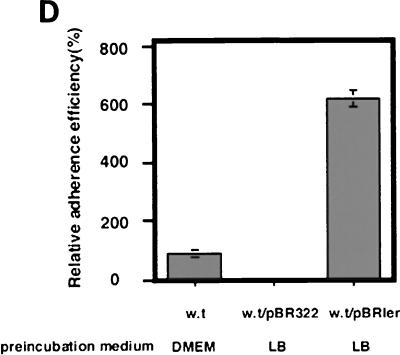
Elliott et al. previously reported that Ler stimulated the level of expression of LEE-encoded genes and bacterial adherence (6); these authors examined the effect of ler when bacteria were grown under certain conditions, such as incubation at 37°C in minimal essential medium. We confirmed that the ler locus enhanced the expression of LEE-encoded genes, including bacterial adherence, in DMEM (data not shown). However, it was not clear whether introduction of the ler gene into EHEC O157:H7 could stimulate adherence when bacteria were grown in LB broth, as it did when bacteria were grown in minimal essential medium or DMEM. The ler mRNA in the bacteria grown in LB broth, LB broth with NaHCO3, or DMEM was examined by Northern blotting with a ler-specific DNA probe. As shown in Fig. 5A, although transcription was barely detected in the absence of NaHCO3, production of ler mRNA could be detected when the bacteria were grown in LB broth with NaHCO3 or in DMEM. The activation of the ler gene by NaHCO3 was further confirmed by constructing a pler-cat transcriptional fusion plasmid (plerCAT) (Fig. 5B). When plerCAT was introduced into O157Sakai and the organism was grown in LB broth, the CAT activity was about three times higher in bacteria grown in the presence of 44 mM NaHCO3 than in bacteria grown in the absence of NaHCO3 (Fig. 5C). Consistent with the results described above, when a cloned ler gene on pBR322 (pBRler) was introduced into O157Sakai, the bacteria adhered more efficiently to Caco-2 cells, even when they were grown in LB broth without NaHCO3, than the mock control adhered (Fig. 5D and E). The levels of production of LEE-encoded genes were more than three times greater in the bacteria carrying pBRler than in the bacteria not carrying pBRler (data not shown).
FIG.5.
HCO3− ion stimulates the expression of LEE-encoded regulator. EHEC O157:H7 was grown in LB broth overnight and then diluted 1:20 with fresh medium. Bacteria were grown for 120 min at 37°C, and 10 ml of the culture was removed to measure optical density and to isolate total cellular RNAs. (A) NaHCO3 stimulated transcription of ler. Total RNAs (10 μg) prepared from EHEC strain O157Sakai grown in LB broth not supplemented with NaHCO3 (lane 1) or supplemented with 44 mM NaHCO3 (lane 2) and in DMEM (lane 3) were subjected to Northern blot analysis. The transcription levels of ler mRNA were determined by using the labeled Sau3AI DNA fragment containing the ler gene as the probe (upper panel). The levels of ihf mRNA were determined as the control by using the labeled PCR fragment containing the ihfα gene as the probe (lower panel). (B) Cloning of the ler regulatory region into the pKK232-8 vector. The blunted 930-bp EcoRI PCR fragment encompassing the ler regulatory region was cloned into the SmaI site between the rrnB T1 terminator and the cat gene. (C) Activation of the ler promoter in response to addition of NaHCO3 to LB broth. EHEC strain O157Sakai containing pKK232-8 (triangles) or plerCAT (squares) was grown in LB broth (open symbols) or in LB broth supplemented with 44 mM NaHCO3 (solid symbols). The solid triangles are not visible due to overlap with the open triangles. CAT activity was determined at different times (38). The data are the means and standard errors of the means (SEM) obtained in triplicate. SEMs are not visible due to the very narrow range. OD600, optical density at 600 nm; w.t, EHEC strain O157Sakai wild type. (D) Effect of ler overproduction on adherence of EHEC to Caco-2 cells. EHEC strain O157Sakai containing pBR322 or pBRler was grown in LB broth. After 120 min, bacterial cultures were allowed to adhere to Caco-2 cells. The adherence assay was performed in triplicate, and values are expressed relative to the value for EHEC preincubated in DMEM for 120 min. The data are the means and standard errors of the means for microscopic fields. (E) Caco-2 cells were infected with EHEC strain O157Sakai containing pBRler preincubated in LB broth for 120 min. Microcolony formation was examined by Giemsa staining and phase-contrast microscopy.
Thus, these results clearly indicated that NaHCO3 could stimulate the expression of ler in O157Sakai, which in turn induced the expression of LEE-encoded genes required for bacterial adherence.
DISCUSSION
In the present study, we investigated the abilities of the components of DMEM to stimulate the adherence of O157Sakai to Caco-2 cells, since it has been noted that both EHEC and EPEC strains grown in a tissue culture medium, such as DMEM, have a remarkably efficient adherence phenotype on epithelial cells compared to the phenotype of EHEC grown in LB broth. In addition, studies have indicated that production and secretion of the type III secreted proteins (LEE gene-encoded secretion proteins), such as EspA, EspB, EspD, and Tir, from EHEC (and Shiga toxin-producing E. coli) or EPEC strains are affected by various environmental stimuli. Recent whole-genome analyses of EHEC strains, including O157Sakai, supported this notion and resulted in the prediction that the pathogens have an elaborate putative sensing system to detect environmental signals, as well as a regulatory system involved in the expression of virulence-associated genes, such as LEE-encoded genes (14, 15, 33). The results of the present study confirmed that O157Sakai grown to the late log phase had a greatly enhanced adherence capacity when it was grown in DMEM rather than in LB broth (Fig. 1A). An examination of the DMEM components responsible for the stimulatory effect on O157Sakai showed that NaHCO3 can greatly enhance the capacity to adhere to Caco-2 cells. The efficiency was increased further when the bacteria were grown in LB broth supplemented with NaHCO3; under these conditions the adherence capacity was more than five times that in DMEM and more than 50 times that in LB broth.
The stimulatory effect of NaHCO3 on EHEC adherence resulted from an increase in expression of the LEE-encoded genes via activation of the ler gene. Importantly, the increase in the amount of protein expressed from O157Sakai accompanied an increase in the concentration of NaHCO3 (from 0 to ∼44 mM) in the medium (Fig. 3), suggesting that NaHCO3 can somehow stimulate the expression of LEE-encoded genes in EHEC. The observation that the NaHCO3 concentration in DMEM affects the secretion of LEE-encoded proteins was originally reported by Kenny et al. (21). In that study, Kenny suggested that secretions produced by small intestine mucosa contain high levels of NaHCO3 that neutralize the acid environment of the stomach and perhaps signal the passage of EPEC to the site of colonization on epithelial cells. This hypothesis is called the high to low gradient hypothesis for EPEC (20). In the present study, we found that although the presence of NaHCO3 in DMEM can affect the expression of LEE-encoded proteins in EHEC (Fig. 3), the effect of NaHCO3 in LB broth on EHEC LEE expression was more dramatic than the effect of NaHCO3 in DMEM, which was never observed to have an effect on EPEC LEE expression (Fig. 4C). Although the mechanisms of the different effects of NaHCO3 on expression in EHEC and EPEC are still unclear, since the effect on EHEC LEE expression was observed when the bacteria were grown in LB broth containing a physiological concentration of NaHCO3, at least the richness of the growth medium could be an important factor in the stimulation of EHEC adherence.
Bacterial growth conditions affect the expression of LEE in a Shiga toxin-producing E. coli O26 strain, as reported by Ebel et al. (5), who found different levels of LEE-encoded secretion proteins, such as EspB, in bacteria grown in LB broth and in bacteria grown in a serum-free tissue culture medium. Later, Beltrametti et al. indicated that the esp promoter in EHEC was activated in response to Ca2+, Mn2+, and high osmolarity in M9 medium or in response to HEPES in DMEM (2). We also examined the capacity of O157Sakai grown in LB broth with Ca2+, Mn2+, or HEPES or in LB broth at high osmolarity to adhere to Caco-2 cells with the capacity of cells grown in plain LB broth to adhere to Caco-2 cells; however, none of these components had an appreciable effect on adherence, including the expression of LEE genes (Fig. 2A and unpublished data), suggesting that they do not affect the adherence capacity of EHEC in rich medium. Therefore, to our knowledge, NaHCO3 is the first reported compound capable of stimulating EHEC adherence, including expression of LEE-encoded genes under nutrient-rich conditions.
As an environmental signal, the HCO3− ion affects the expression of various virulence-associated genes in many pathogenic bacteria (29). For example, the expression of anthrax toxin genes (pag, lef, and cya) in Bacillus anthracis is enhanced by two physiological signals, CO2-HCO3− and temperature (24, 26). The production of several other bacterial virulence factors, including cholera toxin (16) and toxic shock syndrome toxin 1 of Staphylococcus aureus (36), was shown to be stimulated by CO2. Although the target tissues for infection by the pathogens differ, such studies have strongly indicated that the HCO3− ion present in the microenvironment is an important signal not only in promoting infection but also for evading host defense systems. For the stimulatory effect of NaHCO3 in a rich medium on the adherence of EHEC, we hypothesized that the nutrient-rich medium containing the HCO3− ion mimics a growth environment, such as the human intestine. The HCO3− ion in the intestinal tract contributes to the buffering of extracellular fluids in equilibrium with CO2; the concentration of the ion in the ileum is greater than that in the jejunum, and thus the HCO3− ion concentration increases along the intestinal tract (37, 45). Consistent with this notion, HCO3− at a concentration of 44 mM, which is close to the concentration in the ileum (9, 37), can strongly stimulate LEE-encoded gene expression in O157Sakai, thus promoting adherence to epithelial cells.
The effect of the HCO3− ion on EHEC adherence seemed to depend on the growth phase. Transcriptional fusion with cat as a reporter gene demonstrated that ler was activated at the transcriptional level upon addition of NaHCO3 to LB broth (Fig. 5C), in which activation of ler expression was dependent on the bacterial growth phase. Indeed, the ler promoter activity increased greatly from the mid-exponential phase to the stationary phase. Very recently, two kinds of quorum-sensing systems have been reported to be involved in the expression of LEE-encoded genes in EHEC (19, 39). Sperandio et al. (39) showed that expression of LEE-encoded genes in EHEC and EPEC is activated by one of the systems, termed quorum-sensing system 2, and an autoinducer called AI-2, which has been shown to be involved during the transition from the late exponential phase to the stationary phase, although the effect of AI-2 in LB broth seems not to be as strong as the effect in DMEM. Thus, it is intriguing to speculate that the observed stimulation of expression of LEE-encoded genes in O157Sakai from the mid-exponential phase to the stationary phase of growth in the presence of HCO3− ions is due to the synergistic effects of NaHCO3 and AI-2. In fact, it has been proposed that the autoinducer is synthesized by the normal E. coli flora residing in the large intestine (39).
The intestinal tissues targeted by EPEC and EHEC seem to differ. An intestinal biopsy of a patient infected with EPEC showed that the bacteria preferentially adhered to the small intestine epithelium (31). In a piglet infection model, EPEC was shown to adhere to the small intestine, while EHEC colonized the large intestine epithelium (10, 43, 44). Although the host factors and the bacterial factors affecting the preference for attachment sites on the intestinal epithelium remain to be elucidated, environmental regulation of LEE-encoded gene expression in EHEC and EPEC is an important factor. Indeed, Kenny et al. have recently indicated that the secretion of Esp proteins from EPEC is maximized when bacteria are grown under physiological conditions similar to those in the intestine (21). Thus, although the mechanisms underlying the stimulation of ler gene expression in EHEC grown in rich medium in the presence of NaHCO3 remain unclear, our findings provide insight into the strategy employed by the bacteria when they target the site of attachment in the human intestine. At present, although we cannot rule out the possibility that some unknown adherence factor(s) stimulated by NaHCO3 is involved in colonization of the intestine by EHEC, we believe that expression of LEE-encoded genes in EHEC stimulated in response to conditions similar to those in the lower intestinal tract environment plays a crucial role in the initial attachment, including the development of microcolonies on the intestinal epithelium.
Acknowledgments
We thank Toshihiko Suzuki and Asaomi Kuwae for technical advice and helpful discussions. We thank Chizu Sasako and Yukie Sameshima for technical assistance. We thank Akio Abe for providing clinically isolated EHEC strains.
This work was supported by the Research for the Future Program of the Japanese Society for the Promotion of Science.
Editor: V. J. DiRita
REFERENCES
- 1.Abe, H., T. Abo, and H. Aiba. 1999. Regulation of intrinsic terminator by translation in Escherichia coli: transcription termination at a distance downstream. Genes Cells 4:87-97. [DOI] [PubMed] [Google Scholar]
- 2.Beltrametti, F., A. U. Kresse, and C. A. Guzman. 1999. Transcription regulation of the esp genes of enterohemorrhagic Escherichia coli. J. Bacteriol. 181:3409-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Devinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowski, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donnenberg, M. S., J. A. Giron, J. P. Nataro, and J. B. Kaper. 1992. A plasmid-encoded type IV fimbrial gene of enteropathogenic Escherichia coli associated with localized adherence. Mol. Microbiol. 6:3427-3437. [DOI] [PubMed] [Google Scholar]
- 5.Ebel, F., C. Deibel, A. U. Kresse, C. A. Guzman, and T. Chakraborty. 1996. Temperature- and medium-dependent secretion of proteins by Shiga toxin-producing Escherichia coli. Infect. Immun. 64:4472-4479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elliott, S. J., V. Sperandio, J. A. Giron, S. Shin, J. L. Mellies, L. Wainwright, S. W. Hutcheson, T. K. McDaniel, and J. B. Kaper. 2000. The locus of enterocyte effacement (LEE)-encoded regulator controls expression of both LEE- and non-LEE-encoded virulence factors in enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 68:6115-6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliot, S. J., L. A. Wainwright, T. K. McDaniel, K. G. Jarvis, Y.-K. Deng, L.-C. Lai, B. P. McNamara, M. S. Donnenberg, and J. B. Kaper. 1998. The complete sequence of the locus of enterocyte effacement (LEE) from enteropathogenic Escherichia coli E2348/69. Mol. Microbiol. 28:1-4. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, S. J., J. Yu, and J. B. Kaper. 1999. The cloned locus of enterocyte effacement from enterohemorrhagic Escherichia coli O157:H7 is unable to confer the attaching and effacing phenotype upon E. coli K-12. Infect. Immun. 67:4260-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fordtran, J. S., and T. W. Locklear. 1966. Ionic constituents and osmolality of gastric and small-intestinal fluids after eating. Am. J. Dig. Dis. 11:503-521. [DOI] [PubMed] [Google Scholar]
- 10.Francis, D. H., J. E. Collins, and J. R. Duimstra. 1986. Infection of gnotobiotic pigs with an Escherichia coli O157:H7 strain associated with an outbreak of hemorrhagic colitis. Infect. Immun. 51:953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frankel, G., A. D. Phillips, I. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 12.Gomez-Duarte, O. G., and J. B. Kaper. 1995. A plasmid-encoded regulatory region activates chromosomal eaeA expression in enteropathogenic Escherichia coli. Infect. Immun. 63:1767-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grado, M., A. Abe, A. Gauthier, O. S. Mortimer, R. Devinney, and B. B. Finlay. 1999. Identification of the intimin-binding domain of Tir of enteropathogenic Escherichia coli. Cell. Microbiol. 1:7-17. [DOI] [PubMed] [Google Scholar]
- 14.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12 (supplement). DNA Res. 8:47-52. [DOI] [PubMed] [Google Scholar]
- 15.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 16.Iwanaga, M., and K. Yamamoto. 1985. New medium for the production of cholera toxin by Vibrio cholerae O1 biotype El Tor. J. Clin. Microbiol. 22:405-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jarvis, K. G., J. A. Giron, A. E. Jerse, T. K. McDaniel, M. S. Donnenberg, and J. B. Kaper. 1995. Enteropathogenic Escherichia coli contains a putative type III secretion system necessary for the export of proteins involved in attaching and effacing lesion formation. Proc. Natl. Acad. Sci. USA 92:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jerse, A. E., and J. B. Kaper. 1991. The eae gene of enteropathogenic Escherichia coli encodes a 94-kilodalton membrane protein, the expression of which is influenced by the EAF plasmid. Infect. Immun. 59:4302-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanamaru, K., K. Kanamaru, I. Tatsuno, T. Tobe, and C. Sasakawa. 2000. SdiA, an Escherichia coli homologue of quorum-sensing regulators, controls the expression of virulence factors in enterohemorrhagic Escherichia coli O157:H7. Mol. Microbiol. 38:805-816. [DOI] [PubMed] [Google Scholar]
- 20.Kenny, B. 1999. Phosphorylation of tyrosine 474 of the enteropathogenic Escherichia coli (EPEC) Tir receptor molecule is essential for actin nucleateing activity and is preceded by additional host modifications. Mol. Microbiol. 31:1229-1241. [DOI] [PubMed] [Google Scholar]
- 21.Kenny, B., A. Abe, M. Stein, and B. B. Finlay. 1997. Enteropathogenic Escherichia coli protein secretion is induced in response to conditions similar to those in the gastrointestinal tract. Infect. Immun. 65:2606-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny, B., and B. B. Finlay. 1995. Protein secretion by enteropathogenic Escherichia coli is essential for transducing signals to epithelial cells. Proc. Natl. Acad. Sci. USA 92:7991-7995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutton, S., I. Rosenshine, M. J. Pallen, I. Nisan, B. C. Neves, C. Bain, C. Wolff, G. Dougan, and G. Frankel. 1998. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 17:2166-2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koehler, T. M., Z. Dai, and M. Kaufman-Yarbray. 1994. Regulation of the Bacillus anthracis protective antigen gene: CO2 and a trans-acting element activate transcription from one of two promoters. J. Bacteriol. 176:586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kresse, A. U., M. Rohde, and C. A. Guzman. 1999. The EspD protein of enterohemorrhagic Escherichia coli is required for the formation of bacterial surface appendages and is incorporated in the cytoplasmic membranes of target cells. Infect. Immun. 67:4834-4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makino, S., C. Sasakawa, I. Uchida, N. Terakado, and M. Yoshikawa. 1988. Cloning and CO2-dependent expression of the genetic region for encapsulation from Bacillus anthracis. Mol. Microbiol. 2:371-376. [DOI] [PubMed] [Google Scholar]
- 27.McDaniel, T. K., K. G. Jarvis, M. S. Donnenberg, and J. B. Kaper. 1995. A genetic locus of enterocyte effacement conserved among diverse entrobacterial pathogenes. Proc. Natl. Acad. Sci. USA 92:1664-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel, T. K., and J. B. Kaper. 1997. A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23:399-407. [DOI] [PubMed] [Google Scholar]
- 29.Mekalanos, J. J. 1992. Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mellies, J. L., S. J. Elliott, V. Sperandio, M. S. Donnenberg, and J. B. Kaper. 1999. The Per regulon of enteropathogenic Escherichia coli: identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler). Mol. Microbiol. 33:296-306. [DOI] [PubMed] [Google Scholar]
- 31.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perna, N. T., G. F. Mayhew, G. Posfai, S. Elliott, M. S. Donnenberg, J. B. Kaper, and F. R. Blattner. 1998. Molecular evolution of a pathogenicity island from enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 66:3810-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2000. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 34.Phillips, A. D., S. Navabpour, S. Hicks, G. Dougan, and G. Frankel. 2000. Enterohaemorrhagic Escherichia coli O157:H7 target Peyer's patches in humans and cause attaching/effacing lesions in both human and bovine intestine. Gut 47:377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reece, S., C. P. Simmons, R. J. Fitzhenry, S. Matthews, A. D. Phillips, G. Dougan, and G. Frankel. 2001. Site-directed mutagenesis of intimin α modulates intimin-mediated tissue tropism and host specificity. Mol. Microbiol. 40:86-98. [DOI] [PubMed] [Google Scholar]
- 36.Ross, R. A., and A. B. Onderdonk. 2000. Production of toxic shock syndrome toxin 1 by Staphylococcus aureus requires both oxygen and carbon dioxide. Infect. Immun. 68:5205-5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruppin, H., W. Domschke, and K. H. Soergel. 1981. Diarrhea in disorders of intestinal transport. Georg Thieme Verlag Stuttgart, Germany.
- 38.Shaw, W. V. 1975. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 43:737-755. [DOI] [PubMed] [Google Scholar]
- 39.Sperandio, B., J. L. Mellies, W. Nguyen, S. Shin, and J. B. Kaper. 1999. Quorum sensing controls expression of the type III secretion gene transcription and protein secretion in enterohemorrhagic and enteropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 96:15196-15201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Sasakawa. 2000. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 68:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tobe, T., G. K. Schoolnik, I. Sohel, V. H. Bustamante, and J. L. Puente. 1996. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 21:963-975. [DOI] [PubMed] [Google Scholar]
- 42.Tuttle, J., T. Gomez, M. P. Doyle, J. G. Wells, T. Zhao, R. V. Tauxe, and P. M. Griffin. 1999. Lesson from a large outbreak of Escherichia coli O157:H7 infections: insight into the infectious dose and method of widespread contamination of hamburger patties. Epidemiol. Infect. 122:185-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzipori, S., I. K. Wachsmuth, C. Chapman, R. Birner, J. Brittingham, C. Jackson, and J. Hogg. 1986. The pathogenesis of hemorrhagic colitis caused by Escherichia coli O157:H7 in gnotobiotic piglets. J. Infect. Dis. 154:712-716. [DOI] [PubMed] [Google Scholar]
- 44.Tzipori, S., R. Gibson, and J. Montanaro. 1989. Nature and distribution of mucosal lesions associated with enteropathogenic and enterohemorrhagic Escherichia coli in piglets and the role of plasmid-mediated factors. Infect. Immun. 57:1142-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wrong, O. M., C. J. Edmonds, and V. S. Chadwick. 1981. The large intestine: its role in mammalian nutrition and homeostasis. MTP Press Limited, Lancaster, England.