Abstract
Nontypeable (NT) strains of Haemophilus influenzae are an important cause of acute otitis media (OM). The pathogenic process by which NT H. influenzae strains cause OM is poorly understood. In order to identify specific virulence factors important for OM pathogenesis, genomic subtraction of the NT H. influenzae middle ear isolate G622 against H. influenzae strain Rd was conducted and the resulting subtraction products were used to screen a panel of H. influenzae isolates. Subtraction identified 36 PCR fragments unique to strain G622, which were used in a preliminary screen of 48 middle ear isolates and 46 nasopharyngeal and throat isolates to identify genes found more frequently among middle ear isolates. These experiments identified a PCR fragment with high homology to the lipooligosaccharide biosynthesis gene lic2B (originally identified in an H. influenzae type b strain) among 52% of the middle ear isolates and 9% of nasopharyngeal and throat isolates. The lic2B gene cloned from NT H. influenzae strain G622 was 99% identical at the amino acid level to that of the H. influenzae type b strain RM7004. The lic2B gene was used to screen a larger panel of H. influenzae isolates including the original 48 middle ear isolates, 40 invasive type b isolates, 90 NT H. influenzae throat isolates from children attending day care, and 32 NT H. influenzae nasopharyngeal clinical isolates. The lic2B gene was found 3.7 times more frequently among middle ear isolates than in throat isolates from children attending day care. These data suggest that a specific NT H. influenzae gene is associated with OM.
Haemophilus influenzae is a gram-negative bacterium that colonizes the upper respiratory tract of humans. In addition to asymptomatic colonization, H. influenzae also causes significant infection. Encapsulated H. influenzae serotype b strains cause invasive diseases such as bacteremia, septic arthritis, and meningitis. Widespread vaccination with type b polysaccharide conjugate vaccines has greatly reduced the incidence of severe disease due to H. influenzae type b; these vaccines, however, do not protect against infection with non-type b encapsulated or nontypeable (NT) H. influenzae strains.
Although infections with NT H. influenzae are rarely fatal, these strains cause significant morbidity and are isolated from up to half of middle ear aspirates from children with otitis media (OM) (6). OM is an infection of the middle ear resulting in middle ear effusion, fever, irritability, and inflammation of the tympanic membrane and is the most common bacterial infection in infants and young children (17). It is generally assumed that middle ear infection occurs when bacteria colonizing the nasopharynx enter the middle ear space through the Eustachian tube (21). Epidemiologic factors known to be associated with the development of OM include a genetic predisposition, preceding viral respiratory infection, attendance of day care, lack of breast feeding, and young age (1, 2, 28). While these factors have been well studied, the specific bacterial virulence factors important for the invasion of the middle ear have not been well defined.
Multiple studies have shown that NT H. influenzae strains are very diverse (4, 10, 23), and children may be colonized with multiple strains at one time (26), leading us to question whether all strains of H. influenzae are capable of causing OM or if only a particular subset of strains is associated with infection. Animal models have shown that some NT H. influenzae strains are more virulent for OM than others and that the increased virulence may be related to the amount and character of lipooligosaccharide (LOS) produced (20). Long et al. demonstrated that the majority of NT H. influenzae middle ear isolates belonged to biotype I, compared to isolates from healthy individuals, which were rarely biotype I (19). Furthermore, multilocus enzyme electrophoresis analysis of 80 NT H. influenzae isolates provided evidence that the genetic diversity of OM strains is lower than the overall diversity of strains from healthy carriers (29).
While these studies suggest that a subset of nasopharyngeal NT H. influenzae strains may cause OM, the specific virulence genes associated with OM remain unknown. We used a molecular epidemiologic approach (33) involving genomic subtraction followed by a dot blot screen of a panel of H. influenzae isolates to identify virulence genes that might play a role in OM. These experiments were based on the hypothesis that genes found more frequently among NT H. influenzae middle ear isolates than among NT H. influenzae throat isolates from healthy children would be important for OM pathogenesis.
MATERIALS AND METHODS
H. influenzae collections.
Bacterial strains used in this study included H. influenzae strain Rd (8), 48 middle ear NT H. influenzae isolates, 32 NT H. influenzae nasopharyngeal isolates from ill children with respiratory symptoms for which H. influenzae may or may not have been the cause of infection, 90 throat isolates from children attending day care, and 40 H. influenzae type b invasive strains. With the exception of H. influenzae strain Rd, the isolates used in this study were collected from sites in Minnesota (9); Ann Arbor, Mich. (26); Battle Creek, Mich. (26); and Bardstown, Ky. (18), between 1980 and 2001.
Biotyping of H. influenzae isolates.
The biotype of H. influenzae strains was defined based on the production of indole, urease, and ornithine decarboxylase as described by Kilian (16).
Differential cloning by subtraction PCR.
The H. influenzae middle ear strain G622 was used as the tester strain for subtractive hybridization experiments. This strain was chosen from a collection of 17 middle ear strains (18) because of the large number of bands held in common with other middle ear strains as determined by pulsed-field gel electrophoresis (data not shown). H. influenzae strain Rd was used as the driver strain and was chosen for initial subtraction experiments because the genomic sequence of this H. influenzae strain has been fully determined (8).
Genomic subtraction was conducted by using a commercially available kit from Clontech (PCR-Select bacterial genome subtraction kit; Palo Alto, Calif.) based on the suppressive subtractive hybridization method (5, 11). DNA sequences unique to the middle ear strain G622 were identified by subtraction according to the manufacturer's instructions with two modifications. First, plasmid DNA unique to strain G622 was isolated with a Qiagen miniprep kit and added to the driver DNA pool. This step was added because the equalization step within the genomic subtraction is imperfect, and previous subtractions with H. influenzae have resulted in an overrepresentation of cryptic plasmid sequences among the subtraction products (data not shown). A small, 3-kb plasmid was identified within strain G622 and was added to the driver DNA in order to suppress its copy number in the final pool of tester specific sequences. Second, genomic tester, G622 plasmid DNA, and H. influenzae Rd driver DNA were digested with the restriction enzyme AluI from Gibco BRL (Rockville, Md.) instead of RsaI. Secondary PCR products identified by subtractive hybridization (sPCR) fragments were cloned into the plasmid TOPO vector pCR2.1 from Invitrogen (Carlsbad, Calif.). One hundred seventy clones were selected for further analysis.
Determination of tester and driver specificity.
The sPCR inserts were amplified from the 170 clones by using T7 and M13 reverse primers (35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min) followed by a nested PCR with primers provided with the subtraction kit (25 cycles of 94°C for 30 s, 68°C for 30 s, and 72°C for 2 min). To confirm tester specificity, nested PCR products were blotted onto a Hybond N+ membrane (Amersham Pharmacia Biotech, Piscataway, N.J.) with a Bio-Dot microfiltration apparatus (Bio-Rad Laboratories, Hercules, Calif.) and probed separately with AluI-digested tester and driver genomic DNA labeled with the ECF random labeling module (Amersham Pharmacia Biotech). The hybridization signal was detected by using the fluorescein-based ECF detection system (Amersham Pharmacia Biotech) and the STORM 860 PhosphorImager from Molecular Dynamics (Sunnyvale, Calif.). DNA sequence analysis of the tester-specific sPCR fragments was performed at the University of Michigan Molecular Biology Core facility on an Applied Biosystems Model 3700 DNA sequencer.
Screening of H. influenzae isolates.
The presence or absence of each sPCR fragment within an H. influenzae isolate was determined by dot blot hybridization as previously described (34) with the exception that H. influenzae strains were grown in 800 μl of brain heart infusion broth supplemented with NAD and hemin. Each blot was hybridized with unique, tester-specific, sPCR fragments labeled as described above. Images were analyzed with ImageQuaNT version 5.0 (Molecular Dynamics), and the signal was expressed as a percentage of the signal obtained for strain G622 (positive control) after correcting for the background. All isolates were screened independently with each sPCR probe at least twice, and discrepancies were resolved by Southern blot hybridization (22).
Cloning and sequencing of lic genes.
The lic2A gene was amplified from strain G622 with the primers lic2AF, 5′ ATG AGT GCT ATT GAA AAT ATT GTC ATT 3′, and lic2AR, 5′ CTA CAT AAA ACG AAC AAT TTC TTT ACC TTG C 3′ (35 cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min). The complete lic2B gene was amplified with the primers lic2BF, 5′ T AAG TAT GAT CCT CAA ATG CAT 3′, and lic2BR, 5′ CCA TTA ACA ATA TCA AGA AGA TAT C 3′, and the PCR conditions outlined above. The lic2A-lic2B intergenic region was amplified with lic2AF and lic2BF primers and the PCR conditions outlined above, with the exception that the extension time was increased to 4 min. Sequencing was conducted at the University of Michigan Molecular Biology Core facility as described above.
Statistical analyses.
Prevalence ratios were calculated as the ratio of the proportion of isolates with the gene in the collection of interest to the proportion with the gene in the healthy carriers (referent group). Differences in the proportions among each collection were calculated by the χ2 test. Statistical calculations were done in SAS (SAS Institute, Cary, N.C.).
Nucleotide sequence accession number.
The sequence determined in this work was deposited in the GenBank database under accession no. AY091470.
RESULTS
Biotype results.
In order to test the association of biotype of H. influenzae strains with OM virulence, we biotyped our H. influenzae collection (Table 1). The distribution of biotypes among NT H. influenzae OM strains differed from that of the NT H. influenzae strains from healthy children in day care (Fisher's exact test, P value of 0.0031). The prevalence of NT H. influenzae biotype I strains among middle ear strains was 2.9 times higher (11 out of 48 strains compared to 7 out of 89 strains) than that among NT H. influenzae strains from healthy children in day care (P value, 0.013).
TABLE 1.
Biotypes of H. influenzae isolates
| Collectionb (n) | Biotypea
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I
|
II
|
III
|
V
|
VI
|
VII
|
VIII
|
||||||||
| n | % | n | % | n | % | n | % | n | % | n | % | n | % | |
| Healthy (89)c | 7 | 8 | 39 | 44 | 15 | 17 | 11 | 12 | 0 | 0 | 12 | 13 | 5 | 6 |
| Clinical (32) | 6 | 19 | 13 | 41 | 5 | 16 | 5 | 16 | 2 | 6 | 0 | 0 | 1 | 3 |
| Middle ear (48) | 11 | 23 | 23 | 48 | 12 | 25 | 1 | 2 | 0 | 0 | 1 | 2 | 0 | 0 |
| Type b (40) | 35 | 88 | 5 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Total (209) | 59 | 28 | 110 | 53 | 32 | 15 | 17 | 14 | 2 | 1 | 13 | 6 | 6 | 3 |
No biotype IV strains were found in our collection.
Collections represent NT H. influenzae isolates unless they are specifically designated as type b.
One isolate from healthy carriers was lost.
Genomic subtraction.
Forty sPCR clones ranging in size between 248 and 825 bp were shown to be tester specific and underwent DNA sequence analysis. Of these, four were duplicates of the same sPCR fragment, leaving 36 distinct sPCR clones. A search of the GenBank database was conducted for each of the clones, one of which provided no match to currently known sequenced genes or proteins (Table 2). Seventeen of the sPCR clones showed similarity to phage-associated proteins. These included open reading frames from known H. influenzae bacteriophage HP-1 (7) and HP-2. Two sPCR fragments with high similarity to LOS-encoding genes were also identified, one isolated from Haemophilus ducreyi (24) and the other from the H. influenzae type b strain RM7004 (14). We also identified sPCR fragments with 96% similarity at the amino acid level to the high-molecular-weight C protein (HMWC) and 61% amino acid identity to the high-molecular-weight A protein (HMWA) (30). HMWA is thought to mediate adherence to respiratory epithelial cells, and HMWC is thought to help localize the adhesin to the outer membrane. The sPCR fragments with similarity to HmwA and HmwC genes were not analyzed further in this study. Others have shown that these adhesins are present in up to 80% of NT H. influenzae strains (18, 25, 30).
TABLE 2.
Dot blot hybridization results for 48 middle ear isolates and 46 nasopharyngeal or throat isolates probed with 34 unique sPCR fragments
| sPCR probe | No. (%) of typea
|
% Amino acid identity and potential function | |
|---|---|---|---|
| ME | NP | ||
| Phage-related proteins | |||
| A6O2 | 3 (6) | 4 (9) | 26; putative bacteriophage protein of Salmonella enterica |
| A6O8 | 18 (38) | 16 (35) | 41; C5 methylase MarMP of Mycoplasma arthritidis bacteriophage MA |
| A6O25 | 4 (8) | 6 (13) | 37; putative bacteriophage protein of Salmonella enterica |
| A6O26 | 4 (8) | 6 (13) | 39; replication O protein of phage lambda |
| A6N38 | 8 (17) | 9 (20) | 23; putative bacteriophage protein of Salmonella enterica |
| A6N42 | 3 (6) | 4 (9) | 37; Fels-1 prophage transcriptional regulator of Salmonella enterica serovar Typhimurium |
| A6O42 | 3 (6) | 4 (9) | 33; replication O protein of phage H19B |
| A6O43 | 8 (17) | 9 (20) | 33; Orf82 of Pseudomonas phage D3 |
| A6N44 | 1 (2) | 2 (4) | 43; putative phage-related protein of Yersinia pestis |
| A6N60 | 7 (15) | 7 (15) | 50; putative bacteriophage late gene regulator of Salmonella enterica |
| A6O62 | 15 (31) | 18 (39) | 46; replication initiation gene A protein of H. influenzae phage HP-1 |
| A6O70 | 22 (46) | 22 (48) | 84; putative methylase of bacteriophage Tuc2009 |
| A6N81 | —b | —b | 47; regulatory phage antirepressor of phage 434 |
| A6O83 | 8 (17) | 12 (26) | 46; phage-related protein XF2294 of Xylella fastidiosa |
| A6O111 | 13 (27) | 21 (46) | 84; Orf32 of H. influenzae phage HP-2 |
| A6O120 | 15 (31) | 13 (28) | 40; CP4-57 integrase-like protein of H. influenzae |
| A6O129 | 23 (48) | 27 (59) | 40; phage-related protein of Bacillus halodurans |
| Known Haemophilus genes | |||
| A6N17 | 11 (23) | 7 (15) | 51; HINDVIP methyltransferase of H. influenzae Rd |
| A6O22 | 19 (40) | 16 (35) | 50; LOS biosynthesis enzyme LBGB of H. ducreyi |
| A6N36 | 25 (52) | 4 (9) | 95; lipopolysaccharide biosynthesis lic2B-encoded protein of H. influenzae |
| Other | |||
| A6O18 | 4 (8) | 3 (7) | 31; hypothetical protein of E. coli O157:H7 |
| A6O24 | 15 (31) | 14 (30) | 25; vasopressin-activated calcium-mobilizing receptor of Homo sapien |
| A6O28 | 22 (46) | 17 (37) | 31; conserved protein of Streptococcus coelicolor |
| A6N29 | 5 (10) | 7 (15) | 27; transmembrane protein of Methanothermobacter thermautotrophic |
| A6N40 | 6 (15) | 7 (15) | 33; conserved hypothetical protein of Xylella fastidiosa XF0161 |
| A6O55 | 2 (4) | 1 (2) | 48; hypothetical proteins of Listeria innocua |
| A6O73 | 5 (10) | 6 (13) | 35; sodium potassium pump of Drosophila melanogaster |
| A6N95 | 4 (8) | 6 (13) | No match |
| A6O95 | 4 (8) | 6 (13) | 33; glucan endo-1,3-β-d-glucosidase of Tritium aestivium |
| A6O98 | 4 (8) | 5 (11) | 27; hypothetical protein of Mesorhizobium loti |
| A6O127 | 10 (21) | 12 (26) | 25; dual-specificity phosphatase of Arabidopsis thaliane |
| A6O132 | 7 (15) | 2 (4) | 37; DNA repair MMS21 of Saccharomyces cerevisiae |
| A6O139 | 8 (17) | 9 (20) | 54; hypothetical 9.8-kDa protein of E. coli K-12 |
| A6O143 | 3 (6) | 1 (2) | 56; conserved hypothetical protein of Ureaplasma urealyticum |
ME, middle ear; NP, nasopharyngeal or throat.
This probe exhibited high levels of nonspecific binding, perhaps due to the presence of similar sequences among the H. influenzae strains.
Frequency of lic2b in H. influenzae strains.
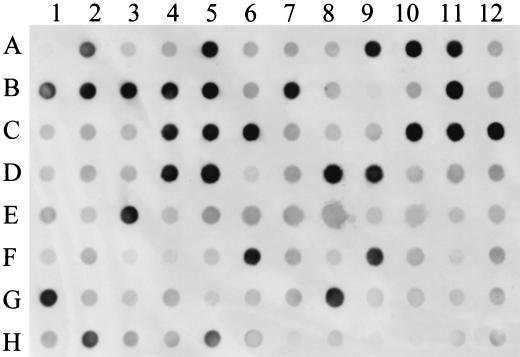
Thirty-four of the sPCR probes that were shown to be both unique and tester specific were used to screen a collection of 48 NT H. influenzae middle ear isolates and 46 NT H. influenzae strains isolated from either the throat or the nasopharynx. One of the clones, number A6N36, hybridized more frequently with middle ear isolates (25 out of 48 isolates) than with NT H. influenzae throat or nasopharyngeal isolates (4 out of 46 isolates) (Fig. 1). Probe A6N36 is a 357-bp fragment with 96% similarity at the nucleotide level to a lipopolysaccharide biosynthesis gene, lic2B, originally described for the H. influenzae type b strain RM7004 (13, 14). Because of the apparent link of sPCR probe A6N36 with middle ear isolates, additional H. influenzae isolates were screened to confirm the association. Additional H. influenzae isolates from 90 healthy children in day care, 32 NT H. influenzae clinical isolates, and 40 H. influenzae type b strains were screened. Probe A6N36 hybridized more frequently with H. influenzae type b strains, NT H. influenzae middle ear isolates, and NT H. influenzae clinical isolates than with isolates from healthy carriers (Table 3).
FIG. 1.
Dot blot hybridization of sPCR fragment A6N36 with 48 H. influenzae middle ear and 46 throat or nasopharyngeal isolates. Dot A1 is blank, dot A5 represents the tester strain G622, and dot E5 represents the driver strain Rd. The top half of the blot represents middle ear isolates (except dots D1, D2, and D3, which are throat isolates), and the bottom half of the blot represents nasopharyngeal and throat isolates with the exception of dots F6, G8, G10, and H5, which are middle ear strains.
TABLE 3.
Distribution of lic2A and lic2B genes by H. influenzae collectiona
| Collectionb (n) |
lic2A
|
lic2B
|
||||||
|---|---|---|---|---|---|---|---|---|
| n | % | PR | P | n | % | PR | P | |
| Healthy (90) | 69 | 77 | Referent | 13 | 14 | Referent | ||
| Clinical (32) | 32 | 100 | 1.30 | 0.002 | 13 | 39 | 2.93 | 2.8 × 10−3 |
| Middle ear (48) | 47 | 98 | 1.27 | 0.001 | 25 | 52 | 3.71 | 2.6 × 10−5 |
| Type b (40) | 40 | 100 | 1.30 | 8.8 × 10−4 | 35 | 88 | 6.29 | <1 × 10−7 |
The prevalence ratios (PR) and χ2 P values of the lic2A and lic2B genes relative to their presence in throat isolates from healthy children (referent) are shown.
Collections represent NT H. influenzae isolates unless they are specifically designated as type b.
Cloning of the lic2A-lic2B cluster.
Because the lic2B gene was originally described for a type b strain as part of a gene cluster, we attempted to determine whether the gene within our NT tester strain was present in a similar gene cluster. PCR with the lic2AF primer and the lic2BF primer amplified a 3.7-kb PCR fragment that was subsequently cloned into the TOPO vector pCR2.1, and DNA sequence was obtained. The lic2B gene, identified in NT H. influenzae strain G622, encodes a protein that is 99% identical at the amino acid level to that found in the type b strain. The sequence and arrangement of the lic2A-lic2B gene cluster are virtually identical to those found in the type b strain (14) The middle ear isolate G622 contains proteins that are 94, 93, and 96% identical to the lic2A-, orf 3-, and ksgA-encoded proteins from the type b strain RM7004, respectively (14). Like the majority of H. influenzae isolates surveyed, the lic2A gene from our middle ear strain contains 19 CAAT repeats (14), indicating that it can undergo phase variation. The complete lic2A gene was amplified from strain G622 and used to probe our collection of H. influenzae isolates. The majority of H. influenzae strains contain the lic2A gene (Table 3). The lic2B gene was present only in strains that also contained the lic2A gene.
DISCUSSION
We hypothesized that some NT H. influenzae strains have virulence characteristics that enhance their ability to cause OM. This paper provides evidence that a specific gene, lic2B, is found significantly more frequently among NT H. influenzae strains that cause OM than among those found in the throats of healthy children. This suggests not only that OM is a disease of opportunity but that special characteristics may be required for throat isolates to invade the middle ear. A potentially analogous model is urinary tract infections caused by Escherichia coli. Urinary tract isolates may originate in the bowel flora, but only a subset of gastrointestinal isolates is found among those causing urinary tract infections (15).
Long et al. previously showed that 67% of NT H. influenzae strains isolated from children with OM were biotype I (19); however, only 14 NT H. influenzae strains were examined in this study. Almost half (48%) of the middle ear isolates belonged to biotype II, but, the higher prevalence of biotype I strains among our middle ear isolates than among isolates from healthy children in day care further supports our hypothesis.
The results from our genomic subtraction experiments identified the lic2B gene as potentially important for OM pathogenesis. The lic2B gene was originally identified in the type b strain RM7004. While the precise function of this gene is not known, lic2B is 58.9% identical at the nucleotide level to lic2A (12). The lic2A gene is thought to encode a galactosyltransferase that adds the terminal galactose on Galα1-4Gal structures of LOS. H. influenzae LOS is composed of a lipid A portion which is anchored into the membrane, linked by a single 2-keto-3-deoxyoctulosonic acid and three molecules of heptose and an outer core of glucose and galactose.
The structure of H. influenzae LOS has been implicated in virulence. Expression of phosphorylcholine through the phase-variable lic1 locus has been shown previously to enhance nasopharyngeal colonization and the development of OM in the chinchilla model of OM (27). Transformation of a virulence-deficient LOS mutant of H. influenzae type b with plasmids from an H. influenzae type b genomic library resulted in transformants with structurally altered LOS molecules that exhibited increased virulence in an animal model for invasive H. influenzae type b disease (3). Observed intrastrain variations in pathogenicity of an NT H. influenzae strain have been postulated to be due to changes in LOS (20).
Several of the surface-exposed epitopes of H. influenzae strains undergo high-frequency phase variation, thought to be important for survival in different microenvironments within the host (31). The lic1A, lic2A, and lic3A genes each contain multiple copies of the tetrameric repeat 5′-CAAT-3′ within the 5′ end of the coding sequence. The number of copies of the repeat can vary due to slipped-strand mispairing, which places the initiation codons in or out of frame with respect to the open reading frame, resulting in important LOS variations.
A mechanism for the influence of lic2A on virulence has also been proposed (32). Galα1-4Gal is found on the surface of human glycolipids, and it has been suggested that the decoration of the bacterial cell surface may allow these cells to evade antibody-mediated killing (32). Furthermore, Galα1-4Gal may not be expressed during carriage but may be important in pneumonia (32). The lic2A gene was expressed in five of five isolates from patients with pneumonia and was not expressed in four of five isolates from individuals without pneumonia.
Unlike the lic2A gene, lic2B does not contain a repeat region and does not undergo phase variation. The lic2A gene was present in the vast majority of H. influenzae strains, and the lic2B gene was present in 14 to 88% of H. influenzae strains, depending on the collection surveyed. lic2B was never found in an H. influenzae strain where lic2A was absent. While the lic2A gene is present in the vast majority of H. influenzae strains, the lic2b gene provides a stronger marker (prevalence ratio, 3.79) for differentiating between middle ear strains and those strains obtained from healthy children. This gene is also found more frequently among NT H. influenzae clinical isolates from sick children (prevalence ratio, 2.93) than among isolates from healthy children. This difference in prevalence is also striking considering the relatively even distribution of other genomic subtraction products among the middle ear and throat isolates.
In summary, lic2B, originally identified in H. influenzae type b strain RM7004 (13, 14), was present in more than half of NT H. influenzae middle ear isolates and only 14% of isolates from children attending day care. lic2B was also present in the majority of type b isolates screened (88%). Although lic2B may be important for the pathogenesis of OM, we cannot rule out the possibility that lic2B is a marker for other virulence determinants. Future mechanistic studies should shed light on this question.
Acknowledgments
We thank May Patel for assistance with the biotyping of H. influenzae strains and both Rand Farjo and Jennifer St. Sauver for the isolation and identification of H. influenzae strains from the day care studies. We also thank Graham Krasan, Stan Block, and Joseph W. St. Geme III for the use of their OM strains.
This work was supported by an award from the National Institute of Allergy and Infectious Diseases (RO1-AI25630 to J.G.), the C. S. Mott Children's Hospital Fund for Research (J.G.), and the Deafness Research Foundation (to M.M.P.). M.M.P. was supported by supplement NIAID-CO36919 (to J.G.).
Editor: A. D. O'Brien
REFERENCES
- 1.Berman, S. 1995. Current concepts: otitis media in children. N. Engl. J. Med. 332:1560-1565. [DOI] [PubMed] [Google Scholar]
- 2.Casselbrant, M. L., E. M. Mandel, P. A. Fall, H. E. Rockette, M. Kurs-Lasky, C. D. Bluestone, and R. E. Ferrell. 1999. The heritability of otitis media: a twin and triplet study. JAMA 282:2125-2130. [DOI] [PubMed] [Google Scholar]
- 3.Cope, L. D., R. Yogev, J. Mertsola, J. L. Latimer, M. S. Hanson, G. H. McCracken, Jr., and E. J. Hansen. 1991. Molecular cloning of a gene involved in lipooligosaccharide biosynthesis and virulence expression by Haemophilus influenzae type B. Mol. Microbiol. 5:1113-1124. [DOI] [PubMed] [Google Scholar]
- 4.Dhooge, I., M. Vaneechoutte, G. Claeys, G. Verschraegen, and P. Van Cauwenberge. 2000. Turnover of Haemophilus influenzae isolates in otitis-prone children. Int. J. Pediatr. Otorhinolaryngol. 54:7-12. [DOI] [PubMed] [Google Scholar]
- 5.Diatchenko, L., Y. F. Lau, A. P. Campbell, A. Chenchik, F. Moqadam, B. Huang, S. Lukyanov, K. Lukyanov, N. Gurskaya, E. D. Sverdlov, and P. D. Siebert. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl. Acad. Sci. USA 93:6025-6030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eskola, J., and T. Kilpi. 2000. Potential of bacterial vaccines in the prevention of acute otitis media. Pediatr. Infect. Dis. J. 19S:S72-S78. [DOI] [PubMed] [Google Scholar]
- 7.Esposito, D., W. P. Fitzmaurice, R. C. Benjamin, S. D. Goodman, A. S. Waldman, and J. J. Scocca. 1996. The complete nucleotide sequence of bacteriophage HP1 DNA. Nucleic Acids Res. 24:2360-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fleischmann, R. D., M. D. Adams, O. White, R. A. Clayton, E. F. Kirkness, A. R. Kerlavage, C. J. Bult, J. F. Tomb, B. A. Dougherty, J. M. Merrick, K. McKenny, G. Sutton, W. FitzHugh, C. Fields, J. D. Gocayne, J. Scott, R. Shirely, L.-I. Liu, A. Glodek, J. M. Kelley, J. F. Weidman, C. A. Phillips, T. Spriggs, E. Hedblom, M. D. Cotton, T. R. Utterback, M. C. Hanna, D. T. Nguyen, D. M. Saudek, R. C. Brandon, L. D. Fine, J. L. Fritchman, J. L. Fuhrmann, N. S. M. Geoghagen, C. L. Gnehm, L. A. McDonald, K. V. Small, C. M. Fraser, H. O. Smith, and J. C. Venter. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496-512. [DOI] [PubMed] [Google Scholar]
- 9.Gilsdorf, J. R., H. Y. Chang, K. W. McCrea, and L. O. Bakaletz. 1992. Comparison of hemagglutinating pili of Haemophilus influenzae type b with similar structures of nontypeable H. influenzae. Infect. Immun. 60:374-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomez-De-Leon, P., J. I. Santos, J. Caballero, D. Gomez, L. E. Espinosa, I. Moreno, D. Pinero, and A. Cravioto. 2000. Genomic variability of Haemophilus influenzae isolated from Mexican children determined by using enterobacterial repetitive intergenic consensus sequences and PCR. J. Clin. Microbiol. 38:2504-2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gurskaya, N. G., L. Diatchenko, A. Chenchik, P. D. Siebert, G. L. Khaspekov, K. A. Lukyanov, L. L. Vagner, O. D. Ermolaeva, S. A. Lukyanov, and E. D. Sverdlov. 1996. Equalizing cDNA subtraction based on selective suppression of polymerase chain reaction: cloning of Jurkat cell transcripts induced by phytohemagglutinin and phorbol 12-myristate 13-acetate. Anal. Biochem. 240:90-97. [DOI] [PubMed] [Google Scholar]
- 12.High, N. J., M. E. Deadman, D. W. Hood, and E. R. Moxon. 1996. The identification of a novel gene required for lipopolysaccharide biosynthesis by Haemophilus influenzae RM7004, using transposon Tn916 mutagenesis. FEMS Microbiol. Lett. 145:325-331. [DOI] [PubMed] [Google Scholar]
- 13.High, N. J., M. E. Deadman, and E. R. Moxon. 1993. The role of a repetitive DNA motif (5′-CAAT-3′) in the variable expression of the Haemophilus influenzae lipopolysaccharide epitope α Gal(1-4)β Gal. Mol. Microbiol. 9:1275-1282. [DOI] [PubMed] [Google Scholar]
- 14.High, N. J., M. P. Jennings, and E. R. Moxon. 1996. Tandem repeats of the tetramer 5′-CAAT-3′ present in lic2A are required for phase variation but not lipopolysaccharide biosynthesis in Haemophilus influenzae. Mol. Microbiol. 20:165-174. [DOI] [PubMed] [Google Scholar]
- 15.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilian, M. 1991. Haemophilus, p. 463-470. In A. Balows, W. J. Hausler, Jr., K. L. Herrmann, H. D. Isenberg, and H. J. Shadomy (ed.), Manual of clinical microbiology, 5th ed. American Society for Microbiology, Washington, D.C.
- 17.Klein, J. O. 1997. Role of nontypeable Haemophilus influenzae in pediatric respiratory tract infections. Pediatr. Infect. Dis. J. 16:S5-S8. [DOI] [PubMed] [Google Scholar]
- 18.Krasan, G. P., D. Cutter, S. L. Block, and J. W. St. Geme III. 1999. Adhesin expression in matched nasopharyngeal and middle ear isolates of nontypeable Haemophilus influenzae from children with acute otitis media. Infect. Immun. 67:449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long, S. S., M. J. Teter, and P. H. Gilligan. 1983. Biotype of Haemophilus influenzae: correlation with virulence and ampicillin resistance. J. Infect. Dis. 147:800-806. [DOI] [PubMed] [Google Scholar]
- 20.Melhus, A., A. Hermansson, A. Forsgren, and K. Prellner. 1998. Intra- and interstrain differences of virulence among nontypeable Haemophilus influenzae strains. APMIS 106:858-868. [PubMed] [Google Scholar]
- 21.Murphy, T. F., J. M. Bernstein, D. M. Dryja, A. A. Campagnari, and M. A. Apicella. 1987. Outer membrane protein and lipooligosaccharide analysis of paired nasopharyngeal and middle ear isolates in otitis media due to nontypable Haemophilus influenzae: pathogenic and epidemiologic observations. J. Infect. Dis. 156:723-731. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 23.Smith-Vaughan, H. C., K. S. Sriprakash, A. J. Leach, J. D. Mathews, and D. J. Kemp. 1998. Low genetic diversity of Haemophilus influenzae type b compared to nonencapsulated H. influenzae in a population in which H. influenzae is highly endemic. Infect. Immun. 66:3403-3409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stevens, M. K., J. Klesney-Tait, S. Lumbley, K. A. Walters, A. M. Joffe, J. D. Radolf, and E. J. Hansen. 1997. Identification of tandem genes involved in lipooligosaccharide expression by Haemophilus ducreyi. Infect. Immun. 65:651-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.St. Geme, J. W., III, V. V. Kumar, D. Cutter, and S. J. Barenkamp. 1998. Prevalence and distribution of the hmw and hia genes and the HMW and Hia adhesins among genetically diverse strains of nontypeable Haemophilus influenzae. Infect. Immun. 66:364-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.St. Sauver, J., C. F. Marrs, B. Foxman, P. Somsel, R. Madera, and J. R. Gilsdorf. 2000. Risk factors for otitis media and carriage of multiple strains of Haemophilus influenzae and Streptococcus pneumoniae. Emerg. Infect. Dis. 6:622-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tong, H. H., L. E. Blue, M. A. James, Y. P. Chen, and T. F. DeMaria. 2000. Evaluation of phase variation of nontypeable Haemophilus influenzae lipooligosaccharide during the development of otitis media in the chinchilla model. Infect. Immun. 68:4593-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhari, M., K. Mantysaari, and M. Niemela. 1996. A meta-analytic review of the risk factors for acute otitis media. Clin. Infect. Dis. 22:1079-1083. [DOI] [PubMed] [Google Scholar]
- 29.van Alphen, L., D. A. Caugant, B. Duim, M. O'Rourke, and L. D. Bowler. 1997. Differences in genetic diversity of nonencapsulated Haemophilus influenzae from various diseases. Microbiology 143:1423-1431. [DOI] [PubMed] [Google Scholar]
- 30.van Schilfgaarde, M., P. van Ulsen, P. Eijk, M. Brand, M. Stam, J. Kouame, L. van Alphen, and J. Dankert. 2000. Characterization of adherence of nontypeable Haemophilus influenzae to human epithelial cells. Infect. Immun. 68:4658-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiser, J. N., J. M. Love, and E. R. Moxon. 1989. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell 59:657-665. [DOI] [PubMed] [Google Scholar]
- 32.Weiser, J. N., and N. Pan. 1998. Adaptation of Haemophilus influenzae to acquired and innate humoral immunity based on phase variation of lipopolysaccharide. Mol. Microbiol. 30:767-775. [DOI] [PubMed] [Google Scholar]
- 33.Zhang, L., B. Foxman, S. D. Manning, P. Tallman, and C. F. Marrs. 2000. Molecular epidemiologic approaches to urinary tract infection gene discovery in uropathogenic Escherichia coli. Infect. Immun. 68:2009-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang, L., B. W. Gillespie, C. F. Marrs, and B. Foxman. 2001. Optimization of a fluorescent-based phosphor imaging dot blot DNA hybridization assay to assess E. coli virulence gene profiles. J. Microbiol. Methods 44:225-233. [DOI] [PubMed] [Google Scholar]