Abstract
The reactivity of the essential element iron necessitates a concerted expression of ferritins, which mediate iron storage in a nonreactive state. Here we have further established the role of the Helicobacter pylori ferritin Pfr in iron metabolism and gastric colonization. Iron stored in Pfr enabled H. pylori to multiply under severe iron starvation and protected the bacteria from acid-amplified iron toxicity, as inactivation of the pfr gene restricted growth of H. pylori under these conditions. The lowered total iron content in the pfr mutant, which is probably caused by decreased iron uptake rates, was also reflected by an increased resistance to superoxide stress. Iron induction of Pfr synthesis was clearly diminished in an H. pylori feoB mutant, which lacked high-affinity ferrous iron transport, confirming that Pfr expression is mediated by changes in the cytoplasmic iron pool and not by extracellular iron. This is well in agreement with the recent discovery that iron induces Pfr synthesis by abolishing Fur-mediated repression of pfr transcription, which was further confirmed here by the observation that iron inhibited the in vitro binding of recombinant H. pylori Fur to the pfr promoter region. The functions of H. pylori Pfr in iron metabolism are essential for survival in the gastric mucosa, as the pfr mutant was unable to colonize in a Mongolian gerbil-based animal model. In summary, the pfr phenotypes observed give new insights into prokaryotic ferritin functions and indicate that iron storage and homeostasis are of extraordinary importance for H. pylori to survive in its hostile natural environment.
The important role of iron in maintaining basic metabolic functions, such as electron transport, on the one hand and its high redox reactivity and toxicity on the other constitute for nearly all living organisms a significant dilemma (8). In most bacteria, the internal concentration of reactive iron is modulated by downregulation of high-affinity iron uptake systems in response to an increased cytoplasmic iron concentration (19) as well as by binding and storage of cytoplasmic iron in a nonreactive state (2). These functions are mostly mediated by the ferric uptake regulator Fur and by bacterial ferritins, respectively.
Helicobacter pylori is a gram-negative, microaerophilic bacterium which colonizes the human gastric mucosa for extended time periods (14). The infection, which concerns about half of the world population and stays lifelong if not treated, is the leading cause of ulcerations and a cofactor for the development of gastric adenocarcinoma and lymphoma (3). Proteins involved in iron metabolism (35) are suggested to represent major virulence determinants of H. pylori (22), and it was shown that ferrous iron uptake mediated by the transport protein FeoB is a prerequisite for the establishment of H. pylori infection in vivo (36). In contrast to other bacteria which use ferric iron as the main iron source (8), H. pylori is heavily dependent on ferrous iron, which is stabilized by the low pH and by the low oxygen concentration of the human stomach (36). As a consequence of the adaptation to the specific environment, these differences in iron metabolism favor H. pylori as an interesting model for the study of iron storage.
In contrast to eukaryotic ferritins, which are well characterized concerning their structure, biocatalytic functions, and regulation, our knowledge of prokaryotic ferritins is still relatively limited (2). Bacterial ferritins mediate storage of iron in cytoplasmic granular structures (2, 7, 38), and the predicted release and reuse of iron from a bacterial ferritin in vivo have also been recently confirmed for the Escherichia coli ferritin FtnA (1). Because free iron ions catalyze the formation of reactive oxygen radicals via the Fenton reaction (24), iron binding by ferritins protects bacteria against oxidative stress (2, 33). In E. coli and Campylobacter jejuni, absence of ferritin increases the sensitivity to oxidative stress (1, 37). The ferritin Pfr, the major iron storage protein of H. pylori (7, 12, 18), was shown to be essential for iron storage in subcellular granules and for iron resistance of H. pylori (7). Preliminary investigations indicated that Pfr may differ from other bacterial ferritins in that it does not protect from superoxide stress mediated by paraquat, as shown by agar dilution and disk diffusion assays, respectively (7). Iron binding properties have also been described for the H. pylori NapA protein (32), a Dps homolog proposed earlier to be a member of the bacterioferritin protein family (15). However, recent studies did not indicate significant NapA functions in iron storage (13).
In H. pylori both iron acquisition and storage are governed by the ferric uptake regulator Fur (4-6, 16, 28, 35). In response to an increased iron concentration, Fur represses transcription of the iron uptake genes fecA2 and frpB1 (10, 11, 16, 35) while transcription of the ferritin gene pfr increases (4, 11, 28, 35). Conversely, under iron-restricted conditions transcription of the iron uptake genes is stimulated and Pfr-mediated iron storage is repressed by Fur. Expression of Pfr is also repressed by Fur upon increased nickel, copper, zinc, and manganese concentrations (4), leading to a model where modulation of Pfr transcription contributes heavily to the maintenance of iron and metal homeostasis in H. pylori. Taken together, these findings suggested that ferritin-mediated iron storage is required for adaptation of H. pylori to the changes in iron availability occurring in the gastric mucosa. In this study we have extended our characterization of the role of Pfr in H. pylori iron metabolism, and we demonstrate that Pfr-mediated iron storage is essential for survival of lethal iron starvation, for protection against acid-amplified iron resistance, for iron acquisition, and for gastric colonization in a Mongolian gerbil model of infection.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
The construction of isogenic pfr and feoB mutants of H. pylori strains G27 and NCTC11637, respectively, was described earlier (4, 36). The gerbil-adapted H. pylori strain Q1 was constructed by transformation of strain G1.1 (39) with plasmid pEG21 (17) conferring streptomycin resistance upon recombination into the chromosome. A cagA mutation was introduced by transformation of strain Q1 with plasmid pWS30 (29). The pfr gene of strain Q1 was mutagenized by the insertion of a cat gene into the single SphI site of the pfr gene from H. pylori strain P1 as described earlier for construction of the pfr mutant of H. pylori strain G27 (7). The lack of Pfr and CagA proteins in the corresponding knockout mutant was confirmed by immunoblot analysis with specific antisera (data not shown). Furthermore, the pfr mutant of strain Q1 displayed the classical iron-sensitive pfr phenotype in vitro, as determined by growth inhibition experiments with ferrous iron. The MIC of iron was 2.5 mM.
H. pylori was routinely cultured on blood agar plates in a microaerobic atmosphere (4, 34). Growth inhibition experiments were performed in brucella broth with 10% fetal calf serum (BBF). Bacteria were precultured to an optical density at 600 nm (OD600) of 1.0 in BBF and subsequently diluted 1:100 in test media containing defined supplements as indicated below in Results. The influence of test conditions on growth of H. pylori was determined by reading the OD600 after 24 and 48 h.
All growth experiments were performed in triplicate and were repeated a minimum of three times. Iron-rich conditions were generated by supplementation of BBF with freshly prepared ferrous iron chloride (catalog no. F2877; Sigma) at various concentrations, as indicated below in Results. Iron starvation was achieved by addition of the iron chelator desferrioxamine B (desferal; catalog no. D9533; Sigma). Control cultures were supplemented with sodium chloride at the highest iron concentrations to exclude the influence of osmotic stress and unspecific influences of chloride. Conditions of oxidative stress were generated by supplementation of BBF with paraquat (methyl viologen; catalog no. M2254; Sigma). For analysis of the influence of pH on iron sensitivity, the BBF medium was titrated to pH 6.0 with hydrochloric acid.
Radiolabeled iron uptake assays and determination of whole-cell iron content.
Cells were grown to the late exponential phase in broth and, after being harvested by centrifugation, pelleted cells were washed twice in 50 mM phosphate-buffered saline, pH 7.4, resuspended in the same medium at a concentration of 20 to 30 mg of cell protein per ml, and kept under microaerobic conditions for no longer than 4 h. Uptake of iron was determined with the centrifugation through oil method, using 0.5 μM 55FeCl3 (86.7 to 100.32 MBq μmol−1) complexed with either sodium ascorbate or citrate as described earlier (36).
For determinations of whole-cell iron content, the wild-type (wt) strain G27 and the pfr mutant were cultivated to an OD600 of 1.0 in BBF medium supplemented with 100 μM ferrous chloride. The cells were harvested by centrifugation, resuspended in double-distilled water, and then lysed with 0.1% sodium dodecyl sulfate (SDS). The iron content of the lysates was determined in triplicate by flame atomic absorption spectrometry by using routine methods with a Perkin-Elmer Analyst 100 apparatus.
Protein analysis.
H. pylori cultures were grown for 48 h in broth with moderate shaking to an OD600 of 1.0 to 1.2 and harvested by centrifugation for 10 min at 4,000 × g at 4°C. Protein concentration determinations, SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and immunoblotting were performed as described earlier (4). The ferritin Pfr and the CagA protein were detected with the specific antisera AK198 (6) and AK257 (26), respectively. Bound rabbit antibodies were detected with a protein A-alkaline phosphatase conjugate followed by incubation with nitroblue tetrazolium as substrate. The Cat protein was quantitated by enzyme-linked immunosorbent assay (ELISA) using the Cat-ELISA system (catalog no. 1363727; Roche) according to the manufacturer's instructions, as described elsewhere (4). The amount of Cat protein was calculated from a standard curve prepared with purified E. coli Cat protein and normalized to the amount of total protein. The amount of Cat was in the range of 1 to 10 μg/mg of total protein, depending on the environmental conditions.
Analysis of Fur-DNA binding activity.
The expression and purification of H. pylori Fur was performed by using the Strep-Tag protein expression system from IBA (Göttingen, Germany) according to the manufacturer's instructions (http://www.iba-go.de) (30). The fur coding sequence from H. pylori strain 26695 was amplified using the primer FURSK7-L1 (5′-ATGGTAGGTCTCAGCGCATGAAAAGATTAGAAACTTTAGAATCTA) in combination with the primer FURSK7-R1 (5′-ATGGTAGGTCTCATATCAACATTCACTCTCTTGGCATTCTTTA) and cloned via the BsaI restriction sites added as 5′ extensions (underlined) into plasmid pASKIBA-7 (IBA). The recombinant H. pylori Fur protein carrying an N-terminal eight-amino-acid Strep-Tag (30) was expressed in E. coli XL1-Blue and purified to homogeneity on a Streptactin column. The intergenic region in front of the pfr gene was amplified by PCR from DNA of H. pylori strain 26695 with primers BP653-L1 (5′-AGATAAACATTGTAGCATTT) and BP653-R1 (5′-TTTTGATAACATAGTATCTC). Fur-DNA complexes were analyzed by electrophoretic mobility shift assay (EMSA). Therefore, 50 pmol of the 287-bp PCR product was incubated for 1 h with 5.6 nmol of recombinant Fur in binding buffer (10 mM Tris-HCl [pH 8.0], 5 mM dithiothreitol, 5% glycerol). After 1 h the samples were subjected to electrophoresis on a 7% nondenaturing polyacrylamide gel. Free DNA and protein-DNA complexes were visualized by staining with ethidium bromide under UV light at 203 nm.
In vivo studies using the gerbil animal model.
The Mongolian gerbils used in this study originated from the breeding colony of the Max von Pettenkofer Institute (Munich, Germany). The animal model is registered at the Regierung Oberbayern (AZ211-2531-60/98). Three animals were housed in each cage at a constant room temperature of 22°C with a 12-h light-dark cycle. For the infection experiment the H. pylori wt strain Q1 and its isogenic cagA and pfr mutants were grown on serum agar (GC agar, 8% horse serum, IsoVitaleX) supplemented with 250 mg of streptomycin/liter (Str250). The animals were infected orogastrically by feeding 0.3 ml of bacterial suspension in brucella broth (OD550 of 3.3) through a feeding needle, corresponding to a final infection dose of approximately 109 bacteria/gerbil. The infection was performed three times on subsequent days. For each infecting strain, three animals were kept for 3 and for 6 weeks. After sacrificing the gerbils in a CO2 chamber, the stomach was isolated from the animal, dissected, and separated from the gastric contents. The washed stomach specimen was homogenized in 2 ml of brucella broth using a glass homogenizer. One hundred-microliter aliquots of a 1:10 and a 1:100 dilution were plated on serum agar (Str250) in duplicate. After 4 to 5 days, the colonies were counted and evaluated with respect to colony morphology and expression of the cagA and pfr genes.
Statistical analysis.
If not mentioned otherwise, data are mean values from at least three independent experiments and the standard deviations are indicated. All data from wt-mutant comparisons were analyzed for statistical significance with Student's t test. All differences referred to as relevant in Results and in the Discussion had P values of <0.05.
RESULTS
The pfr mutant is iron sensitive at low pH.
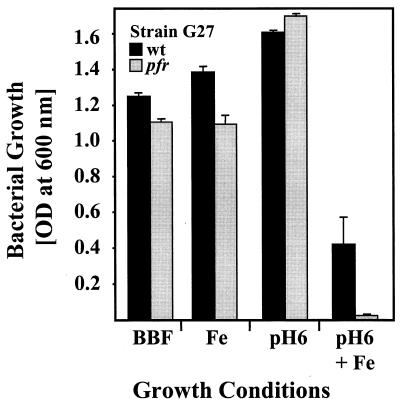
Preliminary analysis of metal resistance by agar dilution assays has shown that Pfr is essential for iron resistance of H. pylori (7). This pfr phenotype was reinvestigated in more detail by growth inhibition experiments with the wt strain G27 and the pfr mutant in broth. In unsupplemented BBF medium, growth of the pfr mutant and the wild-type (wt) strain was identical, indicating that the pfr mutation does not generally limit bacterial fitness (Fig. 1). The MICs for ferrous iron were 4 and 2.5 mM for the wt strain and the pfr mutant, respectively, indicating that iron was clearly more toxic for the pfr mutant than for the wt strain. Sodium chloride applied at 4 mM had no influence on growth, indicating that the iron sensitivity of the pfr mutant was specifically caused by iron and not by osmotic pressure (data not shown).
FIG. 1.
The role of Pfr in acid-amplified iron resistance of H. pylori. The growth of H. pylori strain G27 (black) and of the pfr mutant (gray) in broth (BBF) was determined by measuring the OD600. For high iron (Fe) and acidic conditions the medium was supplemented with 1 mM iron or titrated to pH 6, respectively. The data represent mean values from three independent determinations. Standard deviations are indicated.
Because the acidic conditions predominant in the natural environment of H. pylori increase iron toxicity by stabilizing the more soluble ferrous state, the iron sensitivity of the H. pylori pfr mutant was assessed at pH 6.0 (Fig. 1). Supplementation of BBF (pH 7.5) with 1 mM FeCl2 did not affect growth of the wt and pfr mutant strains. At pH 6.0 without iron supplementation, growth of both strains was also identical, indicating that the pfr mutation does not influence acid resistance (Fig. 1). However, when wt and pfr mutant strains were grown at pH 6.0 with 1 mM FeCl2, the growth reduction of the wt strain demonstrated clearly that iron toxicity is amplified by acid. Growth of the pfr mutant was significantly reduced when compared to the wt strain, indicating that Pfr protects against acid-amplified iron toxicity (Fig. 1).
Iron stored in Pfr supports growth under conditions of iron starvation.
To analyze if iron stored in Pfr under iron-rich conditions can be remobilized to support growth under iron-starved conditions, the effect of iron supplementation of precultures on growth of wt and pfr mutant strains was compared. Preculturing in the presence of 1 mM FeCl2 was not toxic for the pfr mutant but effectively preloaded the cells with iron (see also Fig. 1). The subsequent subcultivation under iron-limited conditions (generated with 40 μM desferal) allowed the wt strain to grow, but the pfr mutant could not, as indicated by OD600 values of 0.188 ± 0.022 and 0.025 ± 0.001, respectively. Control experiments performed without iron preloading showed that both strains were not able to grow in the presence of 40 μM desferal, and the MIC of desferal was 25 μM for both the wt and pfr mutant, indicating that the iron stored under high-iron conditions is indeed essential to overcome conditions of iron starvation. In media without desferal (used as controls), growth of both strains was identical and independent from iron preloading, as demonstrated by OD600 values of 0.5 ± 0.1 and 1.2 ± 0.1 after 24 and 48 h, respectively, indicating that the iron concentrations used were not toxic for the pfr mutant.
The Pfr protein is essential for iron storage and uptake.
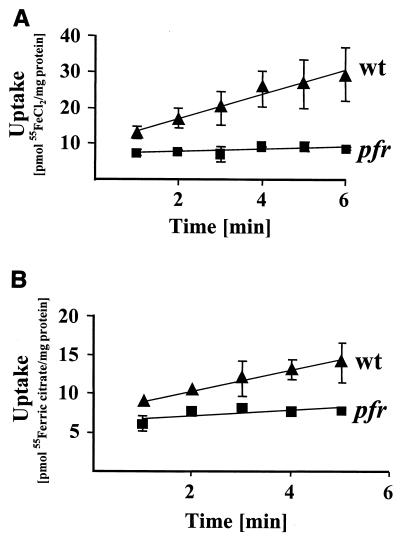
The effect of the pfr mutation on the iron storage capacity of H. pylori was investigated by determination of the total iron content with mass spectrometry (atomic absorption spectrometry). The values obtained showed that the wt strain contains more iron than the pfr mutant, as demonstrated by total iron content of 1.5 ± 0.3 and only 0.73 ± 0.08 ng/μg of total protein, respectively. To test if the reduced iron content of the pfr mutant is associated with a reduction in the iron uptake rates, the transport of 55FeCl2 (Fig. 2A) and 55Fe3+-citrate (Fig. 2B) in the wt strain and the pfr mutant was determined. Uptake rates for 55FeCl2 and 55Fe3+-citrate of 3.32 ± 1.20 and 1.35 ± 0.30 nmol min−1 mg of cell protein−1 in the wt strain were reduced to 0.35 ± 0.21 and 0.38 ± 0.2 nmol min−1 mg of cell protein−1 in the pfr mutant, respectively (Fig. 2).
FIG. 2.
The effect of the pfr mutation on iron uptake rates. The time-dependent uptake of ferrous iron (A) and of ferric citrate (B) by the wt strain G27 (filled triangles) and by the pfr mutant (filled squares) was measured with an uptake assay using the radioactively labeled compounds indicated. The data represent mean values from three independent determinations. Standard deviations are indicated.
The role of Pfr in resistance to superoxide stress.
It was previously reported that in contrast to other bacterial ferritins H. pylori Pfr is not protective against oxidative stress mediated by the superoxide radical generator paraquat, as analyzed by agar dilution and disk diffusion assays (7). This unusual pfr phenotype of H. pylori was reinvestigated by growth inhibition experiments in broth. In BBF supplemented with 10 μM paraquat, the wt strain and the pfr mutant grew to OD600 values of 0.396 ± 0.065 and 1.427 ± 0.095, respectively, indicating that the pfr mutant is clearly more resistant to paraquat than the wt strain G27. The paraquat MICs were 10 and 20 μM for the wt strain and the pfr mutant, respectively. Because iron is known to enhance oxidative stress (24, 33), we performed additional experiments to test if the reduced iron content of the pfr mutant could be involved in causing paraquat resistance. Therefore, the pfr mutant was grown in precultures with and without 1 mM ferrous iron and subsequently cultured in BBF medium supplemented with 10 μM paraquat. Monitoring of growth after 24 h revealed that iron preloading causes a prolongation of the lag growth phase, as indicated by OD600 values of 0.064 ± 0.013 and 0.206 ± 0.049 in cultures with and without iron preloading, respectively. However, this iron-mediated growth inhibition was only transient, as demonstrated by the final OD600 values of 1.2 ± 0.2 reached by the pfr mutant independent from iron preloading after 48 h.
Fur-mediated iron regulation of Pfr depends on iron transport.
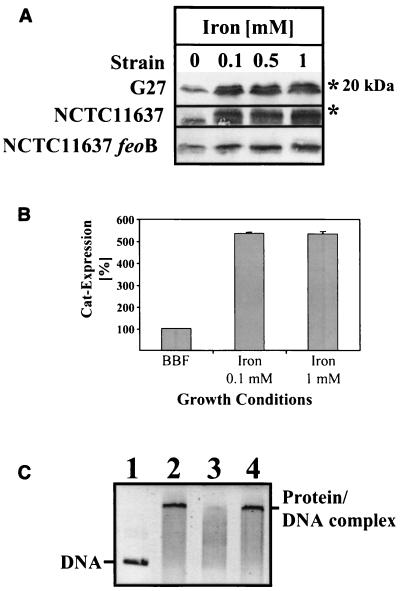
Analysis of Pfr expression in the H. pylori wt strains G27 and NCTC11637 by immunoblotting (Fig. 3A) confirmed the earlier observation that the Pfr protein accumulates in the presence of iron (7). If the proteins were electrophoresed on 17% polyacrylamide gels, a second Pfr protein band with a slightly increased molecular mass (about 1 kDa) could be separated exclusively if the bacteria were cultivated in iron-supplemented medium (Fig. 3A). Similar results were obtained with the H. pylori reference strains NCTC11638, 1061, and 26695 (not shown). The addition of sodium chloride had no influence on Pfr expression, excluding unspecific influences through osmotic stress or chloride ions (results not shown).
FIG. 3.
Iron regulation of Pfr synthesis. (A) The regulation of Pfr synthesis in response to iron. The H. pylori wt strains G27 and NCTC11637 and an isogenic NCTC11637 feoB mutant, respectively (indicated on the left), were grown in BBF without supplementation and with iron at increasing concentrations (indicated on the top). Samples containing 15 μg of total protein were separated by SDS-PAGE, and the Pfr protein was detected by Western immunoblotting with the antiserum AK198. The second Pfr protein bands detected exclusively under iron-rich conditions and the position of the 20-kDa protein marker are indicated by asterisks on the right. (B) Reporter gene analysis of pfr gene expression. The H. pylori strain G27 carrying a transcriptional pfr::cat fusion was grown in broth (BBF) supplemented with iron at the concentrations indicated on the bottom, and the amount of the Cat protein (gray bars) resembling transcriptional activity of the pfr gene was monitored with a Cat-specific ELISA. The amount of Cat produced in unsupplemented BBF medium was set at 100%. The values represent means of two independent determinations. Standard deviations are indicated. The results are representative for two independent experiments. (C) Influence of iron on Fur binding to the pfr promoter. EMSA of the Fur binding activity was performed as described in Materials and Methods. In lanes 1 to 4 the DNA probe containing the intergenic region in front of the pfr gene (lane 1) was incubated with Fur (lanes 2 to 4) without additives (lane 2), with 0.2 mM iron added (lane 3), or with 0.2 mM sodium chloride added (lane 4). The picture represents a black and white image of the ethidium bromide-stained gel visualized under UV light. The positions of the DNA probe and the Fur protein-DNA complexes are indicated.
To investigate if the iron-mediated accumulation of Pfr depends on transport of iron to the cytoplasm, iron regulation of Pfr was analyzed in a mutant of H. pylori strain NCTC11637 lacking FeoB, the major iron uptake system. FeoB transfers ferrous iron from the periplasm to the cytoplasm (36). Compared to the wt strain the iron induction of Pfr expression was clearly less pronounced in the feoB mutant, as indicated by the absence of the iron-induced second Pfr protein band and by the weak intensities of the protein bands when grown under the same conditions (Fig. 3A).
The changes in pfr gene expression in response to iron-rich conditions were investigated using the transcriptional fusion of pfr, with a promoterless pfr::cat gene fusion in the pfr mutant (7) as a reporter. Quantification of the Cat protein by ELISA showed that iron at both 0.1 and 1 mM induced pfr transcription about fivefold (Fig. 3B), indicating that the pfr gene is completely derepressed under these conditions. It was shown previously that regulation of Pfr synthesis occurs at the transcriptional level by iron-mediated removal of the Fur repressor protein from the pfr promoter, resulting in derepression of the pfr gene (4, 11). Direct transcriptional iron regulation of pfr by Fur was demonstrated by EMSA of recombinant H. pylori Fur bound to the pfr promoter region in the presence and absence of iron (Fig. 3C). The results confirmed that the Fur protein binds to the intergenic region in front of the pfr gene and that addition of 0.2 mM ferrous iron diminishes the Fur-DNA interaction, as indicated by the disappearance of the shifted protein-DNA complex (Fig. 3C). Controls performed with identical amounts of sodium chloride had no effect, indicating that the inhibition of Fur binding was specifically caused by iron.
H. pylori pfr mutants do not colonize a gerbil model of infection.
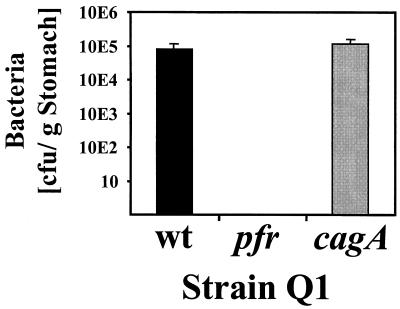
The role of Pfr in colonization of the gastric mucosa was investigated in the gerbil-based animal model (39). For this study, the pfr gene was inactivated in the gerbil-adapted H. pylori strain Q1. To exclude unspecific effects of the mutation protocols, we constructed an isogenic cagA mutant in parallel to serve as a control, because the cagA mutation does not affect the colonization of gerbils (39). Six gerbils were orally infected with the wt Q1 strain and its isogenic pfr and cagA mutants, respectively, keeping three animals for 3 weeks and three animals for 6 weeks. There was no time-dependent difference in the efficiency of colonization within each group, and so the values of each group were averaged. Among the six animals challenged with the wt strain or the cagA mutant, all animals were colonized with an average bacterial load of 8 × 104 (wt) and 1.17 × 105 (cagA mutant) bacteria/g of stomach (Fig. 4). In contrast, all animals challenged with comparable inocula of the pfr mutant were completely free of H. pylori either 3 or 6 weeks after inoculation.
FIG. 4.
Colonization properties of the H. pylori pfr mutant in the gerbil model of infection. The animals were infected with the H. pylori wt strain Q1 or with the pfr and cagA mutants as indicated. The results for bacterial colonization obtained from reisolation and counting of colonies after 3 and 6 weeks are summarized in one column for each strain because there was no significant difference in the bacterial load after the infection periods. The data are representative of two individual experiments performed for 3 and 6 weeks, respectively. Standard deviations are indicated.
DISCUSSION
In the human stomach iron is released from food by peptic degradation, while iron restriction can be encountered via the iron-chelating activity of lactoferrin (25). Therefore, both iron starvation and iron overload may occur in relatively short time intervals. This specific environmental situation necessitates the development of mechanisms which allow H. pylori to adapt its iron metabolism adequately. The complete colonization defect of the pfr mutant in the gerbil stomach indicates that Pfr functions are essential for gastric adaptation and shows for the first time that iron storage is a prerequisite for the successful establishment of H. pylori infection. The possible role of Pfr in the development of H. pylori-associated human diseases has not been investigated in detail. A recent study on the possible role of pfr mutations in the development of iron deficiency anemia did not reveal significant correlations (9).
Phenotypical characterization of the pfr mutant under in vitro conditions mimicking changes in iron availability and acidity thought to occur in vivo suggests that Pfr plays an essential role in gastric adaptation and colonization. The iron storage function of Pfr was found to be essential for adaptation to both high- and low-iron conditions. Iron preloading enabled the wt strain but not the pfr mutant to multiply under iron-restricted conditions, demonstrating that iron stored in Pfr can be remobilized to support growth under conditions of severe iron restriction. The finding that iron toxicity was amplified at low pH might be anticipated because protons increase the availability of the metal for bacteria. However, the fact that low pH as well as changes in iron availability are stress conditions occurring continuously in the gastric mucosa highlights the protective role of Pfr in this specific environment. The results from total cellular iron content determinations suggest that Pfr is responsible for a significant part of the total iron storage capacity of H. pylori. Because iron increases the sensitivity of bacteria to oxygen radicals (24, 33), the low iron content could be responsible for the lowered superoxide stress sensitivity of the pfr mutant. This was further supported by the finding that superoxide sensitivity was transiently increased after iron preloading. The reduced iron uptake rates observed in the pfr mutant suggest that Pfr-mediated removal of iron from the cytoplasm and the storage of the metal in the ferritin cavity are essential for maintaining a full iron uptake capacity. Taken together with the fact that iron uptake is essential for establishment of H. pylori infection (36), the pfr phenotypes observed in vitro indicate that the colonization defect of the pfr mutant is caused by the lack of Pfr functions essential for maintaining iron homeostasis. The expression and activity of the essential H. pylori colonization factor urease (22) was not affected in the pfr mutant (data not shown), and the pfr mutation did not generally limit bacterial fitness in vitro, as the pfr mutation had no effect on H. pylori growth under standard conditions (Fig. 1).
The accumulation of Pfr under iron-rich conditions (7) allows H. pylori to maximize the iron storage capacity in response to an increased iron availability. Similar observations have been made for the bacterioferritins from Rhodobacter capsulatus (27) and Pseudomonas species (21, 23). The second iron-induced Pfr protein band might reflect structural changes of Pfr upon iron binding. Multiple Pfr isoforms were recently also observed in a proteome analysis of H. pylori and it was proposed that Pfr is subject to posttranslational modifications (20). However, because the multimeric structure of ferritins can cause electrophoretic artifacts, Pfr modifications should await further careful analysis at the molecular level. The molecular mechanisms of Fur-mediated iron induction of H. pylori Pfr synthesis have been elucidated recently (4, 11). By using DNase footprinting assays, Delany et al. showed that iron inhibits binding of the Fur protein to multiple operator sites within the pfr promoter (11). To confirm this unique Fur function in derepression of an iron-activated gene, we further analyzed the in vitro DNA binding activity of recombinant H. pylori Fur by EMSA (Fig. 3C). The results demonstrate that iron interferes with Fur binding to the intergenic DNA region in front of the pfr gene. The observation that iron only diminished but did not completely abolish Fur binding is well in agreement with the results from DNase footprinting analysis (11), supporting the hypothesis that upon iron binding Fur does not completely lose its binding activity but undergoes conformational changes (11). This could explain the lack of a single clear band in Fig. 3C, lane 3. Iron-mediated derepression of the pfr gene enables H. pylori to adapt Pfr expression perfectly to a wide range of different iron concentrations, as both low- and high-iron conditions fivefold repressed (4) and induced (Fig. 3B), respectively, the pfr::cat reporter gene fusion. Iron at a concentration of 0.1 mM completely derepressed pfr expression. Higher iron concentrations did not further increase the Cat reporter protein levels (Fig. 3B). Delany et al. (11) reported that in the H. pylori wt strain G27 pfr mRNA increases 20-fold upon treatment with 5 mM iron for 15 min, indicating that, although the experimental conditions are different, the iron induction of pfr can reach higher levels in a wt genetic background. The reduced iron induction of Pfr synthesis in the H. pylori feoB mutant (Fig. 3A) suggests that the cytoplasmic iron transport defect enhances Fur repression of the pfr promoter. This confirms furthermore that Fur regulation of pfr depends on iron uptake and responds to changes in the cytoplasmic iron pool, which is well in agreement with the fact that Fur homologs sense and respond to the cytoplasmic iron concentration (8, 19, 31). Taken together, these results demonstrate that the role of Fur in H. pylori iron storage differs considerably from its classical role in regulation of iron uptake (8, 19, 31). Based on these findings we propose that the complex Fur regulation of Pfr synthesis represents a specific adaptation to the situation in the human stomach, where H. pylori is frequently exposed to the more soluble ferrous iron (36), which is effectively detoxified and stored by Pfr. In conclusion, the results extend our understanding of ferritin functions in prokaryotes, highlight interactions of iron storage with iron uptake, and confirm the importance of H. pylori iron metabolism in gastric colonization.
Acknowledgments
B.W. and S.G. contributed equally to this work.
We thank Tanja Vey for excellent technical assistance.
This work was supported by grants from the Deutsche Forschungsgemeinschaft to M.K. (Ki201/8-2 and Ki201/9-1), the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO 901-14-206 and DN93-340) to A.H.M. V.V. and J.G.K., and the Wellcome Trust to D.J.K. and S.C.A and by a studentship from Catalyst Chemicals Ltd., Kenya, to J.V.
Editor: J. T. Barbieri
REFERENCES
- 1.Abdul-Tehrani, H., A. J. Hudson, Y. S. Chang, A. R. Timms, C. Hawkins, J. M. Williams, P. Harrison, J. R. Guest, and S. C. Andrews. 1999. Ferritin mutants of Escherichia coli are iron deficient and growth impaired, and fur mutants are iron deficient. J. Bacteriol. 181:1415-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrews, S. C. 1998. Iron storage in bacteria. Adv. Microb. Physiol. 40:281-351. [DOI] [PubMed] [Google Scholar]
- 3.Asaka, M., M. Kudo, M. Kato, T. Sugiyama, and H. Takeda. 1998. Long-term Helicobacter pylori infection—from gastritis to gastric cancer. Aliment. Pharmacol. Ther. 12(Suppl. 1):9-15. [DOI] [PubMed] [Google Scholar]
- 4.Bereswill, S., S. Greiner, A. H. van Vliet, B. Waidner, F. Fassbinder, E. Schiltz, J. G. Kusters, and M. Kist. 2000. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J. Bacteriol. 182:5948-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bereswill, S., F. Lichte, S. Greiner, B. Waidner, F. Fassbinder, and M. Kist. 1999. The ferric uptake regulator (Fur) homologue of Helicobacter pylori: functional analysis of the coding gene and controlled production of the recombinant protein in Escherichia coli. Med. Microbiol. Immunol. 188:31-40. [DOI] [PubMed] [Google Scholar]
- 6.Bereswill, S., F. Lichte, T. Vey, F. Fassbinder, and M. Kist. 1998. Cloning and characterization of the fur gene from Helicobacter pylori. FEMS Microbiol. Lett. 159:193-200. [DOI] [PubMed] [Google Scholar]
- 7.Bereswill, S., U. Waidner, S. Odenbreit, F. Lichte, F. Fassbinder, G. Bode, and M. Kist. 1998. Structural, functional and mutational analysis of the pfr gene encoding a ferritin from Helicobacter pylori. Microbiology 144:2505-2516. [DOI] [PubMed] [Google Scholar]
- 8.Braun, V., K. Hantke, and W. Koester. 1998. Bacterial iron transport: mechanisms, genetics, and regulation, p. 67-145. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems. Marcel Dekker, New York, N.Y. [PubMed]
- 9.Choe, Y. H., T. S. Hwang, H. J. Kim, S. H. Shin, S. U. Song, and M. S. Choi. 2001. A possible relation of the Helicobacter pylori pfr gene to iron deficiency anemia? Helicobacter 6:55-59. [DOI] [PubMed] [Google Scholar]
- 10.Delany, I., A. B. Pacheco, G. Spohn, R. Rappuoli, and V. Scarlato. 2001. Iron-dependent transcription of the frpB gene of Helicobacter pylori is controlled by the Fur repressor protein. J. Bacteriol. 183:4932-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1311. [DOI] [PubMed] [Google Scholar]
- 12.Doig, P., J. W. Austin, and T. J. Trust. 1993. The Helicobacter pylori 19.6-kilodalton protein is an iron-containing protein resembling ferritin. J. Bacteriol. 175:557-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dundon, W. G., A. Polenghi, G. Del Guidice, R. Rappuoli, and C. Montecucco. 2001. Neutrophil-activating protein (HP-NAP) versus ferritin (Pfr): comparison of synthesis in Helicobacter pylori. FEMS Microbiol. Lett. 199:143-149. [DOI] [PubMed] [Google Scholar]
- 14.Dunn, B. E., H. Cohen, and M. J. Blaser. 1997. Helicobacter pylori. Clin. Microbiol. Rev. 10:720-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, D. J., D. G. Evans, H. C. Lampert, and H. Nakano. 1995. Identification of four new prokaryotic bacterioferritins, from Helicobacter pylori, Anabaena variabilis, Bacillus subtilis and Treponema pallidum, by analysis of gene sequences. Gene 153:123-127. [DOI] [PubMed] [Google Scholar]
- 16.Fassbinder, F., A. H. M. van Vliet, V. Gimmel, J. G. Kusters, M. Kist, and S. Bereswill. 2000. Identification of iron-regulated genes of Helicobacter pylori by a modified Fur titration assay (FURTA-Hp). FEMS Microbiol. Lett. 184:225-229. [DOI] [PubMed] [Google Scholar]
- 17.Fischer, W., D. Schwan, E. Gerland, G. E. Erlenfeld, S. Odenbreit, and R. Haas. 1999. A plasmid-based vector system for the cloning and expression of Helicobacter pylori genes encoding outer membrane proteins. Mol. Gen. Genet. 262:501-507. [DOI] [PubMed] [Google Scholar]
- 18.Frazier, B. A., J. D. Pfeifer, D. G. Russell, P. Falk, A. N. Olsen, M. Hammar, T. U. Westblom, and S. J. Normark. 1993. Paracrystalline inclusions of a novel ferritin containing nonheme iron, produced by the human gastric pathogen Helicobacter pylori: evidence for a third class of ferritins. J. Bacteriol. 175:966-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantke, K., and V. Braun. 1998. Control of bacterial iron transport by regulatory proteins, p. 11-44. In S. Silver and W. Walden (ed.), Metal ions in gene regulation. Chapman & Hall, New York, N.Y.
- 20.Lock, R. A., S. J. Cordwell, G. W. Coombs, B. J. Walsh, and G. M. Forbes. 2001. Proteome analysis of Helicobacter pylori: major proteins of type strain NCTC11637. Pathology 33:365-374. [PubMed] [Google Scholar]
- 21.Ma, J. F., U. A. Ochsner, M. G. Klotz, V. K. Nanayakkara, M. L. Howell, Z. Johnson, J. E. Posey, M. L. Vasil, J. J. Monaco, and D. J. Hassett. 1999. Bacterioferritin A modulates catalase A (KatA) activity and resistance to hydrogen peroxide in Pseudomonas aeruginosa. J. Bacteriol. 181:3730-3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGee, D. J., and H. L. Mobley. 1999. Mechanisms of Helicobacter pylori infection: bacterial factors. Curr. Top. Microbiol. Immunol. 241:155-180. [DOI] [PubMed] [Google Scholar]
- 23.Miller, C. D., Y. C. Kim, M. K. Walsh, and A. J. Anderson. 2000. Characterization and expression of the Pseudomonas putida bacterioferritin alpha subunit gene. Gene 247:199-207. [DOI] [PubMed] [Google Scholar]
- 24.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nakao, K., I. Imoto, N. Ikemura, T. Shibata, S. Takaji, Y. Taguchi, M. Misaki, K. Yamauchi, and N. Yamazaki. 1997. Relation of lactoferrin levels in gastric mucosa with Helicobacter pylori infection and with the degree of gastric inflammation. Am. J. Gastroenterol. 92:1005-1011. [PubMed] [Google Scholar]
- 26.Odenbreit, S., J. Püls, B. Sedlmaier, E. Gerland, W. Fischer, and R. Haas. 2000. Translocation of Helicobacter pylori CagA into gastric epithelial cells by type IV secretion. Science 287:1497-1500. [DOI] [PubMed] [Google Scholar]
- 27.Penfold, C. N., P. L. Ringeling, S. L. Davy, G. R. Moore, A. G. McEwan, and S. Spiro. 1996. Isolation, characterisation and expression of the bacterioferritin gene of Rhodobacter capsulatus. FEMS Microbiol. Lett. 139:143-148. [DOI] [PubMed] [Google Scholar]
- 28.Scarlato, V., I. Delany, G. Spohn, and D. Beier. 2001. Regulation of transcription in Helicobacter pylori: simple systems or complex circuits? Int. J. Med. Microbiol. 291:107-117. [DOI] [PubMed] [Google Scholar]
- 29.Schmitt, W., and R. Haas. 1994. Genetic analysis of the Helicobacter pylori vacuolating cytotoxin: structural similarities with the IgA protease type of exported protein. Mol. Microbiol. 12:307-319. [DOI] [PubMed] [Google Scholar]
- 30.Skerra, A., and T. G. Schmidt. 2000. Use of the Strep-Tag and streptavidin for detection and purification of recombinant proteins. Methods Enzymol. 326:271-304. [DOI] [PubMed] [Google Scholar]
- 31.Stojiljkovic, I., A. J. Bäumler, and K. Hantke. 1994. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a fur titration assay. J. Mol. Biol. 236:531-545. [DOI] [PubMed] [Google Scholar]
- 32.Tonello, F., W. G. Dundon, B. Satin, M. Molinari, G. Tognon, G. Grandi, G. Del Giudice, R. Rappuoli, and C. Montecucco. 1999. The Helicobacter pylori neutrophil-activating protein is an iron-binding protein with dodecameric structure. Mol. Microbiol. 34:238-246. [DOI] [PubMed] [Google Scholar]
- 33.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta-fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Vliet, A. H. M., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. J. E. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van Vliet, A. H. M., S. Bereswill, and J. G. Kusters. 2001. Ion metabolism and transport, p. 193-206. In H. L. T. Mobley, G. L. Mendz, and S. L. Hazell (ed.), Helicobacter pylori: physiology and genetics. ASM Press, Washington, D.C. [PubMed]
- 36.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 37.Wai, S. N., K. Nakayama, K. Umene, T. Moriya, and K. Amako. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 20:1127-1134. [DOI] [PubMed] [Google Scholar]
- 38.Wai, S. N., K. Nakayama, A. Takade, and K. Amako. 1997. Overproduction of Campylobacter ferritin in Escherichia coli and induction of paracrystalline inclusion by ferrous compound. Microbiol. Immunol. 41:461-467. [DOI] [PubMed] [Google Scholar]
- 39.Wirth, H. P., M. H. Beins, M. Yang, K. T. Tham, and M. J. Blaser. 1998. Experimental infection of Mongolian gerbils with wild-type and mutant Helicobacter pylori strains. Infect. Immun. 66:4856-4866. [DOI] [PMC free article] [PubMed] [Google Scholar]