Abstract
Decay-accelerating factor (DAF), a complement regulatory protein, also serves as a receptor for Dr adhesin-bearing Escherichia coli. The repeat three of DAF was shown to be important in Dr adhesin binding and complement regulation. However, Dr adhesins do not bind to red blood cells with the rare polymorphism of DAF, designated Dr(a−); these cells contain a point mutation (Ser165-Leu) in DAF repeat three. In addition, monoclonal antibody IH4 specific against repeat three was shown to block both Dr adhesin binding and complement regulatory functions of DAF. Therefore, to identify residues important in binding of Dr adhesin and IH4 and in regulating complement, we mutated 11 amino acids—predominantly those in close proximity to Ser165 to alanine—and expressed these mutations in Chinese hamster ovary cells. To map the mutations, we built a homology model of repeat three based on the poxvirus complement inhibitory protein, using the EXDIS, DIAMOD, and FANTOM programs. We show that perhaps Ser155, and not Ser165, is the key amino acid that interacts with the Dr adhesin and amino acids Gly159, Tyr160, and Leu162 and also aids in binding Dr adhesin. The IH4 binding epitope contains residues Phe148, Ser155, and L171. Residues Phe123 and Phe148 at the interface of repeat 2-3, and also Phe154 in the repeat three cavity, were important for complement regulation. Our results show that residues affecting the tested functions are located on the same loop (148 to 171), at the same surface of repeat three, and that the Dr adhesin-binding and complement regulatory epitopes of DAF appear to be distinct and are ≈20 Å apart.
The complement system is an important mediator of the innate immune response that protects the host from foreign particles and invading pathogens (19, 21). The effector functions arising from complement activation, however, contribute directly and/or indirectly to host tissue damage in many clinical conditions (45). Therefore, under physiological conditions, uncontrolled activation of complement is regulated by many membrane-bound and soluble regulatory proteins collectively known as regulators of complement activation (RCA) (29, 31, 36). The RCA gene family encodes four membrane proteins, decay-accelerating factor (DAF; CD55) (44, 48), complement receptor 1 (CR1; CD35), complement receptor 2 (CR2; CD21), and membrane cofactor protein (MCP; CD46), plus two plasma proteins, C4b-binding protein (C4BP) and factor H. The RCA proteins contain short consensus repeats (SCRs), more recently designated complement control protein repeats (CCPs), of ≈60 amino acids arranged in tandem (2, 18, 43). Each SCR contains two disulfide bridges and adopts a β-barrel structure, as shown by nuclear magnetic resonance (NMR) analyses of fragments of factor H (4, 10) and the vaccinia virus complement control protein (55).
In some malignant cells DAF is overexpressed, rendering these cells more resistant to complement attack than their normal counterparts (21). Also, DAF is expressed on all blood cells (23) and in various tissues and organs; its soluble form is present in body fluids (35). A 70-kDa protein, DAF is bound to the cell membrane by a glycosylphosphatidylinositol (GPI) anchor followed by a serine/threonine (ST)-rich region and four consecutive extracellular amino-terminal SCRs (SCR-1 through SCR-4). It has one N-linked oligosaccharide chain between SCR-1 and SCR-2 and multiple O-linked oligosaccharide chains in the ST region (9, 18).
The DAF regulates complement intrinsically by inhibiting the formation and accelerating the decay of C3 and C5 convertases of both the alternate and classical pathways of the complement system (34). The functional domains of DAF have been mapped using SCR deletion mutants. These studies showed that the complement regulatory activity of DAF resides within SCR-2 through SCR-4 (12). The complement regulatory activity of DAF against C4b2a (classical pathway) convertase lies within SCR-2 and SCR-3, whereas its activity against C3bBb (alternate pathway) convertase extends to SCR-4 (7). In the absence of a crystal structure, only limited information is available regarding the interaction of DAF with its ligands.
Based on the previously derived solution structure of human factor H, Kuttner-Kondo et al. proposed a hypothetical model of DAF predicting the potential ligand-binding sites (27, 28). According to this model, the groove at the interface of SCR-2 and SCR-3 and the groove at the interface of SCR-3 and SCR-4 along with its attached cavities were suggested as the most likely candidates for ligand binding (27, 28). In further studies, the combined effect of amino acids Leu147 and Phe148 lining the hydrophobic area of the SCR-3 cavity was shown to be important in regulating C3 convertases of both the classical and alternate pathways (6, 28).
In addition to regulating complement, members of the RCA family also serve as cellular receptors for many bacteria and viruses (29). For example, CD21 and CD46 are used as receptors of the Epstein-Barr virus and the measles virus, respectively (12, 20). Furthermore, DAF serves as a cellular receptor for uropathogenic Escherichia coli expressing Dr family adhesins (39, 42), human picornaviruses (e.g., enterovirus [15, 22, 51]), echovirus (11), and coxsackieviruses (5, 33).
The Dr adhesins of E. coli serve as important virulence factors that facilitate colonization of the human urogenital and gastrointestinal tracts (39, 42). E. coli isolates expressing Dr adhesins were implicated in 30 to 50% of cystitis cases and in 50% of protracted diarrhea cases. The Dr family of adhesins was named on the basis of its ability to recognize the Cromer blood group antigen Dr(a) on DAF (40). The classical pathway convertase and the Dr adhesin-binding regions are both localized on the SCR-2-SCR-3 interface of DAF, suggesting a possible overlap of binding regions. A rare polymorphism of human DAF containing a point mutation in SCR-3 (Ser165-Leu) (32, 39), designated Dr(a−), abolishes binding of Dr adhesins (40).
This study was conducted to identify the important Dr adhesin-binding and complement regulatory residues of human DAF SCR-3. We constructed recombinant GPI-anchored mutant DAF proteins using alanine-scanning mutagenesis to dissect the role of specific amino acids in binding Dr adhesin and complement regulation. We provide evidence that the residues involved in binding of Dr adhesin and the complement regulatory function of DAF appear distinct and are ≈20 Å apart.
MATERIALS AND METHODS
Cell culture.
Chinese hamster ovary (CHO) K-1 cells (ATCC CCL61) obtained from the American Type Culture Collection (Rockville, Md.) were cultured in Ham's nutrient mixture F-12 (GIBCO-BRL, Rockville, Md.) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO-BRL), 50 U of penicillin/ml, 50 μg of streptomycin/ml, 2 mM l-glutamine, nonessential amino acids, and 250 μg of G-418/ml (gentamicin analogue) when indicated.
Bacterial strains.
E. coli EC901 strain BN406 (41) containing plasmid pBJN406 expressing only Dr adhesin (Dr+) and its transposon mutant (BN 17) (41) with a mutation in the draE adhesin subunit gene (Dr−) was grown on Luria agar plates containing 25 μg of chloramphenicol per ml. The Dr+ and Dr− E. coli strains were collected from the plates, suspended in 2% α-methylmannose in phosphate-buffered saline (PBS), and adjusted to an optical density at 600 nm (OD600) of 2. The minimal hemagglutination titer (MHT) for Dr+ E. coli was determined using 3% (vol/vol) human group O erythrocytes in PBS. To eliminate potential differences in attachment to CHO cells due to the variation in expression of adhesins, suspensions of recombinant strains were used at the same MHT. When necessary, before the experiment, the MHT of each strain was adjusted to the same hemagglutinating activity (1/64) by diluting or concentrating the suspension. For example, on average, a suspension of 1.5 × 108 CFU/ml had an MHT of 1/64, and occasionally of 1/32. Therefore, bacterial concentrations were approximately equal and minor adjustments of OD were necessary only occasionally.
Plasmid harboring DAF.
As a DNA template for the site-directed mutagenesis, we used the pcDNA 3.1 plasmid harboring human DAF cDNA (gift from D. M. Lublin, Washington University School of Medicine).
Antibodies.
The following antibodies were used: monoclonal anti-human DAF SCR-1 antibody IA10 (unconjugated) and phycoerythrin (PE)-conjugated IA10 (BD Pharmingen, San Diego, Calif.); monoclonal anti-SCR-3 antibody IH4 (gift from D. M. Lublin); horseradish peroxidase (HRP)-conjugated goat anti-mouse immunoglobulin G (IgG; Bio-Rad Laboratories, Hercules, Calif.); Texas Red-conjugated goat anti-mouse antibody (Molecular Probes, Inc., Eugene, Oreg.); and the IgG fraction of rabbit anti-hamster antibody (Sigma Chemical Co., St. Louis, Mo.).
Source of complement.
Normal human serum (NHS) used in the DAF-mediated complement protection assay was obtained by pooling sera drawn from eight healthy human volunteers. Consent was obtained from each person (Institutional Review Board approval no. 01-210).
Site-directed mutagenesis of DAF.
Site-directed mutagenesis was carried out using the QuickChange kit (Stratagene, La Jolla, Calif.) according to the manufacturer's instructions. The plasmid DNA template (20 ng), 125 ng of each mutagenesis primer, 5 μl of 10× PCR buffer, 1 μl of deoxynucleoside triphosphate (dNTP) mix (10 mM concentration each of dATP, dGTP, dCTP, and dTTP), and 1 μl (2.5 U) of Pfu DNA polymerase were mixed in a final volume of 50 μl. PCR was carried out in a thermal cycler according to the instructions of the mutagenesis kit manufacturer. After PCR, the reaction mixture was placed on ice and DpnI restriction enzyme was added. The mixture was incubated at 37°C for 1 h to digest the methylated DNA strand.
Transformation of E. coli.
Transformation of supercompetent Epicurian E. coli XL1-Blue cells with DpnI-treated DNA was carried out according to the instructions in the mutagenesis kit. The cells and the DNA were incubated on ice for 30 min, followed by heat shock at 42°C for 45 s and incubation on ice for 2 min. Then, 0.5 ml of NZY+ broth was added and the mixture was incubated at 37°C for 1 h with shaking. Finally, the entire volume was spread on L agar plates containing 50 μg of ampicillin and incubated overnight at 37°C. The next day, colonies were isolated and plasmid DNA was prepared by using a mini prep (Qiagen, Valencia, Calif.). We sequenced the complete open reading frame of all the clones with the desired alanine mutations and confirmed that other unexpected mutations were not present.
Transfection of CHO cells.
The CHO cells were transfected with 3 μg of mutated plasmid DNA using a CLONfectin kit (Clontech, Palo Alto, Calif.) according to the method recommended by the manufacturer. Transfected cells were cultured in growth medium (F-12) containing 250 μg of G-418/ml.
Immunofluorescence staining of DAF.
The CHO cells transfected with the wild-type or mutated human DAF were grown on coverslips. For immunostaining, the cells were first incubated with 1% normal goat serum in PBS (blocking solution) for 20 min, washed with PBS, and incubated with IA10 antibody (1:100 dilution in PBS) for 1 h at room temperature. The cells were then washed and incubated with Texas Red-conjugated goat anti-mouse antibody for 1 h at room temperature. Slides were examined using a Nikon Eclipse 600 microscope (Nikon Inc., Melville, N.Y.) equipped with an epifluorescence attachment, Texas Red or double-pass Texas Red-fluorescein isothiocyanate filter, and a U-III camera system.
Isolation of DAF-expressing CHO cells.
CHO cells (5 × 106) in suspension were incubated in blocking solution for 20 min on ice, washed three times with PBS, and incubated with PE-conjugated IA10 antibody for 30 min. The CHO cells were then washed again in PBS (three times) and finally resuspended in 1 ml of PBS. Cells were sorted into 96-well plates in F-12 medium containing 10% fetal bovine serum in 20 mM HEPES buffer by using a fluorescence-activated cell sorter (FACS). Clones expressing DAF, which were identified by immunofluorescence staining, were then subjected to repeated sorting by FACS.
Quantitation of DAF by flow cytometry.
Flow cytometric analysis was conducted to quantitate the level of DAF expression on the surface of the sorted CHO cells with PE-labeled anti-DAF IA10 antibody and QuantiBRITE PE beads (Becton Dickinson and Co., Franklin Lakes, N.J.) as recommended by the manufacturer.
CHO cell binding assay.
CHO cells expressing wild-type or mutated human DAF were grown in F-12 medium. A bacterial suspension in 2% α-methylmannose was prepared and adjusted to an OD600 of 2.0. The bacterial suspension (200 μl) was added to CHO cells, which were seeded on coverslips 24 h before the experiment, in a 12-well plate at a density of 5 × 105 cells per well and incubated at 37°C for 1 h. After washing with PBS and fixing with 1 ml of methanol for 15 min, the monolayer was stained overnight with 1 ml of Giemsa (1:20 dilution). Coverslips were washed with deionized water, mounted on glass slides, and observed under a light microscope with ×400 magnification; 20 CHO cells per field were counted. The results were expressed as the mean number of bacteria bound per CHO cell.
Cytoprotection assay.
DAF-mediated cytoprotection was assayed as previously described by Liszewski and Atkinson (30). Briefly, 10,000 CHO cells per well were incubated overnight in 96-well microtiter plates. The medium was removed the following day, and rabbit anti-hamster antibody was added (13.2 mg/ml) for 1 h at 4°C. The antibody was removed and the cells were incubated with 40% NHS diluted in gelatin Veronal buffer for 3 h at 37°C. The cells were washed twice in Dulbecco's PBS, and complete medium (F-12) was added. Cells were grown for 24 h and assessed with a Cell Titer 96 kit (Promega, Madison, Wis.) according to the manufacturer's protocol. The assay was performed in triplicate in three separate experiments. Untreated controls, antibody-only controls, and serum-only controls were included (30). The results were expressed as percent protection, calculated based on values for untreated controls (antibody-alone and NHS-alone controls were equivalent to untreated controls).
Western blot analysis of mutated DAF proteins.
To analyze the expression of DAF by the CHO cell clones, monolayers of CHO cells expressing wild-type human DAF (positive control) or mutated DAF were lysed by the addition of lysis buffer (10 mM Tris-HCl, 1.0 mM EDTA, 2% Triton X-100, pH 7.5, and protease inhibitor cocktail P1860 [Σ). The lysates were run on a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) under nonreducing conditions and transferred to nitrocellulose membrane (Cosmonics Inc., Westborough, Mass.) in 25 mM Tris, 192 mM glycine, 20% methanol, pH 8.3, using a Trans-blot apparatus (Bio-Rad Laboratories) for 1.5 h at 100 V. The membrane was blocked for 1 h at 4°C in 5% nonfat dry milk (Bio-Rad Laboratories) in PBS, incubated for 1 h at 4°C with anti-DAF IH4 antibody (dilution 1:1,000), and washed three times in PBS. The membrane was then incubated for 1 h at 4°C with a 1:20,000 dilution of HRP-conjugated goat anti-mouse antibody. The unbound conjugate was removed by washing three times in PBS, and the HRP substrate was added. Detection of the protein bands was done using an ECL kit according to the manufacturer's instructions (Amersham Corporation, Piscataway, N.J.).
Molecular modeling of human DAF SCR-3.
A sequence search for closely related DAF proteins in the Protein Data Bank (PDB) was performed using the program BLASTP (3) with the E-value cutoff set to 0.001. The query with DAF SCR-3 yielded a set of proteins containing CCP modules. The best hit (bit score of 62 and E-value of 3 × 10−11) was the central CCP module pair of poxvirus complement inhibitor protein (PDB code 1E5G domain 2) (16). The alignment used for modeling was obtained from the output of the BLASTP program without further modification and it showed an identity of 47% to the target. The functional match, together with a higher degree of identity of the template to DAF, makes the CCP module of poxvirus a better template to model DAF than the previously used template human factor H SCR-16, which showed an identity of 32% with DAF SCR-3 (27). Initial constraints to model DAF SCR-3 were obtained from the template by using our EXDIS program (52). EXDIS extracts distance and dihedral angle constraints from a known template used as the geometric specification for the unknown sequence. The input for EXDIS was alignment in CLUSTALW format (17) and the coordinate file for the template (i.e., 1E5G.pdb). Fragments were defined to exclude loops or gaps in the alignment during the extraction process. For DAF SCR-3, a total of 7,075 distance constraints were used. A tolerance of 0.5 Å was specified to define the upper-bound and lower-bound limits for the distance constraints. The dihedral angles for the side chain of the target sequence, when residues matched in the alignment, were set to values of matching residues of the template. The backbone dihedral angles were extracted from the template in regions defined by the fragment and set to 180° in loop regions where there was a gap in the alignment corresponding to the target. Upper-bound and lower-bound limits of ±10° for psi and phi and ±5° for omega dihedral angle constraints were used. Additional constraints were used to specify disulfide bridges between residues 129 and 170 and between 156 and 186. The distance between SG-SG atoms of two cysteine residues was set to 2.05 ± 0.5 Å (mean ± standard deviation) and the CB-SG atom distance was set to 3.05 ± 0.5 Å. The constraints obtained from the EXDIS program were used to generate 50 structures starting from a random conformation using DIAMOD (37, 47). DIAMOD is a self-correcting distance geometry program that uses a variable target function approach to progressively apply the constraints to an unknown sequence. The best model with the least target function was selected and energy refined using FANTOM (49). FANTOM minimizes constraint energies by successive application of quasi-Newton and Newton-Raphson minimizers. It uses an all-atom ECEPP/2 force field (1). A fourth-power energy function was used for distance constraints, which added kT/2 to the total energy for a violation of 0.2 Å in the regularization stage. The limit was raised in two steps to 1.0 Å toward the end of the minimization. The conformational energy of the model after refinement was −16.7 kcal/mol. Accessible surface area was calculated using the program GETAREA (13). The EXDIS, DIAMOD, FANTOM, and GETAREA programs are available at http://www.scsb.utmb.edu.
RESULTS
Construction of DAF mutants.
Previous data indicated that a naturally occurring mutant Ser165-Leu of DAF abolishes binding of Dr-bearing E. coli (32). The only tyrosine, Tyr160, located in the same repeat, was proposed to be important for binding because modified tyrosines and low concentrations of structurally similar chloramphenicol inhibit the binding of Dr adhesin (40). Furthermore, type IV human collagen, another natural receptor for Dr adhesin, is similarly inhibited by chloramphenicol. Type IV collagen shares homology only with the GlyTyrXXXXSer motif of the SCR-3 region (54). These previous results indicated that the Ser165 region in SCR-3 is involved in binding of Dr adhesin.
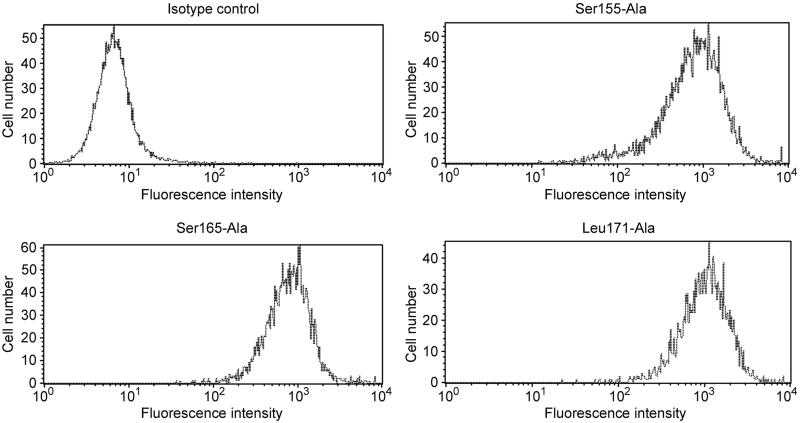
Ten amino acids in SCR-3 (Phe148, Phe154, Ser155, Cys156, Gly159, Tyr160, Leu162, Phe163, Ser165, and Leu171) and one amino acid (Phe123) in the SCR-2-SCR-3 interface were independently mutated to alanine (Ala). As a negative control for Dr adhesin-binding studies, we also constructed the known natural mutation Ser165-Leu. The mutant constructs were confirmed by sequencing and transfected to CHO cells; then, stable transfectants were selected in G-418-containing F-12 medium. Expression of DAF cDNA in each of the G-418-resistant transfectants was confirmed by immunofluorescence staining using anti-SCR-1 antibody IA10 and Texas Red-conjugated goat anti-mouse antibody. The immunofluorescence staining also suggested that the mutations did not grossly affect the conformation of DAF. Stable transfectants were sorted for high DAF expression by FACS with PE-conjugated anti-SCR1 antibody IA10. Individual clones that were obtained and analyzed by flow cytometry with QuantiBRITE beads demonstrated a similar range of expression. Figure 1 shows flow profiles of representative mutants and the isotype control. On average, transfectants differed in DAF expression by not more than twofold from the control (see Table 1 for the range of DAF expression). The binding and cytoprotection results were adjusted to reflect even minor variations in DAF expression. The adjustment formula corrected for binding and cytoprotection by adding or subtracting percent binding or protection relative to control DAF expression by CHO DAF cells.
FIG. 1.
Analysis of DAF expression by flow cytometry. Expression of DAF on the surface of stably transfected clones of CHO cells was quantitated by flow cytometry. Individual clones were analyzed by flow cytometry using PE-conjugated anti-DAF (SCR-1) antibody IA10 and QuantiBRITE PE beads. Flow profiles shown are for the isotype control and three representative clones, Ser155-Ala, Ser165-Ala, and Leu171-Ala.
TABLE 1.
Quantitation of DAF molecules expressed on the surface of CHO cell transfectantsa
| Human (wild-type) or mutant DAF expressed | No. of DAF molecules/cell |
|---|---|
| DAF (control) | 3.4 × 105 |
| Phe123-Ala | 3.6 × 105 |
| Phe148-Ala | 5.1 × 105 |
| Phe154-Ala | 2.4 × 105 |
| Ser155-Ala | 2.9 × 105 |
| Cys156-Ala | 2.0 × 105 |
| Gly159-Ala | 5.2 × 105 |
| Tyr160-Ala | 4.9 × 105 |
| Leu162-Ala | 4.1 × 105 |
| Phe163-Ala | 3.3 × 105 |
| Ser165-Ala | 2.0 × 105 |
| Ser165-Leu | 5.0 × 105 |
| Leu171-Ala | 4.1 × 105 |
DAF expression was measured by using PE-conjugated anti-SCR-1 MAb IA10. All tested clones were measured at the same time and conditions. The receptor density between various clones did not differ more than twofold compared with the control and fulfills the criteria used by Lublin et al (12).
Binding of Dr+ E. coli to CHO cells expressing mutated human DAF.
To analyze the role of specific amino acids of DAF in the interaction with Dr adhesin, CHO cells expressing site-directed mutations of human DAF were incubated with Dr+ E. coli and stained with Giemsa. The experiments were performed at least three times in duplicate, and the number of bacteria bound per cell was counted under a light microscope. The results (Table 2) indicate that, compared with cells expressing wild-type human DAF (positive control), the binding of Dr+ E. coli was completely abolished in cells expressing Ser155-Ala, Cys156-Ala, and Ser165-Leu mutant DAF molecules; cells expressing Gly159-Ala, Tyr160-Ala, and Leu162-Ala mutant DAF showed a relatively decrease in binding (P ≤ 0.05). The results suggest that these amino acids play a critical role in the binding of Dr adhesin to DAF. The CHO cells expressing the remaining DAF mutations did not exhibit significant differences in binding compared with the control.
TABLE 2.
Effect of DAF mutations on binding of Dr+ E. coli and on complement regulation (cytoprotection)
| Mutation | Average no. of bacteria bound/cell ± SDa | % Complement protection of corresponding control ± SDa |
|---|---|---|
| DAF+ | 20 ± 4 | 100 |
| DAF− | 0* | 0.0* |
| Phe123-Ala | 16 ± 2 | 52.4 ± 1.5* |
| Phe148-Ala | 26 ± 5 | 5.3 ± 3.7* |
| Phe154-Ala | 20 ± 9 | 24.9 ± 19.7* |
| Ser155-Ala | 0* | 84.0 ± 19.1 |
| Cys156-Ala | 0* | 35.0 ± 10.2* |
| Gly159-Ala | 8 ± 3* | 116.6 ± 11.0 |
| Tyr160-Ala | 5 ± 2* | 137.3 ± 11.3* |
| Leu162-Ala | 5 ± 2* | 258.3 ± 113.8 |
| Phe163-Ala | 15 ± 2 | 227.0 ± 41.0* |
| Ser165-Ala | 20 ± 8 | 68.6 ± 32.3 |
| Ser165-Leu | 0* | 169.1 ± 63.4 |
| Leu171-Ala | 19 ± 3 | 113.4 ± 55.7 |
*, statistical significance, P ≤ 0.05.
Protection of CHO cells expressing mutated DAF from complement-mediated lysis.
A cytoprotection assay was used to investigate the role of the targeted amino acids in the complement regulatory function of DAF (30). CHO cells expressing mutated DAF or wild-type human DAF (positive control) were incubated with antibodies to the CHO cell surface constituents and challenged with complement (40% NHS). The experiments were performed at least three times in triplicate, and the results shown in Table 2 compare the cytoprotection abilities of the DAF mutants with that of wild-type human DAF. The data indicate that Phe123-Ala, Phe148-Ala, Phe154-Ala, and Cys156-Ala mutants exhibit a significantly reduced (P ≤ 0.05) ability to protect cells from complement-mediated lysis. Additionally, mutant Ser165-Ala may also have some minor effect on complement protection. Among these mutants, the Phe148-Ala mutant is especially notable as there is nearly complete inhibition of the cytoprotective capacity of DAF (approximately 95% reduction compared with control). The data also show that the replacement of hydrophobic residues Tyr160 and Phe163 has a positive affect on DAF protective function (P ≤ 0.05). Although the exact mechanism is not known, one hypothesis is that replacement of these residues may affect the positioning Phe123 and Phe148, thereby increasing the protective function of DAF. Our results suggest that although amino acids Phe123, Phe148, Phe154, Cys156, and perhaps Ser165-Ala are critical for DAF-mediated complement function, Tyr160 and Phe163 may play a secondary role in the complement-protective capacity.
Binding of anti-DAF antibody to mutated DAF proteins.
Monoclonal antibody (MAb) IH4 has been shown to block both the binding of Dr adhesin to DAF and the DAF-mediated protection of cells against complement-mediated lysis (12, 42). IH4 does not recognize DAF that has been linearized in the presence of SDS and disulfide bridge-breaking mercaptoethanol (reducing conditions), suggesting that the epitope of DAF is discontinuous (conformational). We mapped IH4-binding amino acids using lysates of cells expressing DAF mutations. The lysates were resolved by PAGE under nonreducing conditions as previously described (20), followed by Western blot analyses using MAb IH4. The results (Fig. 2) show that the DAF mutants Phe148-Ala, Ser155-Ala, Cys156-Ala, and Leu171-Ala failed to bind IH4, indicating the importance of these amino acids in interaction with MAb IH4. Besides the fact that Cys156 forms the disulfide bridge with Cys186 and hence is important for maintaining the stability of SCR-3 conformation, mutation of other amino acids (Phe148, Ser155, and Leu171) may affect the conformation of SCR-3, further suggesting that IH4 recognizes a conformational epitope.
FIG. 2.
Western blot analyses of the binding of MAb IH4 to mutated DAF proteins. Lysates of the CHO cells expressing the mutated DAF proteins were run in SDS-10% PAGE gels under nonreducing conditions and blotted with IH4 antibody. Note that clones expressing mutant proteins Phe148-Ala, Ser155-Ala, Cys156-Ala, Ser165-Leu, and Leu171-Ala are not recognized by IH4. Wild-type human DAF was used as a positive control, whereas DAF− CHO cells transfected with plasmid only were used as a negative control.
Molecular modeling of the SCR-3 of DAF.
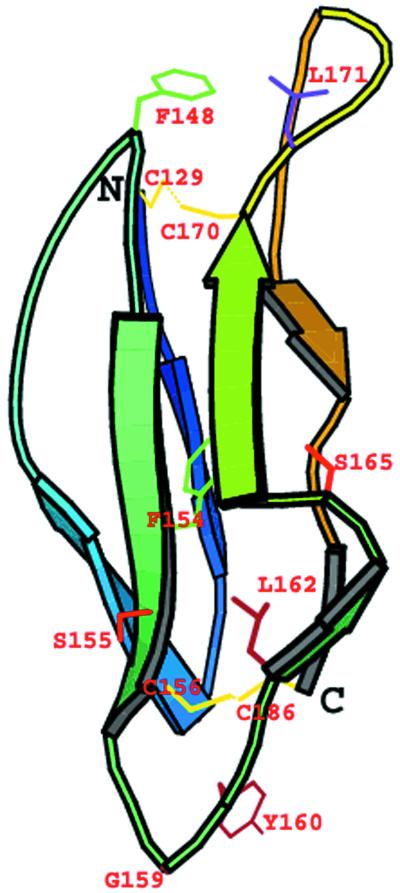
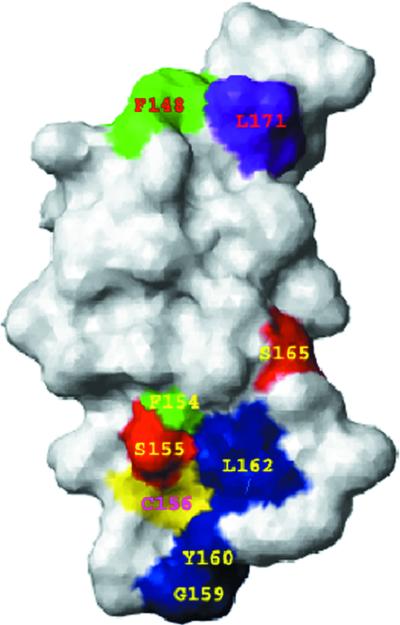
A three-dimensional (3-D) model of human DAF, SCR-3 (residues 129 to 186) was built, and the set of mutations in SCR-3 were mapped on this model. The domain consists of a β-sheet barrel running in an up-and-down fashion and is stabilized by two disulfide bridges between residues 129 and 170 and between 156 and 186 (Fig. 3). The domain is shaped like an ellipsoid tapering towards the outer side of the major axis. The dimensions of SCR-3 measured from the model are approximately 40 by 20 by 20 Å. All mutated residues map on one surface of the SCR-3 (Fig. 4). The side chain of Phe148 is completely surface exposed, whereas the side chain of Phe154 is buried and is part of the hydrophobic core of SCR-3. The lower part of SCR-3 consists of residues that bind to Dr adhesin. This binding surface is characterized by the presence of a loop consisting of Ser155, Gly159, Tyr160, Leu162, and Ser165, with the bulky hydrophobic residues Leu162 and Tyr160 surface exposed. Mutations of these residues to alanine also affected the binding of Dr adhesin to DAF. Details of the stereochemical quality of the model are presented in Table 3.
FIG. 3.
Cartoon model of the SCR-3 of DAF. The model was generated using the EXDIS, DIAMOD, and FANTOM programs. The model shows that the SCR-3 of DAF consists of β-strands running in an up-and-down fashion to form a β-barrel. The model was built for residues 129 to 186 of human DAF. Side chains of mutated residues are also shown. The two disulfide bridges between residues Cys129 and Cys170 and between Cys156 and Cys186 are shown in yellow. Residues Phe148, Tyr160, Ser155, Ser165, and Leu171 are surface exposed, and Phe154 is completely buried. The diagram was generated with the MOLSCRIPT program (26).
FIG. 4.
Surface model of the SCR-3 of human DAF. The surface is shown in the white layer. The mutated residues are shown in different colors. Amino acids Ser155 and Ser165 (shown in red) may affect binding of Dr adhesin. Additional Dr adhesin-binding residues that decrease binding are shown in blue. These include amino acids Gly159, Tyr160, and Leu162. Shown in green are amino acids Phe148 and Phe154, which are required for complement regulation by DAF. Phe154 has minimum surface exposure, and its side chain is completely buried in the model. Phe148 is critical for binding of MAb IH4. Cys156 (shown in yellow) is located next to Ser155, and its side chain is completely buried to form the disulfide bond with Cys186. Amino acid Leu171 (shown in purple), along with Phe148 and Ser155, is part of an epitope for the binding of IH4. Note that residues affecting the tested functions were located on the same loop (171 to 148) of SCR-3. The figure was prepared with the MOLMOL program (24).
TABLE 3.
Summary of the model of the SCR-3 domain of human DAFa
| Parameter | Value |
|---|---|
| Modeling details | |
| Template used | 1E5G.pdb |
| Residues modeled | 129-186 |
| Sequence identity | 47% |
| Disulfide bridges | 2 |
| No. of residues | 58 |
| No. of distance constraints | 7,075 |
| Analysis of model | |
| Backbone RMSD to the template | 0.74 |
| Final conformational energy | −16.7 kcal/mol |
| Residues in the disallowed region in | |
| Ramachandran plot | 0 |
| Total apolar surface area/energy of | |
| the model | 2,585 Å2/kcal |
| Total area/energy of the model | 3,869 Å2/kcal |
This model was generated using the EXDIS, DIAMOD, and FANTOM programs.
DISCUSSION
This study was conducted to enhance the understanding of the structure-function relationship of DAF and to define the importance of the SCR-3 of DAF in the binding of Dr adhesin and the regulation of complement. Twelve mutations of human DAF were constructed and expressed in CHO cells: Phe123-Ala, Phe148-Ala, Phe154-Ala, Ser155-Ala, Cys156-Ala, Gly159-Ala, Tyr160-Ala, Leu162-Ala, Phe163-Ala, Ser165-Ala, Ser165-Leu, and Leu171-Ala. These mutations were predominantly in close proximity to Ser165. Within this group, Ser165-Leu is a rare DAF polymorphism that binds to neither Dr adhesin nor MAb IH4 (40). The stably transfected CHO cell clones expressing mutated DAF were used in three functional assays including binding of E. coli expressing Dr adhesin, complement protection, and binding of MAb IH4 (Table 4). We found that Ser155, and not Ser165, is perhaps the key residue of the SCR-3 surface that binds Dr adhesin; furthermore, distinct regions of SCR-3 in DAF, approximately 20 Å apart, are involved in Dr adhesin binding and complement regulation.
TABLE 4.
Residues affecting binding of mAb IH4, E. coli Dr adhesin and complement protectiona
| Mutation | MAb IH4 | Dr adhesin binding | Protective function |
|---|---|---|---|
| Phe123-Ala | 0 | 0 | I |
| Phe148-Ala | I | 0 | I |
| Phe154-Ala | 0 | 0 | I |
| Ser155-Ala | I | I | 0 |
| Cys156-Ala | I | I | I |
| Gly159-Ala | 0 | [i] | 0 |
| Tyr160-Ala | 0 | [i] | 0* |
| Leu162-Ala | 0 | [i] | 0 |
| Phe163-Ala | 0 | 0 | 0* |
| Ser165-Ala | 0 | 0 | [i] |
| Ser165-Leu | I | I | 0 |
| Leu171-Ala | I | 0 | 0 |
I = mutation completely blocks the binding of MAb IH4 or of Dr adhesin and significantly decreases DAF protective function; [i] = mutation with an intermediary decreasing effect; 0 = mutation does not affect these functions; 0* = mutation has an increased effect on DAF protective function.
DAF modeling.
We have successfully applied our modeling procedure to predict the 3-D structure of the measles virus receptor CD46 (8, 38) and other molecules (14, 50, 53, 56). The recent experimental structure of the central CCP module pair of the poxvirus complement inhibitor protein (3) has a higher degree of template identity (47%) to DAF than the previously used template, human factor H, in modeling of DAF (27). Our model was found to be congruent with the previously published molecular model of DAF showing SCR-3 to be shaped like an ellipsoid measuring 40 by 20 by 20 Å (27). Modeling the individual domain may discount the interactions and/or contribution of the adjacent SCRs to ligand binding but perhaps not direct receptor-ligand interactions. However, one cannot distinguish this possibility until the 3-D structure of DAF in either the unliganded or ligand-bound form is known.
Complement function.
The previous DAF model, based on factor H (27), suggested potential ligand-binding grooves and cavities involving a complement regulatory function of DAF. The SCR 2-3 cavity harboring three lysines—126, 127, and 128—and Leu147, in conjunction with Phe148 at the opening of the cavity, was indicated to be important for binding complement C3b (6, 27, 28). Based on cytoprotection assay results, we pinpointed that the exposed Phe123 of SCR-2 and Phe148 of SCR-3 are important for DAF-mediated complement protection. Our findings supporting the role of Phe123 and Phe148 are further strengthened by two observations: (i) binding of MAb IH4 that recognizes DAF and blocks its complement-protective function is also abolished by mutation of Phe148, and (ii) phenylalanine-containing peptides (25) and rosmarinic acid (which contains two aromatic rings) (46) inhibit complement, possibly by binding to C3b. We therefore propose that phenylalanine-containing peptides and rosmarinic acid conduct their inhibitory actions by binding to C3b via their correctly positioned aromatic groups that mimic the two closely positioned phenylalanines, 123 and 148, of DAF.
Our results show that another phenylalanine, Phe154, of the SCR-3 cavity (27, 28) may also be important in function. Our model (Fig. 4) clearly demonstrates that Phe154 is well buried and forms the core of SCR-3. Thus, we expect any mutations of Phe154 to have a wider influence on function. Surprisingly, a Phe154-Ala mutation disrupted only the complement regulatory function and did not have an effect on the binding of Dr adhesin or MAb IH4. The effect of a Phe154-Ala mutation on DAF regulatory function could be the result of a change in conformation of SCR-2 and SCR-3 with respect to each other. This change would prevent the binding of complement convertase to Phe123 and Phe148 and inhibit the complement regulatory function of DAF.
Mutation Cys156-Ala, however, potentially destroys the secondary structure of DAF by disrupting one of the eight disulfide bridges that support the secondary structure of DAF. Our results show that mutation Cys156-Ala completely abolishes binding of Dr adhesin and MAb IH4 to DAF and significantly affects DAF-mediated complement protection. These results suggest that the epitopes critical for the functions of SCR-3 are conformational.
Comparison of our DAF cytoprotection results with those recently published by Kuttner-Kondo et al. (28) shows that several mutations reported in both studies have a comparable impact on DAF complement regulatory function. However, the two systems differed, particularly in that we used membrane-bound DAF whereas Kuttner-Kondo et al. primarily used soluble yeast-expressed DAF in their cytoprotection assay. Nonetheless, our results for the key mutations affecting complement function, Phe123-Ala and Phe148-Ala, are similar to those reported by Kuttner-Kondo and colleagues. Also, the effect of Ser155-Ala and Ser165-Ala mutation on complement regulatory function falls in the same range of activity as that reported by Kuttner-Kondo et al. Similarly, the mutations Tyr160-Ala and Phe163-Ala had a positive effect on DAF function as reported by Kuttner-Kondo et al. Some mutants appear to differ, including Phe154-Ala and Leu171-Ala; these differences may require additional studies for clarification. We also report three new mutants (Cys156Ala, Gly159-Ala, and Leu162-Ala) that were not tested by the Kuttner-Kondo group; therefore, comparison is not possible at this time.
Binding of Dr adhesin.
We characterized the residues of DAF involved in the binding of Dr adhesin. Mutation Ser155-Ala completely inhibited binding of Dr adhesin and IH4 antibody. This suggests that IH4 blocks the binding of Dr adhesin by sharing a common amino acid in their binding epitopes. Surprisingly, the mutation Ser165-Ala does not abolish binding of Dr adhesin or IH4. This finding contradicts published results on a natural mutant of DAF, Ser165-Leu, which is defective in the binding of both Dr adhesin and IH4. A possible explanation could be that leucine's larger size could sterically hinder the binding of Dr adhesin or IH4 to their respective sites. Alanine is a neutral and small amino acid, almost the same size as serine, but it does not provide a physical or conformational challenge to inhibit the binding of Dr adhesin or IH4.
We also suggest that Ser155 is perhaps the key interacting amino acid in the binding surface for Dr adhesin. Our results demonstrated that mutations of C156, F148, S155, and L171 to alanine abolished binding of IH4 MAb. Accumulated data on these amino acids further indicated that mutation C156A abolished both Dr adhesin and complement binding, and mutation S155A inhibited binding of Dr adhesin only, while mutation F148A resulted in inhibition of complement binding only. Therefore, we propose that in the wild-type DAF, a large conformation of residues C156, F148, S155, and L177 is recognized by MAb IH4 and may accommodate distant regions of SCR-3. This IH4-SCR-3-binding region may consist of two distinct contact points, one for Dr adhesin and the other for complement. As noted above, amino acid C156 forms a cysteine bridge with amino acid C186. The cysteine bridge is important for the stability of the entire SCR-3 conformation. Mutation S155A affects Dr adhesin binding but not complement binding, suggesting that S155 could be the key interacting amino acid in the binding surface for Dr adhesin. Similarly, mutation F148A affects complement binding but not Dr adhesin binding, suggesting that F148 may constitute an important residue in the complement-binding site.
The binding assays performed with DAF mutants and E. coli isolates expressing Dr adhesin also enabled us to identify other important Dr adhesin-binding amino acids of DAF (Table 2). These include the hydrophobic residues Gly159, Tyr160, and Leu162, which are shown to be located downward from the adhesin-binding surface (Fig. 4). Dr adhesin may have some hydrophobic contacts with the residues Leu162 and Tyr160 present in the lower part of the binding site when it docks onto DAF. These contacts may not be critical for Dr adhesin specificity in binding to DAF, but they may enhance the binding. The role of these hydrophobic contacts strengthens our previously reported observations that modified tyrosines inhibit binding of Dr adhesin to DAF (40).
The results and the molecular model produced in this study suggest distinct binding motifs for Dr adhesin and complement convertase. The Dr adhesin binding motif at the lower part of SCR-3 seems to be separated by approximately 20 Å from that of the convertase-binding motif located at the upper part of SCR-3 (closer to SCR-2).
Our results suggest that a loop encompassing residues Ser155 through Ser165, recognized by Dr adhesin as the SCR-3-binding epitope, could potentially inhibit binding of Dr adhesin. We also propose that complement-inhibitory agents can be obtained by screening for molecules with aromatic rings positioned at Phe123 and Phe148 in the SCR-3 of DAF.
Acknowledgments
This work was supported by the National Institutes of Health, grant number DK42029 and HD41687-01 (B. Nowicki, principal investigator [PI]), and by the John Sealy Memorial Endowment Fund for Biomedical Research, grant number 2505-01 (B. Nowicki, PI). The modeling part of the work was funded by grants from the Advanced Research Program of the Texas Higher Education Coordinating Board, grant number 004952-0084-1999 (W. Braun, PI) and the Department of Energy, grant number DE-FG-00ER63041 (W. Braun, PI).
We thank Carla Pairett and Mardelle Susman for help in manuscript preparation. For editorial and graphic assistance, we thank the Ob/Gyn Publications director and staff, R.G. McConnell, Kristi Barrett, John Helms, Traci Morris, and Pam Necessary. We also thank Margaret Das for her helpful comments.
Editor: V. J. DiRita
REFERENCES
- 1.Abe, H., W. Braun, T. Noguti, and N. Go. 1984. Rapid calculation of first and second derivatives of conformational energy with respect to dihedral angles for proteins: general recurrent equations. Comput. Chem. 8:239-247. [Google Scholar]
- 2.Ahearn, J. M., and D. T. Fearon. 1989. Structure and function of the complement receptors, CR1 (CD35) and CR2 (CD21). Adv. Immunol. 46:183-219. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Barlow, P. N., A. Steinkasserer, D. G. Norman, B. Kieffer, A. P. Wiles, R. B. Sim, and I. D. Campbell. 1993. Solution structure of a pair of complement modules by nuclear magnetic resonance. J. Mol. Biol. 232:268-284. [DOI] [PubMed] [Google Scholar]
- 5.Bergelson, J. M., J. G. Mohanty, R. L. Crowell, N. F. St. John, D. M. Lublin, and R. W. Finberg. 1995. Coxsackievirus B3 adapted to growth in RD cells binds to decay-accelerating factor (CD55). J. Virol. 69:1903-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brodbeck, W. G., L. Kuttner-Kondo, C. Mold, and M. E. Medof. 2000. Structure/function studies of human decay-accelerating factor. Immunology. 101:104-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brodbeck, W. G., D. Liu, J. Sperry, C. Mold, and M. E. Medof. 1996. Localization of classical and alternative pathway regulatory activity within the decay-accelerating factor. J. Immunol. 156:2528-2533. [PubMed] [Google Scholar]
- 8.Buchholz, C., D. Koller, P. Devaux, C. Mumenthaler, J. Schneider-Schaulies, W. Braun, D. Gerlier, and R. Cattaneo. 1997. Mapping of the primary binding site of measles virus to its receptor CD46. J. Biol. Chem. 272:22072-22079. [DOI] [PubMed] [Google Scholar]
- 9.Caras, I. W., M. A. Davitz, L. Rhee, G. Weddell, D. W. Martin, and V. Nussenzweig. 1987. Cloning of decay-accelerating factor suggests novel use of splicing to generate two proteins. Nature 325:545-549. [DOI] [PubMed] [Google Scholar]
- 10.Casasnovas, J. M., M. Larvie, and T. Stehle. 1999. Crystal structure of two CD46 domains reveals an extended measles virus-binding surface. EMBO J. 18:2911-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarkson, N. A., R. Kaufman, D. M. Lublin, T. Ward, P. A. Pipkin, P. D. Minor, D. J. Evans, and J. W. Almond. 1995. Characterization of the echovirus 7 receptor: domains of CD55 critical for virus binding. J. Virol. 69:5497-5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coyne, K. E., S. E. Hall, S. Thompson, M. A. Arce, T. Kinoshita, T. Fujita, D. J. Anstee, W. Rosse, and D. M. Lublin. 1992. Mapping of epitopes, glycosylation sites, and complement regulatory domains in human decay accelerating factor. J. Immunol. 149:2906-2913. [PubMed] [Google Scholar]
- 13.Fraczkiewicz, R., and W. Braun. 1998. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 19:319-333. [Google Scholar]
- 14.Friesen, R. H. E., R. J. Castellani, J. C. Lee, and W. Braun. 1998. Allostery in rabbit pyruvate kinase: development of a strategy to elucidate the mechanism. Biochemistry 37:15266-15276. [DOI] [PubMed] [Google Scholar]
- 15.Garratty, G. 1995. Blood group antigens as tumor markers, parasitic/bacterial/viral receptors, and their association with immunologically important proteins. Immunol. Investig. 24:213-232. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, C. E., K. Bromek, N. P. Mullin, B. O. Smith, D. Uhrin, and P. N. Barlow. 2001. Solution structure and dynamics of the central CCP module pair of a poxvirus complement control protein. J. Mol. Biol. 307:323-339. [DOI] [PubMed] [Google Scholar]
- 17.Higgins, D. G., A. J. Bleasy, and R. Fuchs. 1992. CLUSTAL W: improved software for multiple sequence alignment. Comput. Appl. Biosci. 8:189-191. [DOI] [PubMed] [Google Scholar]
- 18.Hourcade, D., V. M. Holers, and J. P. Atkinson. 1989. The regulators of complement activation (RCA) gene cluster. Adv. Immunol. 45:381-416. [DOI] [PubMed] [Google Scholar]
- 19.Hourcade, D., M. K. Liszewski, M. Krych-Goldberg, and J. P. Atkinson. 2000. Functional domains, structural variations and pathogen interactions of MCP, DAF and CR1. Immunopharmacology 49:103-116. [DOI] [PubMed] [Google Scholar]
- 20.Iwata, K., T. Seya, H. Ariga, and S. Nagasawa. 1994. Expression of a hybrid complement regulatory protein, membrane cofactor protein decay accelerating factor on Chinese hamster ovary. Comparison of its regulatory effect with those of decay accelerating factor and membrane cofactor protein. J. Immunol. 152:3436-3444. [PubMed] [Google Scholar]
- 21.Jurianz, K., S. Ziegler, H. Garcia-Schuler, S. Kraus, O. Bohana-Kashtan, Z. Fishelson, and M. Kirschfink. 1999. Complement resistance of tumor cells: basal and induced mechanisms. Mol. Immunol. 36:929-939. [DOI] [PubMed] [Google Scholar]
- 22.Karnauchow, T. M., D. L. Tolson, B. A. Harrison, E. Altman, D. M. Lublin, and K. Dimock. 1996. The HeLa cell receptor for enterovirus 70 is decay-accelerating factor (CD55). J. Virol. 70:5143-5152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kinoshita, T., M. E. Medof, R. Silber, and V. Nussenzweig. 1985. Distributions of decay-accelerating factor in the peripheral blood of normal individuals and patients with paroxysmal nocturnal hemoglobinuria. J. Exp. Med. 162:75-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koradi, R., M. Billeter, and K. Wuthrich. 1996. MOLMOL: a program for display and analysis of macromolecular structure. J. Mol. Graph. 14:51-53. [DOI] [PubMed] [Google Scholar]
- 25.Kossorowtow, A., W. Opitz, E. Etschenberg, and U. Hadding. 1977. Studies on C3 convertases. Inhibition of C5 convertase formation by peptides containing aromatic amino acids. Biochem. J. 167:377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kraulis, P. J. 1991. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 24:946-950. [Google Scholar]
- 27.Kuttner-Kondo, L., M. E. Medof, W. Brodbeck, and M. Shoham. 1996. Molecular modeling and mechanism of action of human decay-accelerating factor. Protein Eng. 9:1143-1149. [DOI] [PubMed] [Google Scholar]
- 28.Kuttner-Kondo, L., L. Mitchell, D. E. Hourcade, and M. E. Medof. 2001. Characterization of the active sites in decay-accelerating factor. J. Immunol. 167:2164-2171. [DOI] [PubMed] [Google Scholar]
- 29.Lindahl, G., U. Sjobring, and J. Eohnsson. 2000. Human complement regulators: a major target for pathogenic microorganisms. Curr. Opin. Immunol. 12:44-51. [DOI] [PubMed] [Google Scholar]
- 30.Liszewski, M. K., and J. P. Atkinson. 1996. Membrane cofactor protein (MCP; CD46) isoforms differ in protection against the classical pathway of complement. J. Immunol. 156:4415-4421. [PubMed] [Google Scholar]
- 31.Liszewski, M. K., T. C. Farries, D. M. Lublin, I. A. Rooney, and J. P. Atkinson. 1996. Control of the complement system. Adv. Immunol. 61:201-283. [DOI] [PubMed] [Google Scholar]
- 32.Lublin, D. M., E. S. Thompson, A. M. Green, C. Levene, and M. J. Telen. 1991. Dr(a−) polymorphism of decay accelerating factor: biochemical, functional, and molecular characterization and production of allele-specific transfectants. J. Clin. Investig. 87:1945-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martino, T. A., M. Petric, M. Brown, K. Aitken, C. J. Gauntt, C. D. Richardson, L. H. Chow, and P. P. Liu. 1998. Cardiovirulent coxsackieviruses and the decay-accelerating factor (CD55) receptor. Virology 244:302-314. [DOI] [PubMed] [Google Scholar]
- 34.Medof, M. E., T. Kinoshita, and V. Nussenzweig. 1984. Inhibition of complement activation on the surface of cells after incorporation of decay accelerating factor (DAF) into their membrane. J. Exp. Med. 160:1558-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medof, M. E., E. I. Walter, J. L. Rutgers, D. M. Knowles, and V. Nussenzweig. 1987. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J. Exp. Med. 165:848-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morgan, B. P., and C. L. Harris. 1999. Complement regulatory proteins. Academic Press, San Diego, Calif.
- 37.Mumenthaler, C. H., and W. Braun. 1995. Automated assignment of simulated and experimental NOESY spectra of proteins by feedback filtering and self-correcting distance geometry. J. Mol. Biol. 254:465-480. [DOI] [PubMed] [Google Scholar]
- 38.Mumenthaler, C. H., U. Schneider, C. J. Buchholz, D. Koller, W. Braun, and R. Cattaneo. 1997. A 3D model for the measles virus receptor CD46 based on homology modeling, Monte Carlo simulations and hemagglutinin binding studies. Protein Sci. 6:588-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowicki, B., A. Hart, K. E. Coyne, D. M. Lublin, and S. Nowicki. 1993. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J. Exp. Med. 178:2115-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowicki, B., J. Moulds, R. Hull, and S. Hull. 1988. A hemagglutinin of uropathogenic Escherichia coli recognizes the Dr blood group antigen. Infect. Immun. 56:1057-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nowicki, B., C. Svanborg-Eden, R. Hull, and S. Hull. 1989. Molecular analysis and epidemiology of the Dr hemagglutinin of uropathogenic Escherichia coli. Infect. Immun. 57:446-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham, T., A. Kaul, A. Hart, P. Goluszko, J. Moulds, S. Nowicki, D. M. Lublin, and B. J. Nowicki. 1995. dra-related X adhesins of gestational pyelonephritis-associated Escherichia coli recognize SCR-3 and SCR-4 domains of recombinant decay-accelerating factor. Infect. Immun. 63:1663-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reid, K. B., and A. J. Day. 1989. Structure-function relationships of the complement components. Immunol. Today 10:177-180. [DOI] [PubMed] [Google Scholar]
- 44.Reid, K. B. M., and S. K. A. Law. 1995. Complement, 2nd ed. IRL Press, Oxford, United Kingdom.
- 45.Sahu, A., and J. D. Lambris. 2000. Complement inhibitors: a resurgent concept in anti-inflammatory therapeutics. Immunopharmacology 49:133-148. [DOI] [PubMed] [Google Scholar]
- 46.Sahu, A., N. Rawal, and M. K. Pangburn. 1999. Inhibition of complement by covalent attachment of rosmarinic acid to activated C3b. Biochem. Pharmacol. 57:1439-1446. [DOI] [PubMed] [Google Scholar]
- 47.Sanner, M., A. Widmer, H. Senn, and W. Braun. 1989. GEOM, a new tool for molecular modelling based on distance geometry calculations with NMR data. J. Comput. Aided Mol. Des. 3:195-210. [DOI] [PubMed] [Google Scholar]
- 48.Seya, T. 1995. Human regulator of complement activation (RCA) gene family proteins and their relationship to microbial infection. Microbiol. Immunol. 39:295-305. [DOI] [PubMed] [Google Scholar]
- 49.Schaumann, T., W. Braun, and K. Wuthrich. 1990. The program FANTOM for energy refinement of polypeptides and proteins using a Newton-Raphson minimizer in torsion angle space. Biopolymers 29:679-694. [Google Scholar]
- 50.Schein, C. H., G. T. Nagle, J. S. Page, J. V. Sweedler, Y. Xu, S. D. Painter, and W. Braun. 2001. Aplysia attractin: biophysical characterization and modeling of a water-borne pheromone. Biophys. J. 81:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shafren, D. R., D. J. Dorahy, R. F. Thorne, T. Kinoshita, R. D. Barry, and G. F. Burns. 1998. Antibody binding to individual short consensus repeats of decay-accelerating factor enhances enterovirus cell attachment and infectivity. J. Immunol. 160:2318-2323. [PubMed] [Google Scholar]
- 52.Soman, K. V., C. H. Schein, H. Zhu, and W. Braun. 2001. Homology modeling and simulations of nuclease structures. Methods Mol. Biol. 160:263-286. [DOI] [PubMed] [Google Scholar]
- 53.Soman, K. V., T. Midoro-Horiuti, J. C. Ferreon, R. M. Goldblum, E. G. Brooks, A. Kurosky, W. A. Braun, and C. H. Schein. 2000. Homology modeling and epitope characterization of mountain cedar allergen ‘JUN A 3’ Biophys. J. 79:1601-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Westerlund, B., P. Kuusela, and J. Ristelli. 1989. The 075X adhesin of uropathogenic E. coli is a type IV collagen binding protein. Mol. Microbiol. 3:329-337. [DOI] [PubMed] [Google Scholar]
- 55.Wiles, A. P., G. Shaw, J. Bright, A. Perczel, I. D. Campbell, and P. N. Barlow. 1997. NMR studies of a viral protein that mimics the regulators of complement activation. J. Mol. Biol. 272:253-265. [DOI] [PubMed] [Google Scholar]
- 56.Zhu, H., C. H. Schein, and W. Braun. 1999. Homology modeling and molecular dynamics simulations of PBCV-1 UV glycosylase complexed with DNA containing a thymidine dimer. J. Mol. Model. 5:302-316. [Google Scholar]