Abstract
The internal dynamics of a 20-residue polyalanine helix was investigated by molecular dynamics simulations. Special attention was paid to the collective vibrations of the helix backbone. The stretch and bend vibrations could be assigned unambiguously to oscillations with periods of 1.4 and 4.3 ps, respectively. The influence of the environment on the dynamics of the collective vibrations was studied by coupling the helix to a heat bath and by adding water molecules. In the presence of water, the stretch vibration becomes more strongly dampened, but still exists as a vibration , while the bend vibration becomes overdamped and degenerates into a relaxation process. The results are compared with available experimental data.
Full text
PDF

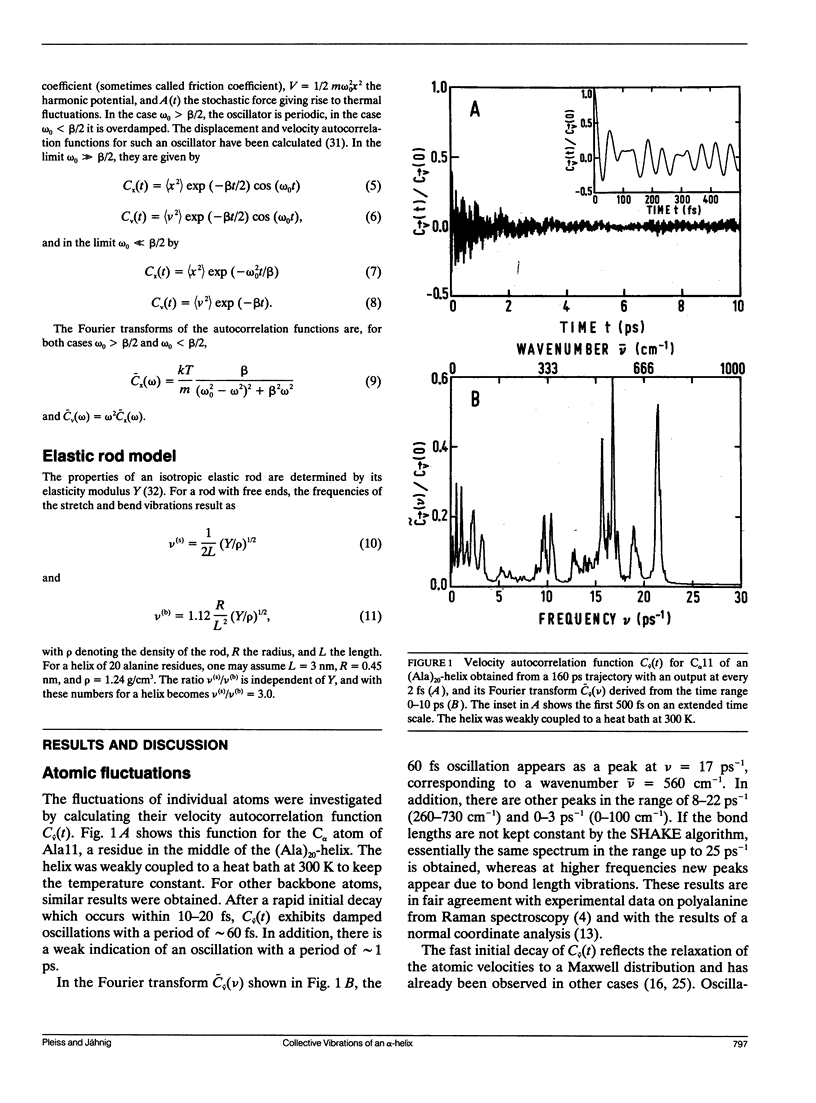




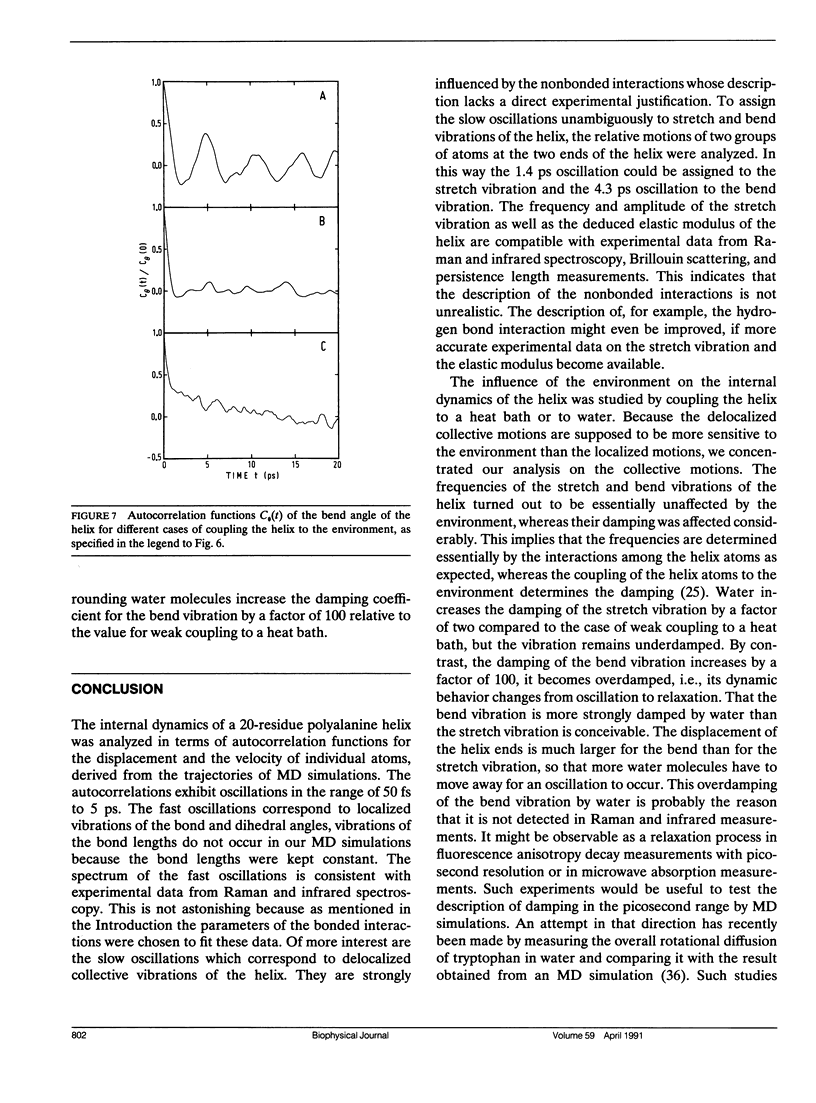


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansari A., Berendzen J., Bowne S. F., Frauenfelder H., Iben I. E., Sauke T. B., Shyamsunder E., Young R. D. Protein states and proteinquakes. Proc Natl Acad Sci U S A. 1985 Aug;82(15):5000–5004. doi: 10.1073/pnas.82.15.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aqvist J., van Gunsteren W. F., Leijonmarck M., Tapia O. A molecular dynamics study of the C-terminal fragment of the L7/L12 ribosomal protein. Secondary structure motion in a 150 picosecond trajectory. J Mol Biol. 1985 Jun 5;183(3):461–477. doi: 10.1016/0022-2836(85)90014-2. [DOI] [PubMed] [Google Scholar]
- Beechem J. M., Brand L. Time-resolved fluorescence of proteins. Annu Rev Biochem. 1985;54:43–71. doi: 10.1146/annurev.bi.54.070185.000355. [DOI] [PubMed] [Google Scholar]
- Brooks C. L., 3rd, Karplus M. Solvent effects on protein motion and protein effects on solvent motion. Dynamics of the active site region of lysozyme. J Mol Biol. 1989 Jul 5;208(1):159–181. doi: 10.1016/0022-2836(89)90093-4. [DOI] [PubMed] [Google Scholar]
- Cusack S., Smith J., Finney J., Tidor B., Karplus M. Inelastic neutron scattering analysis of picosecond internal protein dynamics. Comparison of harmonic theory with experiment. J Mol Biol. 1988 Aug 20;202(4):903–908. doi: 10.1016/0022-2836(88)90566-9. [DOI] [PubMed] [Google Scholar]
- Dornmair K., Jähnig F. Internal dynamics of lactose permease. Proc Natl Acad Sci U S A. 1989 Dec;86(24):9827–9831. doi: 10.1073/pnas.86.24.9827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edholm O., Jähnig F. The structure of a membrane-spanning polypeptide studied by molecular dynamics. Biophys Chem. 1988 Jul 15;30(3):279–292. doi: 10.1016/0301-4622(88)85023-3. [DOI] [PubMed] [Google Scholar]
- Fanconi B., Small E. W., Peticolas W. L. Phonon dispersion curves and normal coordinate analysis of -poly-L-alanine. Biopolymers. 1971;10(8):1277–1298. doi: 10.1002/bip.360100804. [DOI] [PubMed] [Google Scholar]
- Frauenfelder H., Parak F., Young R. D. Conformational substates in proteins. Annu Rev Biophys Biophys Chem. 1988;17:451–479. doi: 10.1146/annurev.bb.17.060188.002315. [DOI] [PubMed] [Google Scholar]
- Genzel L., Kremer F., Poglitsch A., Bechtold G. Relaxation process on a picosecond time scale in hemoglobin and poly(L-alanine) observed by millimeter-wave spectroscopy. Biopolymers. 1983 Jul;22(7):1715–1729. doi: 10.1002/bip.360220708. [DOI] [PubMed] [Google Scholar]
- Ghosh I., McCammon J. A. Sidechain rotational isomerization in proteins. Dynamic simulation with solvent surroundings. Biophys J. 1987 Apr;51(4):637–641. doi: 10.1016/S0006-3495(87)83388-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. H. The dielectric constant of a folded protein. Biopolymers. 1986 Nov;25(11):2097–2119. doi: 10.1002/bip.360251106. [DOI] [PubMed] [Google Scholar]
- Go N., Noguti T., Nishikawa T. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc Natl Acad Sci U S A. 1983 Jun;80(12):3696–3700. doi: 10.1073/pnas.80.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley R., James D., Miller A., White J. W. Phonons and the elastic moduli of collagen and muscle. Nature. 1977 May 19;267(5608):285–287. doi: 10.1038/267285a0. [DOI] [PubMed] [Google Scholar]
- Hofmann H., Voss T., Kühn K., Engel J. Localization of flexible sites in thread-like molecules from electron micrographs. Comparison of interstitial, basement membrane and intima collagens. J Mol Biol. 1984 Jan 25;172(3):325–343. doi: 10.1016/s0022-2836(84)80029-7. [DOI] [PubMed] [Google Scholar]
- Ito K., Shimanouchi T. Vibrational frequencies and modes of alpha-helix. Biopolymers. 1970;9(4):383–399. doi: 10.1002/bip.1970.360090402. [DOI] [PubMed] [Google Scholar]
- Karplus M., McCammon J. A. Dynamics of proteins: elements and function. Annu Rev Biochem. 1983;52:263–300. doi: 10.1146/annurev.bi.52.070183.001403. [DOI] [PubMed] [Google Scholar]
- Levitt M. Protein conformation, dynamics, and folding by computer simulation. Annu Rev Biophys Bioeng. 1982;11:251–271. doi: 10.1146/annurev.bb.11.060182.001343. [DOI] [PubMed] [Google Scholar]
- Levy R. M., Perahia D., Karplus M. Molecular dynamics of an alpha-helical polypeptide: Temperature dependence and deviation from harmonic behavior. Proc Natl Acad Sci U S A. 1982 Feb;79(4):1346–1350. doi: 10.1073/pnas.79.4.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marqusee S., Robbins V. H., Baldwin R. L. Unusually stable helix formation in short alanine-based peptides. Proc Natl Acad Sci U S A. 1989 Jul;86(14):5286–5290. doi: 10.1073/pnas.86.14.5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler W., Brünger A. T., Schulten K., Karplus M. Molecular and stochastic dynamics of proteins. Proc Natl Acad Sci U S A. 1987 Nov;84(22):7933–7937. doi: 10.1073/pnas.84.22.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parak F., Knapp E. W. A consistent picture of protein dynamics. Proc Natl Acad Sci U S A. 1984 Nov;81(22):7088–7092. doi: 10.1073/pnas.81.22.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parak F., Reinisch L. Mössbauer effect in the study of structure dynamics. Methods Enzymol. 1986;131:568–607. doi: 10.1016/0076-6879(86)31055-3. [DOI] [PubMed] [Google Scholar]
- Russell S. T., Warshel A. Calculations of electrostatic energies in proteins. The energetics of ionized groups in bovine pancreatic trypsin inhibitor. J Mol Biol. 1985 Sep 20;185(2):389–404. doi: 10.1016/0022-2836(85)90411-5. [DOI] [PubMed] [Google Scholar]
- Swaminathan S., Ichiye T., van Gunsteren W., Karplus M. Time dependence of atomic fluctuations in proteins: analysis of local and collective motions in bovine pancreatic trypsin inhibitor. Biochemistry. 1982 Oct 12;21(21):5230–5241. doi: 10.1021/bi00264a019. [DOI] [PubMed] [Google Scholar]


