Abstract
Bacteria frequently have multiple mechanisms for acquiring iron, an essential micronutrient, from the environment. We have identified a four-gene Streptococcus pneumoniae operon, named pit, encoding proteins with similarity to components of a putative Brachyspira hyodysenteriae iron uptake ABC transporter, Bit. An S. pneumoniae strain containing a defined mutation in pit has impaired growth in medium containing the iron chelator ethylenediamine di-o-hydroxyphenylacetic acid, reduced sensitivity to the iron-dependent antibiotic streptonigrin, and impaired virulence in a mouse model of S. pneumoniae systemic infection. Furthermore, addition of a mutation in pit to a strain containing mutations in the two previously described S. pneumoniae iron uptake ABC transporters, piu and pia, resulted in a strain with impaired growth in two types of iron-deficient medium, a high degree of resistance to streptonigrin, and a reduced rate of iron uptake. Comparison of the susceptibilities to streptonigrin of the individual pit, piu, and pia mutant strains and comparison of the growth in iron-deficient medium and virulence of single and double mutant strains suggest that pia is the dominant iron transporter during in vitro and in vivo growth.
Recent genetic screens for virulence determinants of bacterial pathogens have emphasized the importance of nutrient acquisition during bacterial growth in vivo. Many of the genes identified by these screens encode components of nutrient transporters or biosynthetic pathways of general metabolism, indicating that essential biosynthetic components and nutrients are often restricted in availability to microorganisms in mammals (25, 29, 33). A detailed understanding of the nutritional requirements of bacterial pathogens during growth in vivo is providing a better understanding of the physiological stresses placed upon them during the course of infection and should identify potential targets for novel antibiotic treatments or vaccines (6). Iron is one such nutrient which is essential for the growth of most bacteria but whose restricted availability within the host forms a nutritional barrier to infection (47). As a consequence many bacterial pathogens contain specialized iron uptake mechanisms to acquire iron from iron-containing mammalian proteins such as transferrin, hemin, and ferritin, either by direct binding of the iron source to the bacterial surface or through secreted low-molecular-weight, high-affinity iron scavengers called siderophores (9, 37, 46). Multiple and often partially redundant iron acquisition mechanisms are frequently present within a single pathogen, emphasizing the importance of iron acquisition for bacterial growth (1, 2, 5, 16, 37). Although pathogenic bacteria utilize a variety of environmental iron sources, frequently specific iron uptake ABC transporters transport the iron moiety into the cytosol across the membrane of gram-positive bacteria (7, 12) and the inner membrane of gram-negative bacteria (15, 48). A role during in vivo growth has been confirmed for numerous iron transporters of gram-negative pathogenic bacteria, including Escherichia coli (42), Salmonella enterica serovar Typhimurium (22), Legionella pneumophila (44), Helicobacter pylori (43), Yersinia pestis (1, 2), and Neisseria species (37). However, only a small number of iron transporters of gram-positive pathogens have been described previously, and there is limited information on their role in virulence (8, 12, 23, 36, 38).
Streptococcus pneumoniae is the commonest cause of bacterial pneumonia and a frequent cause of septicemia and meningitis (26). Growth of S. pneumoniae in iron-restricted medium is known to be supported by Fe2+, Fe3+, and heme-containing compounds but not by transferrin, lactoferrin, or ferritin (5, 40). There is no biochemical or genetic evidence that S. pneumoniae produces siderophores (40, 41). Recently we have described two S. pneumoniae operons, piuBCDA and piaABCD, encoding proteins which have similarity to iron uptake ABC transporters from both gram-negative and gram-positive bacteria (piuBCDA was previously called pit1BCDA, and piaABCD was previously called pit2ABCD) (5, 6). The phenotypes of the piu and pia mutant strains confirmed that they encode iron uptake systems, possibly utilizing hemoglobin as an iron source. Single mutation of piu or pia resulted in a mild or moderate reduction in virulence, respectively. However, a strain containing mutations in both piu and pia was severely attenuated in both pulmonary and systemic models of infection (5), suggesting that the function of at least one protein, Piu or Pia, is required for in vivo growth. To date piu and pia products are the only S. pneumoniae iron transporters which have been characterized genetically. The recently published S. pneumoniae genome (40) revealed the existence of a third ABC transporter operon encoding proteins with similarity to iron uptake transporters. In this paper we characterize this operon, termed pit, and investigate the in vitro and in vivo phenotypes of strains containing a defined pit mutation. By comparing the in vitro and in vivo phenotypes of strains containing mutations in each S. pneumoniae iron uptake ABC transporter individually or in combination, we demonstrate that the pia product is probably the dominant S. pneumoniae iron transporter during both in vitro and in vivo growth.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
S. pneumoniae strains used for this work are listed in Table 1. All mutant strains are derived from a capsular serotype 3 S. pneumoniae strain, 0100993, isolated from a patient with pneumonia and obtained from SmithKline Beecham plc. Ten S. pneumoniae clinical isolates (representing serotypes 2, 4, 7F, 17, 18C, 19A, 19F, 20, and 22) were obtained from J. Paton for PCR analysis of the distribution of pitA. S. pneumoniae strains were cultured at 37°C and 5% CO2 on Columbia agar supplemented with 5% horse blood, in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY), or with a previously described modified version of RPMI medium, RPMIm (5, 8). THY medium was depleted of cations by continuously agitating THY containing 2% Chelex-100 (Bio-Rad) for 8 h, followed by filter sterilization to remove the Chelex-100 and supplementation with 100 μM CaCl2 and 2 mM MgSO4. The iron content of 1 ml of medium was measured in parts per million with flame atomic absorption spectroscopy (performed by Stephen Bowyer, University of North London, using a Varian SpectrAA 220 spectrometer). When necessary, the following supplements were added to medium: chloramphenicol at 4 μg ml−1, erythromycin at 0.4 μg ml−1, kanamycin at 200 μg ml−1, 10 to 50 μM FeCl3, and the cation chelators 200 μM ethylenediamine di-o-hydroxyphenylacetic acid (EDDA; Sigma) and 400 μM 2,2′-dipyridyl (DIP; Sigma). Data for growth curves were collected either by measuring optical density at 580 nm (OD580) of 1-ml cultures grown in sterilized 1.5-ml cuvettes at 1-h intervals or by using 96-well microtiter dishes (200 μl of culture per well) incubated at 37°C with no added CO2 in a Multiskan Ascent instrument (Labsystems) which had been programmed to measure the OD540 at 1-h intervals. To minimize Fe contamination, stock solutions were made with MilliQ-purified water and disposable plasticware was used for all culture conditions. Strains were stored at −70°C as aliquots of THY broth culture (OD580 of 0.3 to 0.4) containing 10% glycerol. Plasmids were amplified in E. coli strain DH5α, grown at 37°C on Luria-Bertani medium with appropriate selection (35).
TABLE 1.
Strains, plasmids, and primers constructed or used in this study
| Strain plasmid, or primer | Description or sequence | Reference |
|---|---|---|
| Strains | ||
| 0100993 | Serotype III clinical isolate | 25 |
| pitA mutant | 0100993 containing an insertion in pitA made with plasmid pPC16; Cmr | This study |
| piaA mutant | 0100993 containing an insertion in piaA; Cmr | 5 |
| piuB mutant | 0100993 containing an insertion in piuB; Cmr | 5 |
| piuB/piaA mutant | 0100993 containing an insertion in piuB and in piaA; Eryr Cmr | 5 |
| piuB/piaA/pitA mutant | piuB/piaA containing an insertion in pitA made with plasmid pPC32; Eryr Cmr Kanr | This study |
| Plasmids | ||
| pID701 | Disruption vector for S. pneumoniae derived from pEVP3; Ampr Cmr | 25 |
| pucMUT | Disruption vector for S. pneumoniae derived from pUC18 (gift from S. Sriskandan); Kanr | 5 |
| pPC16 | pID701 with IT3.1/IT3.2 PCR product ligated into the XbaI site; Ampr Cmr | This study |
| pPC32 | pucMUT with IT3.5/IT3.6 PCR product ligated into the XbaI site; Ampr Cmr | This study |
| Primers (5′ to 3′) | ||
| IT3.1 | GCT CTA GAC TCC TTA TAC ACT AGA TGG | |
| IT3.2 | CGC TCT AGA CCT TAA TGT TAG CTC CGT C | |
| IT3.5 | GGG GTA CCT CCT TAT ACA CTA GAT GG | |
| IT3.6 | CGG GGT ACC TTA ATG TTA GCT CCG TC | |
| IT3.10 | GGA ATT CCA TAT GAA AAA GAA ATG GAT GTA TTA TG | |
| IT3.11 | GGA ATT CCG TAG TTA CCG AGA GCT AGG AC | |
| Sit3.1 | TGA CGG AGC TAA CAT TAA GG | |
| Sit3.2 | AAC GTT GTC TCG GAC AGT CA | |
| Sit3.5 | ATG TCA GCT CCT TTC GTA GG | |
| Sit3.6 | CAA TTG GAT GGA GCA GAT TC | |
| Sit3.7 | TGA TTA CAG GGA CTG CTT TC | |
| Sit3.8 | GGA AGA TCA AGG TGG AGG TA | |
| Sit3.9 | GGA TGG AAT CAA CGC ACT TC | |
| Sit3.10 | TGC TCC AAT TAA GCC CTC TG | |
| Sit3.14 | GAG ATA GCG TCT ATC TTG G | |
| Sit3.15 | CCC AGG AGT AGG CTC CTA C |
Nucleic acid isolation manipulations and analysis.
Nucleic acids were isolated with the indicated kits: S. pneumoniae chromosomal DNA, Wizard genomic DNA isolation kits (Promega); plasmid DNA from E. coli, Qiagen plasmid kits (Qiagen); and S. pneumoniae RNA, SV Total RNA Isolation System (Promega). Prepared RNA samples were protected from degradation by addition of 0.5% RNasin (Promega) and storage as single-use aliquots at −70°C. The Access RT-PCR System (Promega) and target-specific primers were used to derive and amplify cDNA from RNA. The primer concentration for reverse transcription-PCR (RT-PCR) used for assessing operon structure was 400 pmol, and that for assessing the relative abundance of gene transcripts was 80 pmol. Cloning, transformation, restriction digests, and ligations of plasmid DNA were performed according to standard protocols (35). Nylon membranes for Southern hybridizations were prepared and probed with [32P]dCTP-labeled probes made with the RediPrime random primer labeling kit (Amersham International Ltd.) according to standard protocols. S. pneumoniae sequence data were obtained from The Institute for Genomic Research (TIGR) website (http://www.tigr.org) and analyzed and manipulated with the programs MacVector (International Biotechnologies, Inc.) and Artemis3 (Genome Research Ltd.). Sequence similarity searches of the available nucleotide and protein databases (including unfinished microbial genomes) were performed with the BLAST program, available at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov/blast/), and alignments of pairs of sequences were performed with the BLAST 2 service available on the World Wide Web at www.ncbi.nlm.nih.gov/blast/bl2seq/bl2.html. S. pneumoniae loci involved in iron transport were identified by using the Pedant website (http://pedant.mips.biochem.mpg.de/) and the search item “iron.” RNA secondary structure was analyzed with the program Mfold 3.1 (http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi) (28). Clustal X was used for sequence alignment and phylogenetic analysis applying default parameters (altered gap penalties were not applied), except for the pairwise and multiple alignments where protein weight matrices of the Blosum series were used. Gaps in the alignment were not omitted. The cladogram was built by neighbor joining with the distance matrix generated by Clustal X and was represented with the program TreeViewPPC 1.6.2 available at http://taxonomy.zoology.gla.ac.uk/rod/rod.html. The reliability of each node was established by bootstrap methods. Nucleotide sequences were obtained with Applied Biosystems Dye Terminator Chemistry, and cycle sequencing was performed by the Medical Research Council DNA Sequencing Service, Hammersmith Hospital, London, United Kingdom.
Construction of pitA strains.
Plasmids, primers, and S. pneumoniae strains constructed and used for this work are described in Table 1. The construction of piuB, piaA, and piuB/piaA mutant strains has been described previously (5, 6). An internal portion of pitA (bp 358 to 697) was amplified by PCR and ligated into the suicide vector pID701 (primers IT3.1 and IT3.2, XbaI site) or pucMUT (primers IT3.5 and IT3.6, KpnI site) to make the pitA disruption vectors pPC16 (chloramphenicol resistance) and pPC32 (kanamycin resistance), respectively. Plasmid insert identities were confirmed by DNA sequencing. S. pneumoniae mutant strains containing disrupted copies of pitA were constructed by insertion-duplication mutagenesis with a previously described transformation protocol and competence-stimulating peptide 1 (19, 25). The pitA single mutant strain was made by transforming S. pneumoniae strain 0100993 with pPC16. Transformation with pPC32 of the piuB, piaA, and piuB/piaA mutant strains created the piuB/pitA and piaA/pitA double disruption mutant strains and the piuB/piaA/pitA triple disruption mutant strain, respectively. Disruption of pitA was confirmed by PCR and Southern hybridization. All mutations were stable after two 8-h growth cycles (each representing approximately 10 rounds of cell division) in THY broth without antibiotic selection.
Streptonigrin sensitivity assays and 55Fe transport assays.
Sensitivity to streptonigrin disks was assessed as previously described (5). The data presented are the means from four different disks and are representative of similar results from at least two experiments. 55Fe transport assays were modified from our previously described protocol (5). Quantities of 6 × 107 CFU from −70°C stocks of S. pneumoniae strains were subjected to iron stress by incubation for 1 h at 37°C in 1 ml of RPMIm. From this bacterial suspension, 300-μl aliquots were added to 400 μl of RPMIm containing 1.0 μCi of 55FeCl3(NEN) ml−1 and incubated at 37°C. After 5, 15, and 30 min 200-μl aliquots were removed and immediately washed by addition of 800 μl of ice-cold RPMI containing 200 μM EDDA and centrifugation at 20,000 × g at 4°C. The bacterial pellet was washed with a further 1 ml of ice-cold RPMI containing 200 μM EDDA and centrifugation and then resuspended in RPMI and added to 5 ml of Optisafe scintillation fluid (Wallac). The radioactivity was counted with a Beckman LS 1801 scintillation counter. The results presented represent the mean for three separate assays for each strain investigated.
In vivo studies using mouse models of S. pneumoniae infection.
Outbred male white mice (strain CD1; Charles River Breeders) weighing from 18 to 22 g were inoculated with defrosted and appropriately diluted (in 0.9% saline) stocks of S. pneumoniae strains. For mixed infections, inocula consisted of approximately equivalent numbers of cells of two strains. For the pneumonia model, mice were anesthetized by inhalation of halothane (Zeneca) and a 40-μl inoculum containing between 5 × 105 and 5 × 106 bacterial CFU was administered intranasally (i.n.). For the systemic model, mice were given a 100-μl inoculum containing 103 bacterial CFU by intraperitoneal (i.p.) injection. Mice were sacrificed after 24 h (i.p. inoculations) or 48 h (i.n. inoculations), and target organs were recovered and homogenized in 0.5 ml of 0.9% saline. Dilutions of the homogenized organs were plated on nonselective and selective medium for calculation of the competitive index (CI), defined as the ratio of mutant to wild-type strain recovered from the mice divided by the ratio of mutant to wild-type strain in the inoculum (3).
Statistical analysis.
Results for ODs, streptonigrin sensitivity, and 55Fe uptake were compared by two-sample unpaired Student's t tests. CIs were compared by two-sided Student's t test.
RESULTS
Identification and sequence analysis of pitABCD.
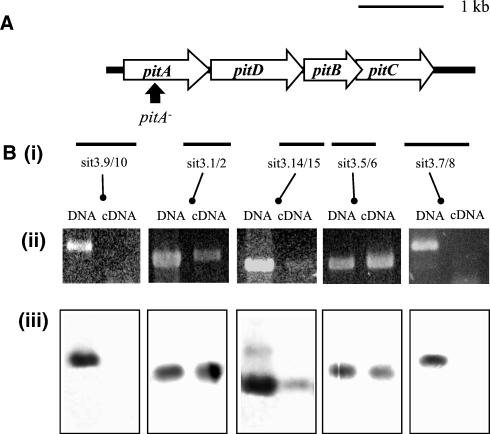
Using the genome analysis and annotation website Pedant (http://pedant.mips.biochem.mpg.de/), we identified three operons, each comprising four genes, encoding probable iron uptake ABC transporters in the genome of the type 4 capsular serotype S. pneumoniae strain sequenced by TIGR. This genome sequence has recently been published, and the annotation describes these three operons as the only genetic loci encoding proteins similar to known bacterial iron transporters (41). Two of these operons, pia (SP1869-1872) and piu (SP1032-1035), encode the two previously described S. pneumoniae iron transporters (5). The third operon we have designated pit (pneumococcal iron transporter), and it is 3,551 bp in length, corresponding to ORFs SP0243 (pitA), SP0242 (pitD), and SP0241 (pitB and pitC) of the TIGR serotype 4 annotated genome (Fig. 1A).
FIG. 1.
(A) Genetic organization of the pit locus. Thick black line, chromosomal DNA; open boxes, pit ORFs (A, putative iron-binding lipoprotein receptor gene; B and C, putative transmembrane genes; D, putative ATPase gene); arrows, site of insertion in mutant strains. (B) Transcriptional analysis of the pit locus. Ethidium bromide-stained agarose gels containing products with the same primer pairs for PCR with S. pneumoniae chromosomal DNA as a template (on the left) and RT-PCR with S. pneumoniae RNA as the template (on the right) are shown in subpanel ii. RT-PCR mixtures containing no reverse transcriptase generated no products. Bars marked in subpanel i represent the corresponding target products for each pair of primers used. Boxes in subpanel iii show the results of Southern hybridizations of the corresponding PCR products when probed with a product representing the whole pit locus.
The published amino acid sequence for the TIGR serotype 4 S. pneumoniae ORF SP0241 is interrupted by a stop codon (TGA) after 141 residues (41), and yet the BLAST alignment of PitB to BitE continues 3′ to this stop codon for a further 47 residues. Hence, in the TIGR strain PitB appears to be translated as a shortened protein, or the stop codon could have been introduced due to a sequencing error. To analyze this further, we determined the nucleotide sequence of a PCR fragment (amplified with primers Sit3.5 and Sit3.6) containing the 3′ end of pitB from the capsular serotype 2 S. pneumoniae strain D39. This sequence contains a G instead of a T as the initial base of the stop codon at residue 141, converting it to a glycine codon. As a result the two smaller ORFs are fused to form a single pitB ORF of 208 amino acid residues. Confirming our result, the recently published sequence of a laboratory strain originally derived from strain D39, R6, also encodes a glycine at this position and has a full-length pitB ORF (21). Finally, PCR with cDNA derived from the serotype 3 S. pneumoniae strain 0100993 used in this work as the template and the primers Sit3.5-Sit3.6 amplified a product spanning the position of the TIGR serotype 4 stop codon at residue 142, confirming that this region is transcribed in strain 0100993 (Fig. 1B).
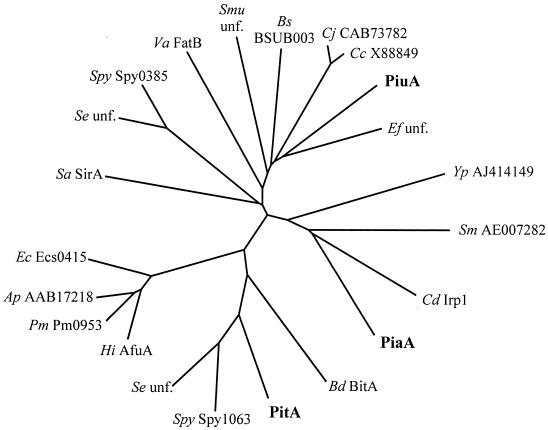
Sequence similarity searches with BLAST of the derived amino acid sequences of the pit operon genes predict that one gene encodes a lipoprotein iron receptor (PitA, 339 residues), one gene encodes an ATPase (PitD, 363 residues), and two genes encode transmembrane permease proteins (PitB, 208 residues, and PitC, 274 residues). The closest homologs for these proteins are putative iron transporter proteins encoded by the bit locus of Brachyspira hyodysenteriae (13) and the product of an ORF present in the recently published Streptococcus pyogenes genome (14) (Table 2). PitA may contain an atypical lipoprotein peptidase cleavage site signal sequence (residues 8 to 12), but this is not recognized by the SignalP bioinformatics program (39); PitD contains Walker A (residues 36 to 47) and B (residues 154 to 161) motifs and the ABC signature characteristic of ATPases (residues 134 to 137) (27), and PitB and PitC both contain sequences matching the permease EAA motif (residues 175 to 194 and 241 to 260, respectively) (24). The pitADBC operon is flanked at the 5′ end by an ORF encoding a protein of unknown function and at the 3′ end by an ORF encoding a protein with 34% identity to a probable pyruvate formate-lyase-activating enzyme of Archaeoglobus fulgidus. In contrast to the pia operon, which is contained within a pathogenicity island (5), the G+C content of the pit operon is 38.2%, similar to the overall G+C content of the S. pneumoniae chromosome (38.9%) (5, 41), and searches of the region adjacent to the pit genes did not identify any genetic mobility genes. PCR with primers IT3.1 and IT3.2 amplified an internal portion of pitA from all S. pneumoniae strains investigated (11 strains representing 10 different serotypes) (data not shown), demonstrating that pit is widely distributed within S. pneumoniae strains. PitA and the two previously described lipoprotein iron receptors of S. pneumoniae, PiuA and PiaA, have no significant similarity when aligned with the BLAST 2 sequence program. Comparison of PiuA, PiaA, and PitA with closely related amino acid sequences (identified by BLAST searches of finished and unfinished microbial genomes) indicates that these three proteins belong to distinct subgroups of lipoprotein iron receptors, as shown by the cladogram presented in Fig. 2. PitA is clustered with proteins predicted from the Streptococcus equi and S. pyogenes genome sequences, as well as BitA of B. hyodysenteriae.
TABLE 2.
Proteins to which PitADBC have close identity and similarity
| Pit protein | Protein name or accession no. | Function | Organism | % Identity/similarity | Length of amino acids compared |
|---|---|---|---|---|---|
| PitA | AE006550 | Putative iron uptake lipoprotein | Streptococcus pyogenes | 52/73 | 323 |
| BitA | Putative iron uptake lipoprotein | B. hyodysenteriae | 42/63 | 306 | |
| BitB | Putative iron uptake lipoprotein | B. hyodysenteriae | 42/60 | 306 | |
| BitC | Putative iron uptake lipoprotein | B. hyodysenteriae | 42/61 | 306 | |
| AfuA | Putative iron uptake lipoprotein | Actinobacillus pleuropneumoniae | 30/46 | 316 | |
| AfuA | Putative iron uptake lipoprotein | Haemophilus influenzae | 28/48 | 284 | |
| PitD | BitD | ATPase | B. hyodysenteriae | 51/68 | 366 |
| MalK | ATPase | Vibrio cholerae | 38/59 | 359 | |
| PotA | ATPase | Borrelia burgdorferi | 42/65 | 294 | |
| PitB | BitE | Transmembrane protein | B. hyodysenteriae | 47/68 | 191 |
| HitB | Transmembrane protein | Haemophilus influenzae | 26/44 | 166 | |
| AfuB | Transmembrane protein | Actinobacillus pleuropneumoniae | 25/42 | 181 | |
| PitC | BitF | Transmembrane protein | B. hyodysenteriae | 48/74 | 189 |
| AfuB | Transmembrane protein | Haemophilus influenzae | 31/51 | 188 | |
| AfuB | Transmembrane protein | Actinobacillus pleuropneumoniae | 31/51 | 188 |
FIG. 2.
Cladogram showing the relationship of PitA, PiuA, and PiaA to each other and their close homologs in the finished and unfinished bacterial genomes (marked by their name if published or accession or annotated genome number if not). unf., unfinished genome data; Ap, Actinobacillus pleuropneumoniae; Bd, B. hyodysenteriae; Bs, Bacillus subtilis; Cc, Campylobacter coli; Cj, Campylobacter jejuni; Cd, C. diphtheriae; Ef, Enterococcus faecalis; Ec, E. coli; Hi, Haemophilus influenzae; Pm, Pasteurella multocida; Se, S. equi; Sa, S. aureus; Sm, Sinorhizobium meliloti; Smu, Streptococcus mutans; Spy, S. pyogenes; Va, Vibrio anguillarum; Yp, Y. pestis.
In order to analyze the function of Pit, strains containing disrupted copies of pitA were constructed by insertional mutagenesis in wild-type and piuB, piaA, and piuB/piaA mutant backgrounds. Strains containing insertions in piuB, piaA, and both piuB and piaA have been described previously (5). Mutations were confirmed by Southern hybridization and by PCR (data not shown).
Transcriptional analysis of pitADBC.
The pitD ORF starts 15 bp after the stop codon of pitA, pitB overlaps pitD by 4 bp, and pitC overlaps pitB by 8 bp, suggesting that pitADB and pitC are transcribed as a single operon. In addition, there is a hairpin loop 216 bp 3′ to pitC (AAAAACAGCCGAAAGGAGTGCCCTCGGCTGTTTTT; ΔG, −17.0 kcal mol−1) but none 3′ to the other pit genes. In order to confirm that pitADBC are transcribed as a single operon, the mRNA structure of the pit genes was analyzed by RT-PCR with combinations of primers designed to amplify across the junctions of the pitADBC ORFs. Control reactions containing heat-killed reverse transcriptase did not generate any products, demonstrating that there was no DNA contamination of the RNA samples. As expected, RT-PCRs across the pitA/pitD, pitD/pitB, and pitB/pitC junctions gave products identical in size to those of the positive control reactions with the same primers and genomic DNA as the template (Fig. 1B). For reasons which are unclear, RT-PCRs with three different pairs of primers designed to amplify the junction of pitD and pitB consistently resulted in only a low yield of product (results for one pair of primers presented in Fig. 1B). However, Southern hybridization with a probe made from a PCR product containing all four pit genes (amplified with primers IT3.10 and IT3.11 and genomic DNA as the template) demonstrated that the RT-PCR products, including those for the pitD/pitB junction, represented amplified portions of the pit transcript (Fig. 1B). RT-PCR with primers which bind 672 bp 5′ or 519 bp 3′ to the pit locus matched with a primer within pitA or pitC, respectively, resulted in no product (Fig. 1B), confirming that the pit genes are not transcriptionally linked to genes 5′ or 3′ to the pit locus. Hence, a phenotype exhibited by an insertional mutation of pitA should be due to disruption of the pit operon alone and should not affect transcription of genes 3′ to the pit locus.
Effect of pitA mutations on growth in iron-deficient medium.
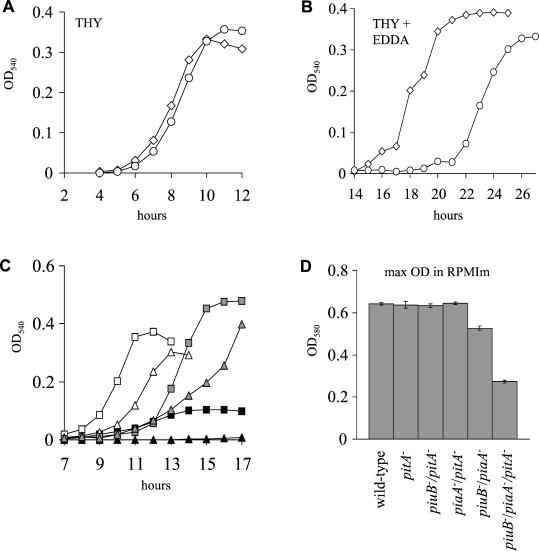
Growth of pitA mutant strains was compared to that of their parent strains (wild type and piuB/piaA mutant) in an undefined complete medium, THY (iron content, 0.78 ppm ± standard deviation [SD] of 0.01), and in three iron-deficient media: Chelex-THY (THY which has been treated with Chelex-100 to remove cations; iron content, 0.17 ppm ± SD of 0.01), THY containing EDDA (a cation-chelating agent with relative specificity for iron), and RPMIm (a defined medium based on RPMI containing no added iron; iron content, 0.30 ppm ± SD of 0.01). As previously described for the piuB and piaA single mutant strains, growth of the pitA mutant strain was not significantly different from that of the wild-type strain in THY, RPMIm, or Chelex-THY (Fig. 3A and D and data not shown). However, growth of the pitA mutant strain in THY containing 200 μM EDDA was delayed and reached a lower maximum OD than did the wild-type strain (maximum OD580 for the wild-type strain was 0.54 ± SD of 0.03 and for the pitA mutant strain was 0.37 ± SD of 0.05, P = 0.02) (Fig. 3B). The impaired growth of the pitA mutant strain relative to the wild-type strain in THY-EDDA medium was partially restored by supplementing the medium with 50 μM FeCl3 (maximum OD580 for the wild-type strain was 0.77 ± SD of 0.04 and for the pitA mutant strain was 0.69 ± SD of 0.04, P = 0.07). The results of growth of the wild-type and pitA mutant strains in THY-EDDA medium when supplemented with MnCl2 or ZnCl2 were variable and inconsistent (data not shown).
FIG. 3.
(A and B) Growth curves as measured by OD of the pitA mutant (circles) and wild-type (diamonds) strains in THY (A) and in THY plus 200 μM EDDA (B). (C) Growth curves as measured by OD of the piuB/piaA (squares) and the piuB/piaA/pitA (triangles) mutant strains in THY (open symbols), Chelex-THY (black symbols), and Chelex-THY-50 μm FeCl3 (grey symbols). Results for panels A to C are representative curves from experiments performed three times. (D) Maximum OD during growth in RPMIm broth for pitA mutant strains and strains containing double mutations in two of the three genes piuB, piaA, and pitA compared to the wild-type strain. Results are the means of three separate samples, and the bars represent SDs. For the difference in maximum OD between the piuB/piaA and the piuB/piaA/pitA mutant strains, P was <0.001.
To investigate further whether pit encodes an iron transporter, we examined the additive effects of mutation of pitA on the in vitro growth phenotype of a strain already containing mutations in both the piu and pia loci. Addition of a mutation in pit to the double mutant piuB/piaA strain increased the growth defect of this strain in the iron-deficient media Chelex-THY and RPMIm (Fig. 3C and D). Growth of the triple mutant strain in Chelex-THY was particularly poor but was partially restored by the addition of FeCl3 to the medium (Fig. 3C). In addition, this strain consistently reached a lower maximum OD580 in the complete medium THY than did the wild-type and piuB/piaA double mutant strains (Fig 3C). The relative contribution of each ABC transporter system to the growth of S. pneumoniae in iron-deficient medium was investigated by comparison of the maximum ODs obtained in RPMIm for the three strains containing each double mutation combination. Of the three double mutation strains, only the growth of the piuB/piaA mutant strain had a lower maximum OD than did the wild-type strain in RPMIm (Fig. 3), indicating that the presence of either a functioning Piu or a functioning Pia is sufficient for growth under these conditions.
Effect of pitA mutations on streptonigrin sensitivity.
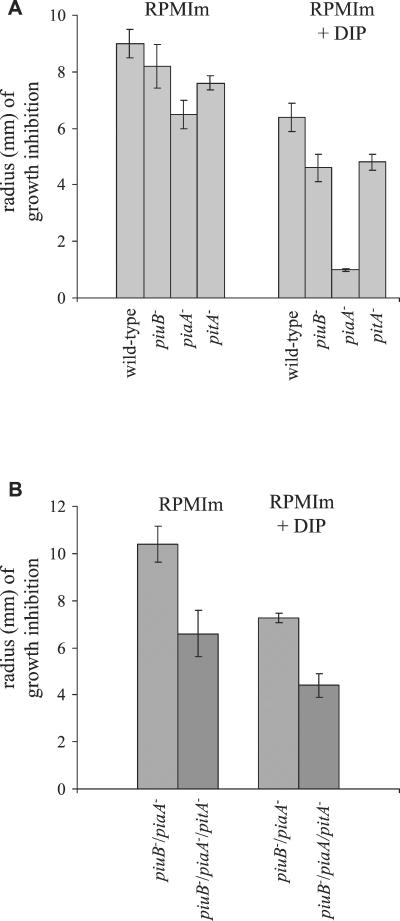
The antibiotic streptonigrin requires intracellular iron for its bactericidal effect. Hence, the degree of susceptibility to streptonigrin is a sensitive measure of intracellular levels of iron and can be used to identify bacterial strains with a reduced ability to acquire environmental iron (4, 34). The sensitivity to streptonigrin of the pitA single mutant strain was compared to those of the wild-type and piuB and piaA mutant strains by measuring the zone of growth inhibition surrounding an antibiotic disk impregnated with 5 μg of streptonigrin on an RPMIm plate with and without supplementation with the iron chelator DIP. The pitA mutant strain had increased resistance to streptonigrin compared to the wild-type strain, indicating that this strain has a lower level of intracellular iron than does the wild-type strain (Fig. 4A). The piuB and pitA mutant strains had similar levels of resistance to streptonigrin, but the piaA mutant strain was markedly more resistant to streptonigrin when grown on RPMIm containing DIP than either the piuB or pitA mutant strains, suggesting that under these conditions Pia is the dominant iron transporter (Fig. 4A). The addition of a mutation in pit to a piuB/piaA mutant strain resulted in increased resistance to high-dose (20-μg) streptonigrin disks, providing further evidence that pit is required for acquisition of intracellular iron by S. pneumoniae (Fig. 4B).
FIG. 4.
Sensitivity to streptonigrin of the pitA mutant strains. Results are expressed as the radius of growth inhibition (millimeters) surrounding an antibiotic disk impregnated with streptonigrin. (A) Comparison of the results with 5-μg streptonigrin disks for the piuB, piaA, and pitA single mutant strains cultured on RPMIm plates and on RPMIm plates containing 400 μM DIP. For the pitA mutant strain versus the wild-type strain, P was <0.001 (on RPMIm) and 0.001 (on RPMIm plus DIP); for the piaA mutant strain versus the piuB or pitA mutant strain, P was <0.05 (on RPMIm) and <0.001 (on RPMIm plus DIP). (B) Comparison of the results with 20-μg streptonigrin disks for the piuB/piaA/pitA triple mutant strain and the piuB/piaA double mutant strain cultured on RPMIm plates and on RPMIm plates containing 400 μM DIP. For the piuB/piaA mutant strain versus the piuB/piaA/pitA mutant strain, P was 0.09 (on RPMIm) and <0.001 (on RPMIm plus DIP). Error bars represent the SDs.
55FeCl3 uptake by pitA mutant strains.
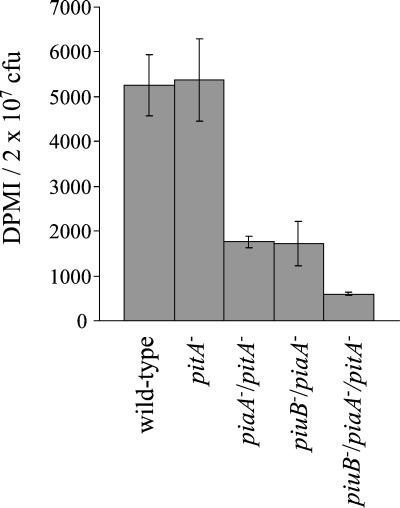
The effects of disruption of the pit locus on iron acquisition were measured directly with an 55FeCl3 uptake assay. After 30 min of incubation in the presence of 55Fe, the accumulation of 55Fe by the pitA single mutant strain was no different from that of the wild-type strain, a result similar to that described for the piuB and piaA single mutant strains (5) (Fig. 5). The piuB/piaA mutant strain has previously been demonstrated to have a strongly reduced rate of 55Fe accumulation compared to that of the wild-type strain (5), and these results were reconfirmed. In addition, the piaA/pitA mutant strain had a much reduced 55Fe content after 30 min compared to the wild-type strain, similar to the 55Fe content of the piuB/piaA mutant strain. The 55Fe content of the triple mutant strain was much reduced compared to those of all the other strains: between 5 and 30 min the 55Fe content of the triple mutant strain increased by only 56%, compared to 544% for the piuB/piaA mutant strain and 1,600% for the wild-type strain (Fig. 5). These results confirm that Pit is required for iron acquisition, at least in the absence of Piu and Pia.
FIG. 5.
Comparison of the 55FeCl3 content after 30 min of incubation for the wild-type, pitA mutant, piaA/pitA and piuB/piaA double mutant, and piuB/piaA/pitA triple mutant strains. Mean data from three assays per strain are presented and normalized for bacterial CFU. For the differences between the wild-type strain and the pitA mutant strain, P was 0.9; for the differences between the wild-type strain and the piaA/pitA, piuB/piaA, and the piuB/piaA/pitA mutant strains, P was ≤0.001; for the differences between the piuB/piaA mutant strain and the piuB/piaA/pitA mutant strain, P was 0.02. Error bars represent the SDs.
Effects of the pitA mutation on virulence.
In order to assess the importance during in vivo growth of Pit, we investigated the virulence of the pitA single mutant strain and the piuB/pitA and piaA/pitA mutant strains by using mixed infections and determining the CI (Table 3). The pitA mutant strain was no less virulent than the wild-type strain for pulmonary infection but was less virulent than the wild-type strain during systemic infection (CI of 0.42), indicating that during in vivo growth iron acquisition by Pit is important for systemic but not intrapulmonary growth (Table 3). This level of attenuation would probably not be detectable by comparison of survival curves for mice given a pure inoculum of the pitA mutant strain or the wild-type strain (5). In contrast to the strain containing mutations in both piuB and piaA, which had CIs of <0.001 in models of systemic infection and pneumonia (5), combining mutations of pitA with those of either piuB or piaA had no additive effect on virulence attenuation during systemic infection (Table 3). The strong attenuation of virulence caused by loss of both piuB and piaA precluded investigation of any additional attenuation conferred by the pitA mutation in the triple mutant strain.
TABLE 3.
Virulence in mice of pitA mutant strains when assessed by mixed infection against the wild-type strain
| Mutant strain used against wild type | Inoculation route | CI (±SD) | n | P value |
|---|---|---|---|---|
| pitA | i.n. | 1.0 (1.1) | 4 | 0.52a |
| pitA | i.p. | 0.42 (0.2) | 5 | 0.001b |
| piuB | i.p. | 1.13 (0.34) | 9 | —c |
| piaA | i.p. | 0.28 (0.11) | 12 | —c |
| piuB/pitA | i.p. | 0.39 (0.1) | 9 | 0.001b |
| piaA/pitA | i.p. | 0.45 (0.2) | 5 | 0.003b |
| piuB/piaA | i.p. | < 0.001 | 4 | —c |
Compared to 1.0, the expected CI for a strain with the same virulence as the wild-type strain.
Compared to 1.2 (SD of 0.2), the CI for a strain containing a mutation in SP1429 (TIGR genome annotation) which is not attenuated after i.p. inoculation.
—, previously reported data (5).
Relative abundance of piu, pia, and pit RNA transcripts.
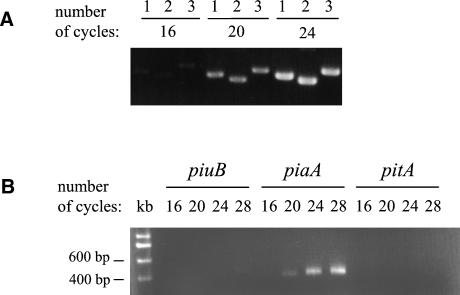
The varying importance of the Piu, Pia, and Pit iron uptake ABC transporters for in vitro and in vivo growth may be due to different levels of expression of their corresponding genes. We therefore assessed the level of expression of the piuB, piaA, and pitA genes (the first genes of each operon) by using RT-PCR and primers for the internal portions of each gene (Smt6.1-Smt6.2, IRP1-IRP2, and IT3.5-IT3.6, respectively) (5). To ensure that PCRs with these primers were of approximately equal efficiency, the quantity of specific PCR product for each primer pair was compared after 12, 16, 20, and 24 PCR cycles with equal quantities of DNA as the template. Each of the three primer pairs resulted in a PCR product of approximately equal intensity when assessed by ethidium bromide staining of an agarose gel (Fig. 6A). However, when cDNA made from RNA extracted from the wild-type strain grown in Chelex-THY to an OD of 0.2 (mid-log growth phase) was used as the target, a strong product was consistently obtained for piaA but only low quantities of products were obtained for piuB and pitA (Fig. 6B). Similar results were obtained with cDNA made from three different RNA preparations extracted from bacteria grown in Chelex-THY and from RNA preparations extracted from wild-type bacteria grown in THY to an OD of 0.5 as the template (data not shown). These results show that pia mRNA transcripts are present at a higher level than are piu and pit mRNA transcripts and provide a partial explanation for the dominance of the Pia iron transporter in the conditions that we have investigated.
FIG. 6.
Relative abundance of piu, pia, and pit mRNA transcripts. (A) Efficiency of PCRs for piuB (lanes 1), piaA (lanes 2), and pitA (lanes 3) with DNA as the template. Product quantity was assessed by running 8 μl of product after 16, 20, and 24 cycles on a 1.5% agarose gel and staining the product with ethidium bromide. (B) PCRs for piuB, piaA, and pitA with cDNA made from RNA obtained from wild-type bacteria grown in Chelex-THY. Product quantity was assessed by running 8 μl of product after 16, 20, 24, and 28 cycles on a 1.5% agarose gel and staining the product with ethidium bromide. The kilobase ladder is presented in the lane marked kb.
DISCUSSION
We have previously described two S. pneumoniae four-gene operons, piu and pia, which encode iron uptake ABC transporters (5). In this paper we have characterized a third four-gene operon, pit, encoding proteins which are similar to components of iron uptake ABC transporters. Redundancy of iron transporters within a species has often hampered investigation of a particular iron transporter's function, with single mutations affecting an iron transporter frequently resulting in no discernible in vitro or in vivo phenotype (5, 16, 31). Hence, it is not surprising that the pitA single mutant strain had no growth defect in RPMIm and Chelex-THY and a similar rate of 55Fe uptake as the wild-type parental strain. However, this mutant strain did have delayed growth in medium containing a cation chelator (possibly a more stringent test of iron transporter function than Chelex-THY medium, as the iron chelator can compete with bacterial iron uptake mechanisms for the available iron during bacterial growth) and increased resistance to the iron-dependent antibiotic streptonigrin, indicating that the pit operon also encodes an iron transporter. Furthermore, addition of a pitA mutation to a strain already carrying mutations in both piu and pia further impaired this strain's ability to grow in two varieties of iron-depleted media, increased its resistance to streptonigrin, and reduced its rate of 55FeCl3 uptake. These results confirm that the S. pneumoniae pit locus is involved in iron acquisition and provide further evidence that the pit homologs in B. hyodysenteriae and S. pyogenes also encode iron transporters (13, 14). Iron uptake transporters, including pia, are frequently encoded on horizontally acquired regions of genomic DNA called either genomic islands or, if important for virulence, pathogenicity islands (5, 17, 18, 22, 45). However, the region of DNA including and surrounding the pit locus has none of the features of a genomic island, such as a G+C content different from the average for the whole genome, the presence of genetic mobility genes, or variable distribution among strains of the same species (18), and there is no evidence to suggest that pit was horizontally acquired by S. pneumoniae.
Multiple iron transporters with different substrates are well described for gram-negative bacteria. For example, the genome of Y. pestis contains seven separate iron uptake operons in addition to those that have been previously reported (1, 2, 16, 32), and Neisseria species have mechanisms for acquiring iron from transferrin and heme compounds as well as producing siderophores (37). The iron acquisition systems of gram-positive pathogens are less well described but include ABC transporters involved in Fe3+ uptake by S. pyogenes (23), siderophore uptake by Staphylococcus aureus (7, 20, 31), and hemin uptake by Corynebacterium diphtheriae (12), siderophores produced by mycobacteria (10, 11), and a transferrin receptor of S. aureus (30). In this and our previous study (5) we have now identified a total of three separate and seemingly unrelated S. pneumoniae iron uptake ABC transporters, piu, pia, and pit. In the published TIGR S. pneumoniae serotype 4 strain genome sequence these three ABC transporters are the only genetic loci with similarity to known iron uptake transporters (41). The reduced ability of the triple mutant strain to grow in a variety of cation-depleted media and its low rate of 55Fe uptake provide additional evidence that these three ABC transporters account for the iron uptake mechanisms of S. pneumoniae. Loss of the majority of its iron uptake transporters might also explain why the piaB/piuA/pitA triple mutant strain has a growth defect even in THY, a medium which is relatively iron replete. However, there may be other S. pneumoniae genes whose products are involved in iron acquisition, either through the Piu, Pia, or Pit transporters (possibly acting as intermediates between the substrate and the ABC transporter lipoprotein) or by a separate mechanism, but which have no homology to known iron uptake proteins.
In general, the relative contribution to growth and for virulence of the various iron transporters present in gram-positive pathogens is unclear. By comparing the phenotypes of the pitA, piuB, and piaA mutant strains, we have been able to assess the relative importance of each S. pneumoniae iron transporter during in vitro and in vivo growth. Three lines of evidence suggest that Piu and especially Pia are the most important S. pneumoniae iron transporters for iron acquisition during both in vitro and in vivo growth: (i) the only double iron transporter mutant strain with impaired growth in RPMIm is the piuB/piaA mutant strain, (ii) in the presence of the chelator DIP the piaA mutant strain has a markedly lower sensitivity to streptonigrin than do the piuB and pitA mutant strains, and (iii) piaA mutant strains have the highest degree of attenuation in virulence in mouse models of nasal and systemic infection (5). Furthermore, the three combinations of double mutations in these genes have strikingly different effects on virulence. Combining a mutation in pit with a mutation in either piu or pia had no additive effect on virulence attenuation when assessed by mixed infection experiments, one of the most sensitive methods available for identifying subtle differences in virulence (3, 5). However, combining disruption of piu with disruption of pia causes a dramatic reduction in S. pneumoniae virulence in mouse models of pneumonia and systemic infection (5). Taken together, these results show that, if either the Piu or Pia iron transporter is present, then Pit has only a relatively small role during growth in vivo, a finding which is consistent with the in vitro growth data. The variable importance of different iron transporters for growth and virulence has previously been described for Y. pestis and is likely to occur in other pathogens containing multiple iron acquisition systems (2, 15, 16). Analysis of the relative abundance of piu, pia, and pit RNA transcripts demonstrated that in vitro there are abundant quantities of pia, but not piu or pit, RNA, hence providing one explanation of why Pia is the dominant iron transporter. Further investigation is required to explain why there are such marked differences in the roles of the S. pneumoniae iron uptake ABC transporters, including identification of their substrates and control of their expression in vivo.
Identification of pit and the demonstration that it encodes an iron transporter provide further evidence that iron acquisition is important for growth in vitro and in vivo of gram-positive as well as gram-negative pathogens. Defining the genes whose products allow S. pneumoniae and other pathogens to acquire micronutrients in vivo will improve our understanding of how microbial pathogens can grow within internal organs and cause disease and may identify potential candidates for new vaccine antigens (6) or novel targets for antimicrobial therapy. Detailed biochemical characterization of Pit, Piu, and Pia function is now required in order to identify their substrates and the exact mechanisms by which iron is transferred into the cell.
Acknowledgments
We are grateful to Stephen Bowyer for performing the flame atomic absorption spectroscopy.
This work was supported by a Wellcome Trust Advanced Research Fellow grant awarded to J. S. Brown.
Editor: E. I. Tuomanen
REFERENCES
- 1.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bearden, S. W., and R. D. Perry. 1999. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol. Microbiol. 32:403-414. [DOI] [PubMed] [Google Scholar]
- 3.Beuzon, C. R., S. Meresse, K. E. Unsworth, J. Ruiz-Albert, S. Garvis, S. R. Waterman, T. A. Ryder, E. Boucrot, and D. W. Holden. 2000. Salmonella maintains the integrity of its intracellular vacuole through the action of SifA. EMBO J. 19:3235-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braun, V., R. Gross, W. Koster, and L. Zimmermann. 1983. Plasmid and chromosomal mutants in the iron(III)-aerobactin transport system of Escherichia coli. Use of streptonigrin for selection. Mol. Gen. Genet. 192:131-139. [DOI] [PubMed] [Google Scholar]
- 5.Brown, J. S., S. M. Gilliland, and D. W. Holden. 2001. A Streptococcus pneumoniae pathogenicity island encoding an ABC transporter involved in iron uptake and virulence. Mol. Microbiol. 40:572-585. [DOI] [PubMed] [Google Scholar]
- 6.Brown, J. S., A. D. Ogunniyi, M. C. Woodrow, D. W. Holden, and J. C. Paton. 2001. Immunization with components of two iron-uptake ABC transporters protects mice against systemic infection with Streptococcus pneumoniae. Infect. Immun. 69:6702-6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cabrera, G., A. Xiong, M. Uebel, V. K. Singh, and R. K. Jayaswal. 2001. Molecular characterization of the iron-hydroxamate uptake system in Staphylococcus aureus. Appl. Environ. Microbiol. 67:1001-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockayne, A., P. J. Hill, N. B. Powell, K. Bishop, C. Sims, and P. Williams. 1998. Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66:3767-3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelissen, C. N., and P. F. Sparling. 1994. Iron piracy: acquisition of transferrin-bound iron by bacterial pathogens. Mol. Microbiol. 14:843-850. [DOI] [PubMed] [Google Scholar]
- 10.De Voss, J. J., K. Rutter, B. G. Schroeder, and C. E. Barry III. 1999. Iron acquisition and metabolism by mycobacteria. J. Bacteriol. 181:4443-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Voss, J. J., K. Rutter, B. G. Schroeder, H. Su, Y. Zhu, and C. E. Barry III. 2000. The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. USA 97:1252-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drazek, E. S., C. A. Hammack, and M. P. Schmitt. 2000. Corynebacterium diphtheriae genes required for acquisition of iron from haemin and haemoglobin are homologous to ABC haemin transporters. Mol. Microbiol. 36:68-84. [DOI] [PubMed] [Google Scholar]
- 13.Dugourd, D., C. Martin, C. R. Rioux, M. Jacques, and J. Harel. 1999. Characterization of a periplasmic ATP-binding cassette iron import system of Brachyspira (Serpulina) hyodysenteriae. J. Bacteriol. 181:6948-6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetherston, J. D., V. J. Bertolino, and R. D. Perry. 1999. YbtP and YbtQ: two ABC transporters required for iron uptake in Yersinia pestis. Mol. Microbiol. 32:289-299. [DOI] [PubMed] [Google Scholar]
- 16.Gong, S., S. W. Bearden, V. A. Geoffroy, J. D. Fetherston, and R. D. Perry. 2001. Characterization of the Yersinia pestis Yfu ABC inorganic iron transport system. Infect. Immun. 69:2829-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guyer, D. M., J. S. Kao, and H. L. Mobley. 1998. Genomic analysis of a pathogenicity island in uropathogenic Escherichia coli CFT073: distribution of homologous sequences among isolates from patients with pyelonephritis, cystitis, and catheter-associated bacteriuria and from fecal samples. Infect. Immun. 66:4411-4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hacker, J., and J. B. Kaper. 2000. Pathogenicity islands and the evolution of microbes. Annu. Rev. Microbiol. 54:641-679. [DOI] [PubMed] [Google Scholar]
- 19.Havarstein, L. S., G. Coomaraswamy, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinrichs, J. H., L. E. Gatlin, C. Kunsch, G. H. Choi, and M. S. Hanson. 1999. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J. Bacteriol. 181:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoskins, J., W. E. Alborn, Jr., J. Arnold, L. C. Blaszczak, S. Burgett, B. S. DeHoff, S. T. Estrem, L. Fritz, D. J. Fu, W. Fuller, C. Geringer, R. Gilmour, J. S. Glass, H. Khoja, A. R. Kraft, R. E. Lagace, D. J. LeBlanc, L. N. Lee, E. J. Lefkowitz, J. Lu, P. Matsushima, S. M. McAhren, M. McHenney, K. McLeaster, C. W. Mundy, T. I. Nicas, F. H. Norris, M. O'Gara, R. B. Peery, G. T. Robertson, P. Rockey, P. M. Sun, M. E. Winkler, Y. Yang, M. Young-Bellido, G. Zhao, C. A. Zook, R. H. Baltz, S. R. Jaskunas, P. R. Rosteck, Jr., P. L. Skatrud, and J. I. Glass. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J. Bacteriol. 183:5709-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Janakiraman, A., and J. M. Slauch. 2000. The putative iron transport system SitABCD encoded on SPI1 is required for full virulence of Salmonella typhimurium. Mol. Microbiol. 35:1146-1155. [DOI] [PubMed] [Google Scholar]
- 23.Janulczyk, R., J. Pallon, and L. Bjorck. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol. Microbiol. 34:596-606. [DOI] [PubMed] [Google Scholar]
- 24.Kerppola, R. E., and G. F. Ames. 1992. Topology of the hydrophobic membrane-bound components of the histidine periplasmic permease. Comparison with other members of the family. J. Biol. Chem. 267:2329-2336. [PubMed] [Google Scholar]
- 25.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic analysis of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 26.Lim, W. S., J. T. Macfarlane, T. C. Boswell, T. G. Harrison, D. Rose, M. Leinonen, and P. Saikku. 2001. Study of community acquired pneumonia aetiology (SCAPA) in adults admitted to hospital: implications for management guidelines. Thorax 56:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Linton, K. J., and C. F. Higgins. 1998. The Escherichia coli ATP-binding cassette (ABC) proteins. Mol. Microbiol. 28:5-13. [DOI] [PubMed] [Google Scholar]
- 28.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 29.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 30.Modun, B., and P. Williams. 1999. The staphylococcal transferrin-binding protein is a cell wall glyceraldehyde-3-phosphate dehydrogenase. Infect. Immun. 67:1086-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68:6281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parkhill, J., B. W. Wren, N. R. Thomson, R. W. Titball, M. T. Holden, M. B. Prentice, M. Sebaihia, K. D. James, C. Churcher, K. L. Mungall, S. Baker, D. Basham, S. D. Bentley, K. Brooks, A. M. Cerdeno-Tarraga, T. Chillingworth, A. Cronin, R. M. Davies, P. Davis, G. Dougan, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, A. V. Karlyshev, S. Leather, S. Moule, P. C. Oyston, M. Quail, K. Rutherford, M. Simmonds, J. Skelton, K. Stevens, S. Whitehead, and B. G. Barrell. 2001. Genome sequence of Yersinia pestis, the causative agent of plague. Nature 413:523-527. [DOI] [PubMed] [Google Scholar]
- 33.Polissi, A., A. Pontiggia, G. Feger, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pope, C. D., W. O'Connell, and N. P. Cianciotto. 1996. Legionella pneumophila mutants that are defective for iron acquisition and assimilation and intracellular infection. Infect. Immun. 64:629-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning. A laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schmitt, M. P., B. G. Talley, and R. K. Holmes. 1997. Characterization of lipoprotein IRP1 from Corynebacterium diphtheriae, which is regulated by the diphtheria toxin repressor (DtxR) and iron. Infect. Immun. 65:5364-5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schryvers, A. B., and I. Stojiljkovic. 1999. Iron acquisition systems in the pathogenic Neisseria. Mol. Microbiol. 32:1117-1123. [DOI] [PubMed] [Google Scholar]
- 38.Sebulsky, M. T., and D. E. Heinrichs. 2001. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutcliffe, I. C., and R. R. Russell. 1995. Lipoproteins of gram-positive bacteria. J. Bacteriol. 177:1123-1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tai, S. S., C. J. Lee, and R. E. Winter. 1993. Hemin utilization is related to virulence of Streptococcus pneumoniae. Infect. Immun. 61:5401-5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 42.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Velayudhan, J., N. J. Hughes, A. A. McColm, J. Bagshaw, C. L. Clayton, S. C. Andrews, and D. J. Kelly. 2000. Iron acquisition and virulence in Helicobacter pylori: a major role for FeoB, a high-affinity ferrous iron transporter. Mol. Microbiol. 37:274-286. [DOI] [PubMed] [Google Scholar]
- 44.Viswanathan, V. K., P. H. Edelstein, C. D. Pope, and N. P. Cianciotto. 2000. The Legionella pneumophila iraAB locus is required for iron assimilation, intracellular infection, and virulence. Infect. Immun. 68:1069-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vokes, S. A., S. A. Reeves, A. G. Torres, and S. M. Payne. 1999. The aerobactin iron transport system genes in Shigella flexneri are present within a pathogenicity island. Mol. Microbiol. 33:63-73. [DOI] [PubMed] [Google Scholar]
- 46.Wandersman, C., and I. Stojiljkovic. 2000. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr. Opin. Microbiol. 3:215-220. [DOI] [PubMed] [Google Scholar]
- 47.Wooldridge, K. G., and P. H. Williams. 1993. Iron uptake mechanisms of pathogenic bacteria. FEMS Microbiol. Rev. 12:325-348. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, D., W. D. Hardt, and J. E. Galan. 1999. Salmonella typhimurium encodes a putative iron transport system within the centisome 63 pathogenicity island. Infect. Immun. 67:1974-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]