Abstract
Enterococci rank among leading causes of nosocomial bacteremia and urinary tract infection and are also a leading cause of community acquired subacute endocarditis. Limited evidence suggests that biological cues in serum and urine may play an important role in modulating enterococcal virulence at sites of infection. To determine the extent to which biological cues affect enterococcal virulence-associated gene expression, we used quantitative real-time PCR to compare mRNA levels in Enterococcus faecalis cultures grown in serum or urine to that achieved in laboratory medium. Both environment- and growth phase-specific variations were observed, demonstrating the occurrence of as-yet-uncharacterized mechanisms for control of gene expression in E. faecalis that may play an important role in vivo.
Enterococci rank among leading causes of nosocomial bacteremia and urinary tract infection (51) and are also a leading cause of community-acquired subacute endocarditis (33). Enterococcal factors that contribute to the pathogenesis of disease have been identified. The cytolysin of Enterococcus faecalis contributes to enterococcal virulence in all models studied (7, 29, 31, 34, 35). It was recently shown to be autoinduced by a quorum-sensing mechanism involving a two-component regulatory system (25). Aggregation substance is an enterococcal surface protein, encoded by several pheromone-responsive plasmids (12), that aids in the formation of mating aggregates during bacterial conjugation. Aggregation substance also mediates adherence to renal epithelial cells (40), internalization (52, 65, 76), and intracellular survival of E. faecalis in eukaryotic cells in vitro (57, 74) and also contributes to cardiac vegetation size in vivo (7, 66). The enterococcal adhesin Esp bears structural similarity to the Rib and C-alpha proteins of group B streptococci (69). In addition to enrichment among clinical isolates of E. faecalis (69) and both vancomycin-resistant (77) and vancomycin-susceptible clinical isolates of Enterococcus faecium (L. Baldassarri, L. Bertuccini, M. G. Ammendolia, G. Gherardi, and R. Creti, Letter, Lancet 357:1802, 2001; N. Woodford, M. Soltani, and K. J. Hardy, Letter, Lancet 358:584, 2001), Esp contributes to the colonization and persistence of E. faecalis during ascending urinary tract infection (68).
Ace is a microbial surface component recognizing adhesive matrix molecule specific to E. faecalis (62). The identification of Ace-specific antibodies in sera collected from patients following enterococcal infection (50) provides evidence that the protein is produced under physiologic conditions, but the contribution of Ace or Ace-specific antibodies to the pathogenesis of infection and conditions that may occur physiologically that regulate its expression are unknown. The endocarditis antigen (EfaA) of E. faecalis bears similarity to adhesins encoded by genes in other streptococci (43). Although the biological role of EfaA and the regulation of its expression are relatively unknown, a potential role of the protein in vivo was demonstrated in a murine model of peritonitis (72). Gelatinase is an extracellular zinc metalloproteinase secreted by E. faecalis that has been shown to potentially contribute to the virulence of E. faecalis in some animal models (13, 18, 73). The production of gelatinase appears to be regulated in a cell-density-dependent manner (49, 55) by the products of fsrA, fsrB, and fsrC, which show similarity to elements of bacterial two-component regulatory systems (56). In a murine peritonitis model, the products of each of these genes contributed to the virulence of E. faecalis in a manner consistent with the contribution of gelatinase (56). Gls24 is a functionally novel general stress protein in E. faecalis that is induced during several types of environmental stress, and its inactivation is associated with alterations in growth, cell morphology, and protein expression during stress (16). The molecular mechanisms of Gls24 and the regulators of its expression are unknown (16).
Although enterococci are leading causes of nosocomial infections of the bloodstream and urinary tract, comparatively little is known of how these environments affect enterococcal gene expression. Urinary tract isolates of E. faecalis show an eightfold increase in adherence to Girardi heart cells following growth in pooled human serum (22). Growth in serum reduces adherence of enterococci to polymorphonuclear cells by two- to fivefold (22). Moreover, growth of either endocarditis or urinary tract isolates of E. faecalis in serum induces the expression of carbohydrate ligands responsible for adhesion to Girardi heart cells (23). Serum is known to induce the expression of aggregation substance (40), but its effect on the expression of other known and suspected enterococcal virulence traits is unexplored. Growth in urine is known to modulate the expression of genes important for uropathogenesis by other organisms (63, 64), but its effect on expression of known and suspected enterococcal virulence factors is unexplored.
The emergence of enterococcal strains with resistance to a wide variety of antimicrobial agents has precipitated a need for new therapeutic strategies for treating enterococcal infection, possibly targeting gene products involved in colonization and disease (48, 70). To determine the extent to which E. faecalis virulence-associated gene expression is influenced by infection-relevant environmental cues, we quantified enterococcal virulence factor-encoding mRNA following growth in serum or urine, and compared it to mRNA abundance in laboratory medium-grown cultures, using quantitative real-time PCR (26-28).
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Laboratory medium 2×YT (composed of yeast extract, tryptone, and sodium chloride) was selected because it contained a minimum of potentially infection-relevant mammalian cues, as occurs for example in brain heart infusion. For growth of E. faecalis in serum, sterile pooled heat-inactivated rabbit serum (Pel Freez Biologicals, Rogers, Ark.) was used as a lot-controlled source. For E. faecalis culture in urine, human urine was collected from healthy volunteers (i.e., without a history of recent or chronic urinary tract infection or antibiotic use) by midstream clean catch (10), pooled, and sterilized by filtration through a 0.2-μm-pore-size membrane.
An E. faecalis clinical isolate, MMH594, which caused multiple infections in a hospital ward outbreak (30), was used to study differential gene expression in each environment. To quantify virulence factor mRNA expression, 10-ml cultures of E. faecalis were grown in each medium for 17 h. To rejuvenate and synchronize cultures to an active mode of growth, and to eliminate potential quorum signals that may have accumulated, 1 ml of culture was centrifuged (2,500 × g for 10 min at 25°C), the supernatant medium was discarded, and the bacterial pellet was resuspended in 40 ml of prewarmed media. Following subculture for 30 min at 37°C without aeration, cells were again collected by centrifugation, the supernatant was discarded, and the pellet was resuspended in 40 ml of fresh prewarmed medium and subcultured for an additional 30 min. Finally, actively growing cells were collected by centrifugation as described above and resuspended in 1 ml of prewarmed medium. Fifty milliliters of prewarmed medium was inoculated with 20 μl of this actively dividing culture and incubated at 37°C without aeration to the desired phase of growth, as determined in preliminary experiments that characterized growth rates in serum, urine, and 2×YT.
RNA extraction and preparation.
Total RNA was isolated as previously described with minor modifications (71). Briefly, E. faecalis cells from 50 ml of serum, urine, or 2×YT were collected by centrifugation (2,500 × g for 2 min at 4°C). The bacterial pellet was washed twice with 5 ml of ice-cold phosphate-buffered saline prior to lysis, which was found to be essential for maximum RNA yield from urine grown cultures. Subsequent steps were essentially as described previously (71), and the purified RNA was resuspended in 200 μl of diethyl pyrocarbonate-treated water.
Because of the sensitivity of real-time PCR, residual contaminating DNA was removed in the following manner. To the 200-μl RNA preparation, 0.25 volume of transcription-optimized buffer containing 200 mM Tris-HCl (pH 7.9), 30 mM MgCl2, 10 mM spermidine, and 50 mM NaCl (Promega, Madison, Wis.) was added. Contaminating DNA was hydrolyzed with 10 U of RQ1 RNase-free DNase (Promega) at 37°C for 15 min. Following DNase treatment, 250 μl of phenol-water (3.75:1, vol/vol; Life Technologies, Grand Island, N.Y.) was added and the RNA was extracted and precipitated as described previously (71) and resuspended in 50 μl of diethyl pyrocarbonate-treated water containing 0.1 mM EDTA. RNA integrity was globally assessed by electrophoresis through 1.2% agarose-0.66 M formaldehyde gel in MOPS running buffer (20 mM MOPS [morpholinepropanesulfonic acid] [pH 7.0], 8 mM sodium acetate, 1 mM EDTA [pH 8.0]) at a power of 3 to 4 V/cm.
Real-time quantitative PCR.
Amplification, detection, and real-time analysis were performed using the ABI Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, Calif.). SYBR Green I (Applied Biosystems) was used for detection of the amplified product. Sequence data for primer design was obtained from both prior investigations in our laboratory and the Entrez database at the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov). Primers designed to produce amplicons of equivalent length (approximately 100 bp), were selected using Primer Express software (Applied Biosystems), and the nucleotide sequences of primers used in these studies are listed in Table 1.
TABLE 1.
Targets and primers used for real-time quantitative PCR
| Target (reference) | Forward primer | Reverse primer |
|---|---|---|
| cylR2 (25) | CCAAAGTGAATTAGCTGCTTTATTAGAA | TTAATGCTAACTGTAAAGAAGGGTTATATTTATT |
| cylR1 (25) | TTTATTTTTTTATTGGATATCATTTCTGTAGTC | TTCGCTCATCTTTTTTTGAATCAG |
| cylLL (17) | CTGTTGCGGCGACAGCT | CCACCAACCCAGCCACAA |
| cylLS (17) | GCTAAATAAGGAAAATCAAGAAAACTATTACTC | CAAAAGAAGGACCAACAAQGTTCTAATT |
| cylM (17) | TCGGACACGGTATATATAGCTATGT | GTTCTTCTTCAAAGTATGATTCTTTAAT |
| cylB (17) | GAAAAGATTGAAGTACGTTGCG | TTCTACTAGTGTACTTTGATTACCATAATAATT |
| cylA (17) | GGTTATGCATCAGATCTCTCAA | CTGTATATAATCTACTTTTTCAGAAGATAATTC |
| cylI (8) | ACTTCTCGTAATCTTTACTCTTGTTTTTG | CTAAGTGCCCTAACTCATGTACAA |
| gls24 (16) | TAACAGTCGATGGCGGCTTT | CAGCGACTTGTTTTTTACCAACTTC |
| fsrB (55) | TGCTCAAAAAGCAAAGCCTTATAA | GATGACGAGACCGTAGAGTATTACTGAA |
| fsrC (55) | GCTTATTTGGAAGAACAACGTATCAA | CGAAACATCGCTAGCTCTTCGT |
| gelE (55) | CGGAACATACTGCCGGTTTAGA | TGGATTAGATGCACCCGAAAT |
| inl-like gene | GTGACAGTATTAGAGATCCGAGATTTG | ATACGCAGGTGCTGTCTTAGATAA |
| esp (69) | GGAACGCCTTGGTATGCTAAC | GCCACTTTATCAGCCTGAACC |
| ace (62) | CGGCGACTCAACGTTTGAC | TCCAGCCAAATCGCCTACTT |
| asa (11) | GATACAAAGCCAATGTCGTTCCT | TAAAGAGTCGCCACGTTTCACA |
| efa (43) | TGGGACAGACCCTCACGAATA | CGCCTGTTTCTAAGTTCAAGCC |
| 23S rRNA | CCTATCGGCCTCGGCTTAG | AGCGAAAGACAGGTGAGAATCC |
Preliminary quantification of 23S rRNA levels was performed and used as an internal control to normalize RNA concentration. Approximately 10 ng of normalized RNA was used to generate cDNA by reverse transcription with the appropriate specific reverse primer (added to a final concentration of 2.5 μM) using reagents supplied in the TaqMan Reverse Transcription Reagents kit (Applied Biosystems) according to the manufacturer's recommendations. Five microliters of the resulting cDNA synthesis reaction mixture was used for subsequent PCR amplification with the appropriate forward and reverse primers and the SYBR Green PCR Master Mix kit (Applied Biosystems). Preliminary experiments testing a range of concentrations for both the forward and reverse primers in PCR amplification demonstrated that a final concentration of 300 nM for each specific forward primer and reverse primer (with approximately 250 nM reverse primer being carried over from the reverse transcription reaction) yielded optimum amplification without interference from primer dimer formation, as determined in control reactions lacking the cDNA template. Primer pairs that yielded primer dimers at these concentrations were eliminated from further use. An additional control for each primer pair and RNA sample included sham cDNA synthesis reactions that lacked reverse transcriptase, followed by PCR amplification, to identify RNA preparations contaminated by residual genomic DNA.
The value used for comparison of gene expression in various environments was the number of PCR cycles required to reach the midpoint of the amplification curve, or threshold cycle (CT). CT was determined by monitoring incorporation of SYBR Green I into the amplified product with fluorescent detection. SYBR Green I fluorescence was normalized to a passive reference dye (carboxy-x-rhodamine [ROX]) included in each reaction. For specific products amplified from RNA derived from serum and urine cultures, CT values were compared to the CT values of the amplified product derived from 2×YT cultures. Preliminary control experiments showed that each cycle of difference in CT value represented a precise doubling (2.07 ± 0.04). Therefore, to relate CT value back to abundance of an mRNA species, CT was converted to “n-fold difference” by comparing mRNA abundance in 2×YT to that under one of the experimental conditions (serum or urine). The n-fold difference was calculated by the formula y = 2−x, where x = (CT in serum or urine − CT in 2×YT) and y = (n-fold difference in mRNA abundance). The n-fold differences in mRNA abundance for genes expressed in late log versus stationary phase were calculated using the same equation, but with x calculated as CT in exponential phase − CT in stationary phase. For each gene for which mRNA abundance was investigated, real-time quantitative PCR analysis was performed on RNA purified from three independently grown cultures in each environment. Statistical comparison of means was performed using Student's t test.
Because comparisons in most cases were between products of PCRs using identical primers, and not between PCR products derived from different genes, the amplification efficiencies of different primer pairs were eliminated as a variable in the interpretation of the data. However, comparisons of real-time PCR data were made between different genes of the cyl operon. To control for the potential differences in amplification efficiency among primer pairs in this case, several primer pairs for each of the cyl genes were selected following experimental determination that each amplified a DNA template at an identical rate.
Nucleotide sequence comparisons.
Enterococcal genes showing similar patterns of expression were located within the emerging E. faecalis genome database (www.tigr.org), or through the Entrez database. For each gene, 500 nucleotides 5′ to the predicted start codon were aligned and compared. Multiple sequence alignment of the 5′regions was performed using the Clustal X software program (32, 75), and the data were viewed using the SeqVu program.
RESULTS
Growth of E. faecalis and isolation of RNA.
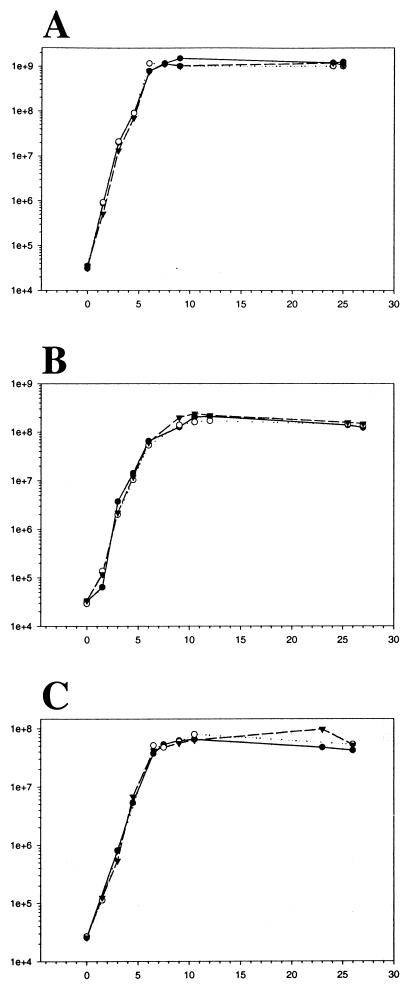
Growth kinetics for E. faecalis MMH594 cultured in 2×YT, serum, and urine were determined in triplicate by serially diluting and plating aliquots at various times after inoculation. Representative growth curves are provided in Fig. 1. E. faecalis grew readily in both serum and urine, without the supplementation of exogenous carbon sources or micronutrients. Maximal growth in serum was comparable to that in urine, yielding approximately 2.0 × 108 and 7.0 × 107 CFU/ml, respectively. However, maximum cell density in both environments was approximately an order of magnitude less than that in 2×YT.
FIG. 1.
Growth of E. faecalis MMH594 in 2×YT (A), serum (B), and urine (C). The x axis shows time (in hours), and the y axis shows cell count (in CFU per milliliter).
RNA was isolated from log- or stationary-phase cultures using the same lot of each medium in which growth was evaluated. For isolation of exponential-phase RNA, cells were harvested in log phase 4.5, 6, and 6 h following subculture in 2×YT, serum, and urine, respectively, at levels of approximately 5 × 107 CFU/ml. For isolation of stationary-phase RNA, cells were grown for 25, 27, and 26 h in 2×YT, serum, and urine, respectively, which represented 18 h following entry into stationary phase as determined in preliminary experiments.
Real-time quantitative PCR.
Genes known or suspected to play a role in the pathogenesis of bloodstream or urinary tract infection are listed in Table 1. For each gene, amplifications were performed on three independent RNA samples from each environment. No statistically significant differences were detected in internal control amplifications of 23S rRNA, regardless of medium used for culture or phase of growth. All comparisons were made between values obtained from amplification of RNA derived from serum or urine relative to those obtained from amplification of RNA from 2×YT cultures. Thus, abundance of mRNA for a particular gene in 2×YT was normalized to a value of 1 as a baseline for comparison.
Differences in abundance of virulence gene mRNA from log-phase cells grown in serum or urine, compared to baseline expression in 2×YT, are shown in Table 2. For genes related to expression of the enterococcal cytolysin (8, 17), small environment-dependent differences in the abundance of mRNA for cytolysin structural (cylLL and cylLS) and regulatory genes (cylR1 and cylR2) were noted (Table 2). RNA encoding functions related to cytolysin maturation (cylM, -B, -A, and -I) was modestly (about fourfold) but significantly more abundant in serum cultures during late exponential phase than in either 2×YT or urine cultures.
TABLE 2.
Change in abundance of E. faecalis virulence factor mRNA when cultured in serum or urine, relative to expression levels in 2 × YTa
| Gene | Fold change during phase when cultured in:
|
|||
|---|---|---|---|---|
| Log
|
Stationary
|
|||
| Serum | Urine | Serum | Urine | |
| cylR2 | 2.1 | 1.8 | 1.5 | 24 |
| cylR1 | 1.6 | 1.5 | 1.6 | 34 |
| cylLL | 1.9 | 1.5 | 1.9 | 8 |
| cylLS | 1.7 | 1.5 | 1.6 | 12 |
| cylM | 3.6 | −2.5 | −2.5 | 1.4 |
| cylB | 3.8 | −2 | −1.4 | 3.4 |
| cylA | 7 | 1.2 | 1.9 | 9 |
| cylI | 3.7 | 1.1 | −1.1 | 8 |
| inl-like gene | 4 | 6 | 1.6 | 162 |
| esp | 10 | 4.9 | 1.2 | 24 |
| ace | 3.3 | 1.9 | 1.1 | 12 |
| asa | 5 | 6 | 1.4 | 19 |
| efa | 66 | 89 | 5 | 2,195 |
| gls24 | 15 | 9 | 2.1 | 17 |
| fsrB | 28 | 16 | −5 | 2.1 |
| fsrC | 26 | 11 | −3 | 1.9 |
| gelE | 52 | 7 | −12.5 | −6.7 |
Values greater than 1 reflect a relative increase in mRNA abundance compared to 2 × YT culture; negative values reflect a relative decrease. Values in boldface type represent changes with a statistical significance (P) of <0.05. Each data point and statistical significance are derived from the mean and standard deviation of three independent experiments.
Expression of genes encoding enterococcal surface proteins related, or potentially related, to enterococcal virulence (11, 43, 62, 69) were also examined. Significant differences in abundance of log-phase mRNA encoding each of the targeted surface genes from E. faecalis grown in serum or urine, relative to 2×YT, were detected (Table 2). The gene encoding the E. faecalis endocarditis antigen (efaA) demonstrated the greatest environment-dependent change in expression. In log-phase cells, the abundance of efaA message increased 66-fold in serum and 89-fold in urine, over levels of expression in 2×YT. The abundance of mRNA from genes encoding the enterococcal surface protein (esp), aggregation substance (asa), and a newly identified homolog of internalin (inl-like gene) increased significantly in both serum and urine (4- to 89-fold [Table 2]). While statistically significant, the increase in abundance of mRNA for the gene encoding the enterococcal collagen-binding adhesin (ace) in serum and urine cultures was modest (3.3- and 1.9-fold, respectively).
The third group of genes analyzed included three associated with cell signaling (49, 56) or stress response (16) in E. faecalis. As with genes encoding surface proteins, significant differences in the abundance of mRNA from each targeted gene were detected in log-phase serum or urine cultures relative to 2×YT (Table 2). The abundance of mRNA from a gene encoding a stress- and starvation-inducible protein in E. faecalis (gls24) exhibited significant increases in both serum (15-fold) and urine (9-fold). The abundance of mRNA from two genes, one encoding one component (fsrC) of an apparent two-component regulatory system (55) and the second encoding the precursor of the signaling molecule (fsrB) (49), increased in parallel and by similar amounts in both serum (26- and 28-fold) and urine (11- and 16-fold). The abundance of mRNA from the gene encoding gelatinase (gelE), which is regulated by the fsr system (49, 55), demonstrated similar patterns of increased abundance in both serum (52-fold) and urine (7-fold), compared to levels in 2×YT.
It was next determined whether cues in serum or urine affected virulence gene expression in stationary phase. As shown in Table 2, there was a significantly greater abundance of cytolysin operon mRNA in stationary-phase cultures in urine compared to those in 2×YT. The greatest change was noted for mRNA encoding the regulatory genes cylR2 and cylR1, which was increased in urine 24- and 34-fold, respectively. This corresponded with an increase in abundance of mRNA from cytolysin subunit and modification genes by 1.4- to 12-fold. In contrast, no significant differences were detected in the abundance of cyl operon mRNA in stationary-phase cells cultured in serum compared to 2×YT.
Growth to stationary phase in urine also resulted in a significant increase in the abundance of mRNA for genes encoding enterococcal surface proteins efa (2,195-fold), esp (24-fold), asa (19-fold), ace (12-fold), and an inl-like protein (162-fold) (Table 2); only efaA mRNA levels were also significantly increased in serum culture (5-fold). Genes encoding proteins associated with cell signaling also demonstrated significant increases in stationary-phase urine culture. The mRNA encoding gls24 was observed to be 17-fold more abundant in stationary urine cultures relative to those in 2×YT. The abundance of fsrB and fsrC mRNA was only slightly but statistically significantly greater in cells cultured in urine than in 2×YT culture (2.1- and 1.9-fold, respectively). Interestingly, despite this small increase in fsrB and fsrC mRNA abundance, the abundance of gelatinase-encoding mRNA was observed to decrease 6.7-fold in stationary-phase urine cultures.
Growth to stationary phase in serum resulted in few statistically significant changes in gene expression compared to stationary-phase 2×YT cultures. These changes included the aforementioned increase in efa message (5-fold), the small increase in gls24 mRNA (2.1-fold), and decreases in fsrB (5-fold reduction), fsrC (3-fold reduction), and gelatinase mRNA (12.5-fold reduction).
The preceding analysis of data compared mRNA abundance in cells cultured in three physiologically distinct environments, within either log or stationary phase. It was also of interest to determine how mRNA abundance is affected by passage from log to stationary phase within a single environment (Table 3). The abundance of nearly all genes decreased significantly by large amounts in going from log- to stationary-phase culture in a given medium, with few exceptions.
TABLE 3.
Change in mRNA in transition from log to stationary phase
| Gene | Fold change when cultured in:
|
||
|---|---|---|---|
| 2×YT | Serum | Urine | |
| cylR2 | −95a | −127 | −7 |
| cylR1 | −136 | −131 | −6 |
| cylLL | −97 | −99 | −17 |
| cylLS | −146 | −156 | −18 |
| cylM | −37 | −315 | −10 |
| cylB | −22 | −123 | −3 |
| cylA | −72 | −247 | −10 |
| cylI | −19 | −79 | −2.5 |
| inl-like gene | −19 | −48 | 1.4 |
| esp | −10 | −91 | −2.1 |
| ace | −3.6 | −11 | 1.7 |
| asa | −2.9 | −10 | 1.1 |
| efa | −352 | −4,973 | −14 |
| gls24 | −2.5 | −18 | −1.7 |
| fsrB | −1.1 | −119 | −8 |
| fsrC | −1.3 | −123 | −7 |
| gelE | 16 | −50 | −3 |
Values in boldface type represent statistically significance differences (P < 0.05).
For each gene of the cyl operon, mRNA decreased from log phase to stationary phase, regardless of the environment in which E. faecalis was cultured (Table 3). All mRNA species encoding surface proteins decreased in abundance in stationary-phase cells grown either in 2×YT or serum but most were statistically unchanged in urine. In urine, only efa exhibited a statistically significant decrease in expression in stationary-phase cells (14-fold [Table 3]) compared to its abundance in log-phase cells. mRNA encoding the stress regulator gls24 was slightly reduced, if at all, in stationary-phase cells in 2×YT and urine but was reduced 18-fold in stationary-phase serum culture. Signaling system genes fsrB and -C were essentially unchanged in stationary phase when grown in 2×YT, but mRNA levels were approximately 100-fold less in stationary-phase serum cells compared to log-phase cells grown in the same medium. In urine, the change was less dramatic, with both being reduced seven- to eightfold (Table 3). The abundance of gelatinase mRNA, which is regulated to some degree by the fsr system, was enigmatically increased in stationary-phase 2×YT as previously reported (55, 56) but was decreased 50-fold in stationary-phase serum cultures and 3-fold in urine cultures, highlighting the importance of context.
Nucleotide sequence comparisons.
Variation in the abundance of mRNA encoding surface proteins appeared to follow similar patterns. The abundance of mRNA from genes encoding the surface proteins was greater in cells grown to log phase in serum or urine than in those grown in 2×YT (Table 2). Further, these mRNA levels also were elevated in stationary-phase cells cultured in urine relative to those grown in 2×YT (although efa demonstrated comparatively modest increases during growth in serum as well [Table 2]). Therefore, it was of interest to examine the nucleotide sequences upstream of these genes for identities in potential regulatory regions that may mediate shared responses to biological cues. For analysis, 500 nucleotides 5′ to the predicted start codon of esp, ace, efa, and the inl-like gene were aligned and compared. Each of these genes was located in the emerging E. faecalis genome database located at The Comprehensive Microbial Resource (www.tigr.org) provided by The Institute for Genomic Research. Confirmation of appropriate sequence was achieved by comparison to sequence data available in our laboratory or through the Entrez database. Multiple sequence alignment was performed using the Clustal X software program (32, 75), and viewing of the data for analysis was facilitated through the use of the SeqVu program.
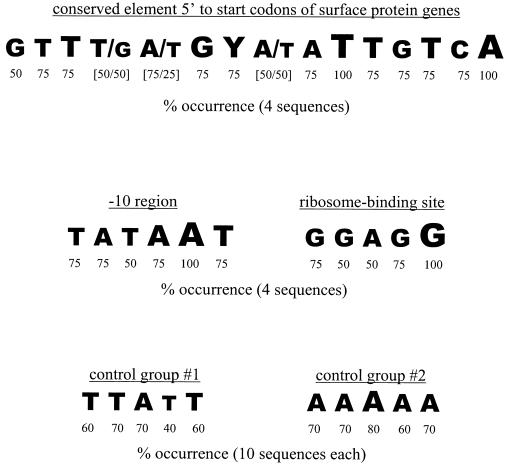
Multiple sequence alignment revealed a region of greatest sequence similarity 5′ to each of the genes (Fig. 2). The consensus sequence spanned 15 bp and was located approximately 200 bp (range of 179 to 219 bp) 5′ to each gene (179 to 219). Within this sequence, two bases were invariant and nine occurred in 75% of sequences (Fig. 2). The extent of conservation within this region was similar to that observed in the putative −10 boxes and ribosome-binding sites of the genes following multiple sequence alignment (Fig. 2). Alignment of 500 nucleotides 5′ to the predicted start codon from each of 20 randomly chosen genes in the E. faecalis genome database failed to reveal a similar sequence motif (Fig. 2).
FIG. 2.
Common sequence of nucleotides identified by multiple sequence alignment of regions 5′ to the start codon of E. faecalis genes encoding surface proteins. For analysis, 500 nucleotides 5′ to the predicted start codon of esp, ace, efa, and the inl-like genes were aligned and compared. The identified consensus sequence is located approximately 200 bp (range, 179 to 219 bp) 5′ to each gene. The percent conservation for each nucleotide is shown and is proportionately represented by font size. Alignment of 500 nucleotides 5′ to the predicted start codon from each of 20 randomly chosen genes (analyzed in two groups of 10 genes each) in the E. faecalis genome failed to reveal a similar sequence motif. From each control group of 10 genes, the alignment yielding the highest degree of conservation over the greatest number of bases, other than that observed in the putative −10 boxes and ribosome-binding sites, is provided for comparison.
DISCUSSION
Real-time quantitative PCR has emerged as a sensitive and quantitative tool for measuring nucleic acid abundance in a wide variety of biological systems. While the technology has been widely applied in a broad range of studies of eukaryotic systems, few studies have used real-time quantitative PCR to study gene expression and transcriptional organization in prokaryotes, with the majority being sharply focused in scope (5, 14, 25, 38, 44, 61). With a view toward verifying targets for the design of new antimicrobials, we applied real-time PCR to examine the expression patterns of enterococcal genes associated with virulence in biological environments that would be expected to occur at the site of common forms of enterococcal infection. Because real-time quantitative PCR used in the present study was based on amplification of mRNA, the results reflect only the processes of transcription and those forces regulating the degradation of mRNA following transcription. As focus was directed toward the environmental regulation of enterococcal gene expression and mRNA turnover, extension of the results to the protein level was beyond the scope of the present study. The present study demonstrated that the mechanisms regulating enterococcal gene expression and mRNA turnover are sensitive to the environment, and identified and quantified environment-dependent changes in mRNA abundance from genes related to toxin production, surface adherence, and cell signaling.
Streptococcal surface proteins that demonstrate increased expression in stationary phase have been identified (15, 61). Several studies have identified proteins and environmental cues that coordinately mediate entry of bacteria into stationary phase and subsequently modulate patterns of gene expression during stationary phase (4, 36, 41, 54). Evidence suggests that quorum-sensing may provide such a cue and thus play a role in the regulation of entry into stationary phase (41).
The abundance of mRNA from genes encoding proteins associated with cell signaling in E. faecalis was affected by growth in either serum or urine, relative to that in 2×YT. Levels of fsrB and fsrC mRNA in exponential phase were higher in serum and urine than in 2×YT (Table 2). However, mRNA levels for each gene were comparable in all media during stationary phase (Table 2). The parallel patterns of mRNA abundance from both genes most likely results from the inferred cotranscription from a single promoter (56). Levels of mRNA from gelE, which is transcriptionally regulated by fsrB and fsrC (55, 56), were higher in exponential phase in both serum and urine than laboratory medium (Table 2), but not in stationary phase (Table 2), and they broadly paralleled the pattern of mRNA abundance from fsrB and fsrC. A recent report suggests that expression of fsrC and gelE is increased postexponentially in laboratory medium (55). In that report, a spike in expression was observed at the 4 h-subculture time point, which appeared to represent the point of inflection between exponential- and stationary-phase growth (55). The present study demonstrated an approximately 16-fold increase in gelE mRNA abundance at a time point approximately 18 h after entry into stationary phase, compared to levels in log-phase cells in laboratory medium, which contrasts with the previously reported observation that following the burst of expression, gelE message became undetectable over the next few hours (55). In the present study, we observed a significant medium-dependent increase in the level of gelE mRNA in exponential-phase cells from either serum or urine, demonstrating that in addition to growth phase, host cues play a role in gelE gene expression and/or mRNA turnover.
Levels of gls24 mRNA were higher during exponential phase in E. faecalis cultured in either serum or urine, relative to that cultured in 2×YT (Table 2). However, during stationary phase, only urine caused a substantial increase over levels obtained in 2×YT (Table 2). gls24 encodes a protein that has been identified as a general stress protein in E. faecalis, due to its induction in starvation and stress responses (16). Inactivation of gls24 has pleiotropic effects on cell physiology, morphology, and gene expression (16). Of interest is the observation in the present work that in both serum and urine, increased levels of gls24 mRNA were observed in log phase, indicating that these environments present E. faecalis with physiologic stresses that do not occur in 2×YT. Interestingly, during both log and stationary phase in each of the three environments, the abundance of gls24 mRNA paralleled that of genes encoding enterococcal surface proteins. Further investigation may define a direct or indirect association between the two observations.
The abundance of cyl operon mRNA showed few medium-dependent changes during exponential phase (Table 2). The expression of cylLL and cylLS was recently shown to be controlled by quorum-sensing autoinduction at cell densities above 107 CFU/ml (25). As cell densities at the time of log-phase RNA isolation in each of the three environments were above this threshold, the operon was most likely maximally induced in each environment. However, mRNA encoding the modification (cylM, cylA), transport (cylB), and immunity (cylI) functions of the cyl operon exhibited increased abundance during log-phase growth in serum relative to 2×YT. Expression of these genes occurs by initiation of transcription from a promoter (PL) 5′ to cylLL, and elongation of transcription through an attenuator between cylLS and cylM (17, 24, 25). This process generates a short transcript encoding cylLL and cylLS and a full-length transcript bearing each gene in the order cylLLLSMBAI. The increased abundance of cylM, cylB, cylA, and cylI in serum may be due to factors in serum that facilitate transcriptional elongation through antitermination at the attenuator, or decreased turnover by endogenous RNases. In Escherichia coli, a protein has been identified that promotes transcript elongation in a subset of operons associated with the production of extracellular components required for virulence (3, 42), including the E. coli hemolysin.
During stationary-phase growth, increased abundance of mRNA from genes within the cyl operon was detected only for E. faecalis cultured in urine relative to 2×YT (Table 2). This suggests either enhanced transcription of the cytolysin operon in urine, the lack of a signal that downregulates expression of the cyl operon in stationary phase in urine, increased stability of cyl mRNA in urine, or a combination of these processes. Examination of the mechanisms regulating cytolysin expression and mRNA turnover in late stationary phase is the subject of ongoing study. As the cyl primer pairs used in real-time PCR amplification were designed and confirmed to amplify without statistically significant difference, direct comparison between different genes of the operon was possible. mRNA encoding the cytolysin maturation functions demonstrated a distinct pattern of abundance when derived from urine-grown cultures. No difference in abundance was detected for cylM, cylB exhibited a threefold increase, and cylA and cylI demonstrated an eight- to ninefold increase. Because cylM, cylB, cylA, and cylI are joined in that order within a polycistronic message that includes cylLL and cylLS at the 5′ end (17, 24), the increase in abundance of the four genes shows 5′→3′ directionality. This may reflect variable mRNA stability or degradation, both of which could be controlled by environmental cues. Independent degradation at different rates of individual segments within a polycistronic message is known to lead to segmental differences in stability of several prokaryotic polycistronic mRNAs, and subsequent differential expression of genes within an operon (2, 21, 58, 60). Moreover, mechanisms that regulate mRNA degradation in a variety of bacteria have been shown to be sensitive to environmental signals (1, 9, 19, 39, 46, 67). Such features of mRNA stability and degradation have yet to be studied in detail in the enterococci.
With only a few exceptions, the abundance of mRNA from each of the analyzed genes decreased from log to stationary phase in any given environment (Table 3). Although the data highlight a general trend toward decreased mRNA abundance in stationary phase versus exponential phase in each of the three environments, the decrease was generally less for E. faecalis cultured in urine than for E. faecalis cultured in either serum or 2×YT. In a relative comparison, increased abundance of a specific mRNA may reflect either an increased level in a given environment (e.g., serum or urine) or a decreased level in the environment used as a baseline for comparison (e.g., 2×YT). In the present work, the abundance of mRNA from efaA provides the most-striking example of the latter. In stationary phase, the abundance of efaA mRNA was 2,195-fold greater in E. faecalis cultured in urine than it was when cultured in 2×YT (Table 2), and this difference was far greater than the 89-fold difference between the two environments during exponential phase (Table 2). Nonetheless, the abundance of efaA mRNA decreased from exponential to stationary phase in both urine and 2×YT (Table 3). However, the decrease in abundance was far greater for E. faecalis cultured in 2×YT than for E. faecalis cultured in urine (Table 3). Thus, the 2,195-fold increase in abundance of efaA mRNA from E. faecalis cultured to stationary phase in urine relative to that when cultured in 2×YT reflects primarily the decreased abundance of efaA mRNA in E. faecalis cultured in 2×YT.
The abundance of mRNA encoding the enterococcal surface proteins studied was observed to vary in similar patterns (Table 2 and 3). To probe for possible evidence of a mechanism of coregulation at the level of transcription, nucleotide sequences 5′ to each of these genes were aligned and examined for identities that may mediate shared responses to biological cues. The region of greatest sequence conservation occurred approximately 200 bp 5′ to the predicted start codon of each gene and represented a consensus sequence spanning 15 bp (Fig. 2). This sequence was notable as the extent of conservation was observed to be similar to that occurring at the putative ribosome-binding sites and −10 boxes of the same genes. Moreover, no similarly conserved motif was observed in control alignments of randomly selected genes, although a consensus putative ribosome-binding site and −10 box were identified in each of the control genes. The importance of the consensus sequence, if any, remains to be proven experimentally. The identified sequence shares no similarity to consensus motifs associated with transcriptional regulation that are located 5′ to several genes encoding surface proteins and other virulence factors in Staphylococcus aureus (6, 37, 53, 59) and Streptococcus pyogenes (20, 45, 47).
In the present study, the abundance of mRNA from enterococcal genes was found to be dependent on biological cues that occur in serum and urine. The identification of such responses demonstrates the occurrence of as-yet-uncharacterized mechanisms for control of gene expression in enterococci that are likely to play an important role in vivo. At any given point in time, the level of mRNA in a bacterial cell is a function of two opposing forces. The abundance of a specific mRNA is determined by both the efficiency of transcription of the corresponding DNA and the degradation of the mRNA, which is primarily determined by the activity of endogenous exo- and endoribonucleases. Thus, the moment of RNA isolation represents a snapshot of the consequence of these processes. This study demonstrates that for E. faecalis, there are both environment-specific and growth phase-specific cues that contribute to regulation of virulence factor gene expression.
Acknowledgments
We thank Phillip Coburn, Lynn Hancock, Michael Engelbert, Chris Cox, Kenneth Hatter, Wolfgang Haas, Chris Pillar, Mark Huycke, Brad Jett, and Keeta Gilmore for technical support and critical review of the manuscript.
This work was supported by grants from the National Institutes of Health and Research to Prevent Blindness.
Editor: E. I Tuomanen
REFERENCES
- 1.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 2.Alifano, P., C. B. Bruni, and M. S. Carlomagno. 1994. Control of mRNA processing and decay in prokaryotes. Genetica 94:157-172. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, M. J., C. Hughes, and V. Koronakis. 1996. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol. Microbiol. 22:729-737. [DOI] [PubMed] [Google Scholar]
- 4.Boylan, S. A., A. Rutherford, S. M. Thomas, and C. W. Price. 1992. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J. Bacteriol. 174:3695-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., R. O. Watson, J. C. Smoot, and J. M. Musser. 2001. Identification of Rgg-regulated exoproteins of Streptococcus pyogenes. Infect. Immun. 69:822-831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chien, Y., A. C. Manna, S. J. Projan, and A. L. Cheung. 1999. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J. Biol. Chem. 274:37169-37176. [DOI] [PubMed] [Google Scholar]
- 7.Chow, J. W., L. A. Thal, M. B. Perri, J. A. Vazquez, S. M. Donabedian, D. B. Clewell, and M. J. Zervos. 1993. Plasmid-associated hemolysin and aggregation substance production contribute to virulence in experimental enterococcal endocarditis. Antimicrob. Agents Chemother. 37:2474-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coburn, P. S., L. E. Hancock, M. C. Booth, and M. S. Gilmore. 1999. A novel means of self-protection, unrelated to toxin activation, confers immunity to the bactericidal effects of the Enterococcus faecalis cytolysin. Infect. Immun. 67:3339-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, J. J., G. P. Roberts, and W. J. Brill. 1986. Posttranscriptional control of Klebsiella pneumoniae nif mRNA stability by the nifL product. J. Bacteriol. 168:173-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drew, W. L., M. A. C. Edelstein, L. S. Shore, and G. D. Roberts. 1986. Selection, collection, and transport of specimens for microbiological examination, p. 53-69. In S. M. Finegold and E. J. Baron (ed.), Bailey and Scott's diagnostic microbiology. The C. V. Mosby Co., St. Louis, Mo.
- 11.Dunny, G. M., R. A. Craig, R. L. Carron, and D. B. Clewell. 1979. Plasmid transfer in Streptococcus faecalis: production of multiple sex pheromones by recipients. Plasmid 2:454-465. [DOI] [PubMed] [Google Scholar]
- 12.Dunny, G. M., B. A. Leonard, and P. J. Hedberg. 1995. Pheromone-inducible conjugation in Enterococcus faecalis: interbacterial and host-parasite chemical communication. J. Bacteriol. 177:871-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupont, H., P. Montravers, J. Mohler, and C. Carbon. 1998. Disparate findings on the role of virulence factors of Enterococcus faecalis in mouse and rat models of peritonitis. Infect. Immun. 66:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, K. J., and N. A. Saunders. 2001. Real-time PCR used to measure stress-induced changes in the expression of the genes of the alginate pathway of Pseudomonas aeruginosa. J. Appl. Microbiol. 91:29-37. [DOI] [PubMed] [Google Scholar]
- 15.El-Sabaeny, A., D. R. Demuth, Y. Park, and R. J. Lamont. 2000. Environmental conditions modulate the expression of the sspA and sspB genes in Streptococcus gordonii. Microb. Pathog. 29:101-113. [DOI] [PubMed] [Google Scholar]
- 16.Giard, J. C., A. Rince, H. Capiaux, Y. Auffray, and A. Hartke. 2000. Inactivation of the stress- and starvation-inducible gls24 operon has a pleiotrophic effect on cell morphology, stress sensitivity, and gene expression in Enterococcus faecalis. J. Bacteriol. 182:4512-4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilmore, M. S., R. A. Segarra, M. C. Booth, C. P. Bogie, L. R. Hall, and D. B. Clewell. 1994. Genetic structure of the Enterococcus faecalis plasmid pAD1-encoded cytolytic toxin system and its relationship to lantibiotic determinants. J. Bacteriol. 176:7335-7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 19.Goldenberg, D., I. Azar, and A. B. Oppenheim. 1996. Differential mRNA stability of the cspA gene in the cold-shock response of Escherichia coli. Mol. Microbiol. 19:241-248. [DOI] [PubMed] [Google Scholar]
- 20.Granok, A. B., D. Parsonage, R. P. Ross, and M. G. Caparon. 2000. The RofA binding site in Streptococcus pyogenes is utilized in multiple transcriptional pathways. J. Bacteriol. 182:1529-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunberg-Manago, M. 1999. Messenger RNA stability and its role in control of gene expression in bacteria and phages. Annu. Rev. Genet. 33:193-227. [DOI] [PubMed] [Google Scholar]
- 22.Guzmàn, C. A., C. Pruzzo, G. LiPira, and L. Calegari. 1989. Role of adherence in pathogenesis of Enterococcus faecalis urinary tract infection and endocarditis. Infect. Immun. 57:1834-1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guzmàn, C. A., C. Pruzzo, M. Plate, M. C. Guardati, and L. Calegari. 1991. Serum dependent expression of Enterococcus faecalis adhesins involved in the colonization of heart cells. Microb. Pathog. 11:399-409. [DOI] [PubMed] [Google Scholar]
- 24.Haas, W., and M. S. Gilmore. 1999. Molecular nature of a novel bacterial toxin: the cytolysin of Enterococcus faecalis. Med. Microbiol. Immunol. 187:183-190. [DOI] [PubMed] [Google Scholar]
- 25.Haas, W., B. D. Shepard, and M. S. Gilmore. 2002. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature 415:84-87. [DOI] [PubMed] [Google Scholar]
- 26.Higuchi, R., G. Dollinger, P. S. Walsh, and R. Griffith. 1992. Simultaneous amplification and detection of specific DNA sequences. Bio/Technology 10:413-417. [DOI] [PubMed] [Google Scholar]
- 27.Higuchi, R., C. Fockler, G. Dollinger, and R. Watson. 1993. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology 11:1026-1030. [DOI] [PubMed] [Google Scholar]
- 28.Holland, P. M., R. D. Abramson, R. Watson, and D. H. Gelfand. 1991. Detection of specific polymerase chain reaction product by utilizing the 5′-3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc. Natl. Acad. Sci. USA 88:7276-7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huycke, M. M., C. A. Spiegel, and M. S. Gilmore. 1991. Bacteremia caused by hemolytic, high-level gentamicin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 35:1626-1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ike, Y., H. Hashimoto, and D. B. Clewell. 1984. Hemolysin of Streptococcus faecalis subspecies zymogenes contributes to virulence in mice. Infect. Immun. 45:528-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgins, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 33.Jett, B. D., M. M. Huycke, and M. S. Gilmore. 1994. Virulence of enterococci. Clin. Microbiol. Rev. 7:462-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jett, B. D., H. G. Jensen, R. V. Atkuri, and M. S. Gilmore. 1995. Evaluation of therapeutic measures for treating endophthalmitis caused by isogenic toxin-producing and toxin-nonproducing Enterococcus faecalis strains. Investig. Ophthalmol. Vis. Sci. 36:9-15. [PubMed] [Google Scholar]
- 35.Jett, B. D., H. G. Jensen, R. E. Nordquist, and M. S. Gilmore. 1992. Contribution of the pAD1-encoded cytolysin to the severity of experimental Enterococcus faecalis endophthalmitis. Infect. Immun. 60:2445-2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang, M., W. Shao, M. Perego, and J. A. Hoch. 2000. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 38:535-542. [DOI] [PubMed] [Google Scholar]
- 37.Jonsson, K., C. Signas, H. P. Muller, and M. Lindberg. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur. J. Biochem. 202:1041-1048. [DOI] [PubMed] [Google Scholar]
- 38.Khun, H. H., V. Deved, H. Wong, and B. C. Lee. 2000. fbpABC gene cluster in Neisseria meningitidis is transcribed as an operon. Infect. Immun. 68:7166-7171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klug, G. 1991. Endonucleolytic degradation of puf mRNA in Rhodobacter capsulatus is influenced by oxygen. Proc. Natl. Acad. Sci. USA 88:1765-1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreft, B., R. Marre, U. Schramm, and R. Wirth. 1992. Aggregation substance of Enterococcus faecalis mediates adhesion to cultured renal tubular cells. Infect. Immun. 60:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lazazzera, B. A. 2000. Quorum sensing and starvation: signals for entry into stationary phase. Curr. Opin. Microbiol. 3:177-182. [DOI] [PubMed] [Google Scholar]
- 42.Leeds, J. A., and R. A. Welch. 1997. Enhancing transcription through the Escherichia coli hemolysin operon, hlyCABD: RfaH and upstream JUMPStart DNA sequences function together via a postinitiation mechanism. J. Bacteriol. 179:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lowe, A. M., P. A. Lambert, and A. W. Smith. 1995. Cloning of an Enterococcus faecalis endocarditis antigen: homology with adhesins from some oral streptococci. Infect. Immun. 63:703-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 45.McIver, K. S., A. S. Heath, B. D. Green, and J. R. Scott. 1995. Specific binding of the activator Mga to promoter sequences of the emm and scpA genes in the group A streptococcus. J. Bacteriol. 177:6619-6624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Melin, L., H. Friden, E. Dehlin, L. Rutberg, and A. von Gabain. 1990. The importance of the 5′-region in regulating the stability of sdh mRNA in Bacillus subtilis. Mol. Microbiol. 4:1881-1889. [DOI] [PubMed] [Google Scholar]
- 47.Miller, A. A., N. C. Engleberg, and V. J. DiRita. 2001. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol. Microbiol. 40:976-990. [DOI] [PubMed] [Google Scholar]
- 48.Mundy, L. M., D. F. Sahm, and M. Gilmore. 2000. Relationships between enterococcal virulence and antimicrobial resistance. Clin. Microbiol. Rev. 13:513-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 50.Nallapareddy, S. R., K. V. Singh, R. W. Duh, G. M. Weinstock, and B. E. Murray. 2000. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of ace during human infections. Infect. Immun. 68:5210-5217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.National Nosocomial Infections Surveillance Syst. 1999. National Nosocomial Infections Surveillance (NNIS) System report, data summary from January 1990-May 1999, issued June 1999. Am. J. Infect. Control 27:520-532. [DOI] [PubMed] [Google Scholar]
- 52.Olmsted, S. B., G. M. Dunny, S. L. Erlandsen, and C. L. Wells. 1994. A plasmid-encoded surface protein on Enterococcus faecalis augments its internalization by cultured intestinal epithelial cells. J. Infect. Dis. 170:1549-1556. [DOI] [PubMed] [Google Scholar]
- 53.Patti, J. M., H. Jonsson, B. Guss, L. M. Switalski, K. Wiberg, M. Lindberg, and M. Hook. 1992. Molecular characterization and expression of a gene encoding a Staphylococcus aureus collagen adhesin. J. Biol. Chem. 267:4766-4772. [PubMed] [Google Scholar]
- 54.Price, G. P., and A. C. St John. 2000. Purification and analysis of expression of the stationary phase-inducible slp lipoprotein in Escherichia coli: role of the Mar system. FEMS Microbiol. Lett. 193:51-56. [DOI] [PubMed] [Google Scholar]
- 55.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rakita, R. M., N. N. Vanek, K. Jacques-Palaz, M. Mee, M. M. Mariscalco, G. M. Dunny, M. Snuggs, W. B. Van Winkle, and S. I. Simon. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 67:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 59.Rechtin, T. M., A. F. Gillaspy, M. A. Schumacher, R. G. Brennan, M. S. Smeltzer, and B. K. Hurlburt. 1999. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol. Microbiol. 33:307-316. [DOI] [PubMed] [Google Scholar]
- 60.Regnier, P., and C. M. Arraiano. 2000. Degradation of mRNA in bacteria: emergence of ubiquitous features. Bioessays 22:235-244. [DOI] [PubMed] [Google Scholar]
- 61.Reid, S. D., N. M. Green, J. K. Buss, B. Lei, and J. M. Musser. 2001. Multilocus analysis of extracellular putative virulence proteins made by group A streptococcus: population genetics, human serologic response, and gene transcription. Proc. Natl. Acad. Sci. USA 98:7552-7557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rich, R. L., B. Kreikemeyer, R. T. Owens, S. LaBrenz, S. V. Narayana, G. M. Weinstock, B. E. Murray, and M. Hook. 1999. Ace is a collagen-binding MSCRAMM from Enterococcus faecalis. J. Biol. Chem. 274:26939-26945. [DOI] [PubMed] [Google Scholar]
- 63.Russo, T. A., U. B. Carlino, A. Mong, and S. T. Jodush. 1999. Identification of genes in an extraintestinal isolate of Escherichia coli with increased expression after exposure to human urine. Infect. Immun. 67:5306-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russo, T. A., S. T. Jodush, J. J. Brown, and J. R. Johnson. 1996. Identification of two previously unrecognized genes (guaA and argC) important for uropathogenesis. Mol. Microbiol. 22:217-229. [DOI] [PubMed] [Google Scholar]
- 65.Sartingen, S., E. Rozdzinski, A. Muscholl-Silberhorn, and R. Marre. 2000. Aggregation substance increases adherence and internalization, but not translocation, of Enterococcus faecalis through different intestinal epithelial cells in vitro. Infect. Immun. 68:6044-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, J. W. Harmala, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Segal, G., and E. Z. Ron. 1995. The groESL operon of Agrobacterium tumefaciens: evidence for heat shock-dependent mRNA cleavage. J. Bacteriol. 177:750-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shankar, N., C. V. Lockatell, A. S. Baghdayan, C. Drachenberg, M. S. Gilmore, and D. E. Johnson. 2001. Role of Enterococcus faecalis surface protein Esp in the pathogenesis of ascending urinary tract infection. Infect. Immun. 69:4366-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shankar, V., A. S. Baghdayan, M. M. Huycke, G. Lindahl, and M. S. Gilmore. 1999. Infection-derived Enterococcus faecalis strains are enriched in esp, a gene encoding a novel surface protein. Infect. Immun. 67:193-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shepard, B. D., and M. S. Gilmore. 2002. Antimicrobial-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 4:215-224. [DOI] [PubMed] [Google Scholar]
- 71.Shepard, B. D., and M. S. Gilmore. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh, K. V., T. M. Coque, G. M. Weinstock, and B. E. Murray. 1998. In vivo testing of an Enterococcus faecalis efaA mutant and use of efaA homologs for species identification. FEMS Immunol. Med. Microbiol. 21:323-331. [DOI] [PubMed] [Google Scholar]
- 73.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 74.Sussmuth, S. D., A. Muscholl-Silberhorn, R. Wirth, M. Susa, R. Marre, and E. Rozdzinski. 2000. Aggregation substance promotes adherence, phagocytosis, and intracellular survival of Enterococcus faecalis within human macrophages and suppresses respiratory burst. Infect. Immun. 68:4900-4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wells, C. L., E. A. Moore, J. A. Hoag, H. Hirt, G. M. Dunny, and S. L. Erlandsen. 2000. Inducible expression of Enterococcus faecalis aggregation substance surface protein facilitates bacterial internalization by cultured enterocytes. Infect. Immun. 68:7190-7194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Willems, R. J., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]