Abstract
Clostridium perfringens type A isolates causing food poisoning have a chromosomal enterotoxin gene (cpe), while C. perfringens type A isolates responsible for non-food-borne human gastrointestinal diseases carry a plasmid cpe gene. In the present study, the plasmid cpe locus of the type A non-food-borne-disease isolate F4969 was sequenced to design primers and probes for comparative PCR and Southern blot studies of the cpe locus in other type A isolates. Those analyses determined that the region upstream of the plasmid cpe gene is highly conserved among type A isolates carrying a cpe plasmid. The organization of the type A plasmid cpe locus was also found to be unique, as it contains IS1469 sequences located similarly to those in the chromosomal cpe locus but lacks the IS1470 sequences found upstream of IS1469 in the chromosomal cpe locus. Instead of those upstream IS1470 sequences, a partial open reading frame potentially encoding cytosine methylase (dcm) was identified upstream of IS1469 in the plasmid cpe locus of all type A isolates tested. Similar dcm sequences were also detected in several cpe-negative C. perfringens isolates carrying plasmids but not in type A isolates carrying a chromosomal cpe gene. Contrary to previous reports, sequences homologous to IS1470, rather than IS1151, were found downstream of the plasmid cpe gene in most type A isolates tested. Those IS1470-like sequences reside in about the same position but are oppositely oriented and defective relative to the IS1470 sequences found downstream of the chromosomal cpe gene. Collectively, these and previous results suggest that the cpe plasmid of many type A isolates originated from integration of a cpe-containing genetic element near the dcm sequences of a C. perfringens plasmid. The similarity of the plasmid cpe locus in many type A isolates is consistent with horizontal transfer of a common cpe plasmid among C. perfringens type A strains.
The gram-positive, anaerobic pathogen Clostridium perfringens is classified into five biotypes (A to E), based upon production of alpha, beta, epsilon, and iota toxins (14). Approximately 5% of C. perfringens isolates, mostly belonging to type A, produce another important toxin named C. perfringens enterotoxin (CPE) (12-14, 22). CPE-producing type A isolates are major human gastrointestinal (GI) pathogens, causing C. perfringens type A food poisoning (14) and such non-food-borne human GI diseases as antibiotic-associated diarrhea and sporadic diarrhea (15, 16). Enterotoxigenic type A isolates may also be responsible for some veterinary GI illnesses (21). Recent studies with cpe knockout mutants confirmed that CPE expression is necessary for the enteric virulence of enterotoxigenic C. perfringens type A isolates (20).
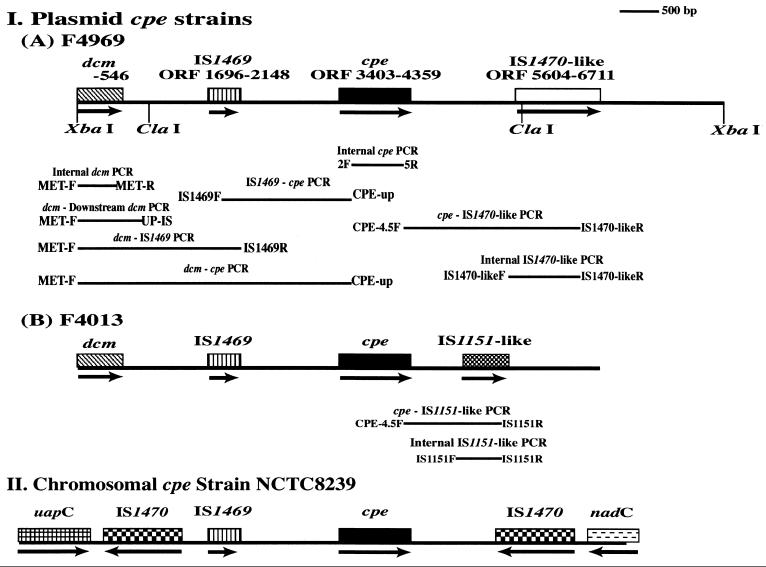
The gene encoding C. perfringens enterotoxin (cpe) can be located on either the chromosome or large plasmids (2-8, 23). Most (if not all) enterotoxigenic C. perfringens type A isolates associated with food poisoning carry their cpe genes on the chromosome (2-8, 23). Sequencing and Southern blot studies demonstrated that the chromosomal cpe gene of type A food poisoning strain NCTC8239 (and, apparently, other C. perfringens type A food poisoning strains) is closely associated with insertion sequences (2, 3, 5, 8). In NCTC8239, those cpe-associated insertion sequences include a single IS1469 sequence present ∼1.3 kbp upstream of the cpe gene and a pair of IS1470 sequences residing ∼3 kb upstream and ∼1.2 kbp downstream of the cpe gene (5, 8). Based upon this arrangement (see Fig. 1), it was proposed that the chromosomal cpe gene of NCTC8239 (and, by extension, other type A food poisoning isolates) is located on a 6.3-kbp transposon with terminal IS1470 elements (5). More recent results (2) suggest that this putative transposon, named Tn5565, may have several circular intermediate forms. However, actual movement of Tn5565 has not yet been demonstrated.
FIG. 1.
Genetic organization of the cpe locus in C. perfringens type A isolates. (I) Type A isolates carrying a cpe plasmid, including the sequenced ∼9-kbp XbaI cpe-containing fragment of the cpe plasmid from sporadic diarrhea isolate F4969 (A) and the deduced plasmid cpe locus of isolates F4013 (B). (II) The previously described (5) chromosomal cpe locus of food poisoning strain NCTC8239 is shown for comparison. Broad bars show ORFs; arrows under ORFs indicate orientation. Numbers shown for F4969 indicate nucleotide bases. Long thin bars depict the PCR products amplified with each primer pair (see the text).
Interestingly, the cpe gene is plasmid localized in most (if not all) enterotoxigenic type A isolates obtained from either CPE-associated non-food-borne human GI disease or veterinary sources (7, 8, 23). The cpe plasmids of those type A isolates are large (∼100 kbp) and present at low copy numbers (7, 8, 20). The cpe plasmid of the type A human sporadic diarrhea isolate F4969 was recently shown to be transferable, via conjugation, to cpe-negative C. perfringens type A strains (4).
The plasmid cpe locus of type A isolates has not yet been sequenced or otherwise studied in comparable detail as the chromosomal cpe locus of NCTC8239. To our knowledge, the only information currently available regarding the plasmid cpe locus of type A isolates was provided by Southern blot analyses (8) of just three such isolates (only one of which, isolate F3686, was associated with human non-food-borne GI disease). Those limited studies revealed that, as for type A food poisoning isolate NCTC8239, which carries a chromosomal cpe gene, IS1469 sequences appear to be present directly upstream of the plasmid cpe gene in the three initially examined type A isolates. However, no IS1470 sequences were detected near the plasmid cpe gene of those isolates. Instead, two of the isolates (including the human non-food-borne-GI-disease isolate F3686) apparently carry IS1151 sequences ∼260 bp downstream of their plasmid cpe gene.
The limited information now available regarding the organization of the plasmid cpe locus suggests that considerable variation exists between the organization of the chromosomal cpe locus and the plasmid cpe locus of C. perfringens type A isolates. To better address that possibility and to assess the diversity of the plasmid cpe locus in different type A isolates, we first cloned and sequenced the plasmid cpe locus of human sporadic diarrhea strain F4969, a well-studied type A isolate (4, 6, 7, 20). The sequencing results obtained for the F4969 plasmid cpe locus were then used to prepare primers and probes for conducting PCR and Southern blot analyses to further explore the organization of the cpe locus in other type A isolates, including a number of plasmid cpe isolates originating from various sources (including non-food-borne human GI diseases).
MATERIALS AND METHODS
Bacterial strains.
The toxin genotype, cpe genotype, and source of each C. perfringens isolate examined in this study are listed in Table 1. Our study used six previously uncharacterized C. perfringens isolates, including normal human fecal isolates (MR1-1, MR2-2, and MR2-4) and veterinary isolates (316366 D3, 315455 N4, and 297442). These six new isolates were first subjected to multiplex PCR analysis (23), which confirmed their identity as type A isolates and revealed whether they carried cpe sequences. As indicated in Table 1, the three new type A isolates identified as cpe positive by multiplex PCR were then subjected to cpe restriction fragment length polymorphism analysis (23) in order to determine whether they carried a chromosomal or plasmid cpe gene.
TABLE 1.
C. perfringens isolates used in this study
| Isolate group | Strain | Type | Location of cpe | Date and source | Reference |
|---|---|---|---|---|---|
| Food poisoning isolates | NCTC10239 | A | Chromosome | 1950s, Europe | 7 |
| NCTC8239 | A | Chromosome | 1950s, Europe | 7 | |
| NCTC8798 | A | Chromosome | 1950s, Europe | 7 | |
| 537-5 | A | Chromosome | Late 1990s, North America | 23 | |
| 538-1 | A | Chromosome | Late 1990s, North America | 23 | |
| Non-food-borne human GI disease isolates | F4969 | A | Plasmid | Early 1990s, Europe, sporadic diarrhea | 7 |
| F4013 | A | Plasmid | Early 1990s, Europe, sporadic diarrhea | 7 | |
| F5603 | A | Plasmid | Early 1990s, Europe, sporadic diarrhea | 7 | |
| B40 | A | Plasmid | 1980s, Europe, antibiotic-associated diarrhea | 7 | |
| Newbury16 (NB 16) | A | Plasmid | 1980s, Europe, antibiotic-associated diarrhea | 7 | |
| T34058 | A | Plasmid | Late 1990s, North America, antibiotic-associated diarrhea | 23 | |
| M26413 | A | Plasmid | Late 1990s, North America, antibiotic-associated diarrhea | 23 | |
| W30554 | A | Plasmid | Late 1990s, North America, antibiotic-associated diarrhea | 23 | |
| Normal human fecal isolates | MR1-1 | A | cpe negative | 2000, Japan | This study |
| MR2-2 | A | cpe negative | 2000, Japan | This study | |
| MR2-4 | A | Plasmid | 2000, Japan | This study | |
| Isolates from veterinary sources | 157 | A | Plasmid | Early 1990s, North America, canine peritoneal fluid | 7 |
| 452 | A | Plasmid | Early 1990s, North America, canine diarrhea | 7 | |
| 315455 N4 | A | Plasmid | Late 1990s, North America, canine diarrhea | This study | |
| 316366 D3 | A | Plasmid | Late 1990s, North America, canine diarrhea | This study | |
| 297442 | A | cpe negative | Late 1990s, North America, canine diarrhea | This study | |
| Other isolates | NCTC8533 | B | cpe negative | 23 | |
| CN5383 | C | cpe negative | 23 | ||
| PS52 | D | cpe negative | 23 | ||
| 853 | E | Plasmida | 23 |
Like most type E isolates (1), 853 carries silent cpe sequences containing nonsense and missense mutations in the cpe ORF.
Escherichia coli strain DH5α was grown in tryptic soy broth medium and used for transformation experiments.
DNA isolation from C. perfringens isolates.
C. perfringens isolates were cultured for 6 to 8 h at 37°C in TGY medium, as described previously (20). Total DNA was extracted from those TGY cultures using a previously described method (23). Plasmid DNA was isolated from TGY cultures by the method of Roberts et al. (18), except that RNase treatment was omitted.
Cloning of ClaI and XbaI fragments containing the plasmid cpe gene of C. perfringens type A isolate F4969.
Plasmid DNA from isolate F4969 was digested overnight at 37°C with either ClaI or XbaI (Roche). That digested plasmid DNA was electrophoresed on 1% agarose gels, and DNA fragments of ∼5 kbp (for ClaI-digested plasmid DNA) or ∼9 kbp (for XbaI-digested plasmid DNA) were then extracted with a DNA purification kit (Bio-Rad). A pBlueScript vector was digested with either ClaI or XbaI and then treated with calf intestinal phosphatase (Roche). That dephosphorylated plasmid was ligated, for 16 h at 16°C, with either the isolated ∼5-kbp ClaI or ∼9-kbp XbaI fragments of F4969 plasmid DNA (19). Ligated plasmids were then transformed into E. coli DH5α host cells by using standard techniques (19).
DNA sequencing of E. coli transformants containing F4969 DNA.
Southern blotting with a cpe-specific probe (7, 20, 23) identified several E. coli transformants carrying F4969 cpe sequences (data not shown). Two transformant clones, named KCpeFC5-1 (containing an ∼5-kbp ClaI insert of F4969 plasmid DNA) and KCpeFX28 (containing an ∼9-kbp XbaI insert of F4969 plasmid DNA), were then used to DNA sequence the plasmid cpe locus of isolate F4969. DNA sequencing was initially performed with universal primers for the pBlueScript vector. Additional sequencing was performed using primers designed from those initial and subsequent sequencing results (see Fig. 1).
IS1469-cpe PCR.
Based upon sequencing results obtained for the cpe-containing XbaI and ClaI fragments of KCpeFX28 and KCpeFC5-1, the following primers complementary to IS1469 and cpe gene sequences were designed: 5′-AAGTGAGATATTAAGGCAGC-3′ (IS1469F) and 5′-CCTAATATCCAACCATCTCC-3′ (CPE-up). These primers (1 μM final concentration) were then included in a PCR utilizing template DNA (total DNA extracted from specified C. perfringens type A isolates), 0.2 mM deoxynucleotide triphosphates (Promega), 2.5 mM MgCl2, and 5 U of Taq polymerase (Promega). The reaction mixtures, in a total volume of 50 μl, were placed in a thermal cycler (Techne) under the following conditions: 1 cycle at 94°C for 5 min; 33 cycles of 94°C for 30 s, 60°C for 60 s, and 68°C for 180 s; and 1 cycle at 94°C for 90 s, 60°C for 90 s, and 68°C for 10 min. The resultant PCR products were run on a 2% agarose gel and stained with ethidium bromide.
Cytosine methyltransferase gene (dcm) PCR.
After sequencing of the cpe-containing XbaI fragment in KCpeFX28 had revealed the presence of sequences that could encode the C-terminal half of cytosine methyltransferase (dcm sequences), the following primer pair was designed to PCR amplify those putative dcm sequences: 5′-CGATAATCT AGTTACACTAGAGAGC-3′ (MET-F) and 5′-CTGGATTACAATACTATTCCCAGC-3′ (MET-R). These primers were added (at a 1 μM final concentration) to a PCR mixture utilizing template DNA (total DNA extracted from specified C. perfringens isolates), 0.1 mM deoxynucleotide triphosphates (Promega), 2.5 mM MgCl2, and 2.5 U of Taq polymerase (Promega). The reaction mixtures, in a total volume of 50 μl, were then placed in a thermal cycler (Techne) under the following conditions; 1 cycle at 94°C for 2 min; 38 cycles at 94°C for 30 s, 50°C for 60 s, and 68°C for 60 s; and 1 cycle at 94°C for 90 s, 50°C for 90 s, and 68°C for 7 min.
To PCR amplify the DNA region lying between dcm sequences and sequences located ∼400 bp downstream from those dcm sequences in F4969, a second PCR was performed with MET-F and a primer named UP-IS (see Fig. 1) (5′-ATATTGGCCACATTCACAGC-3′). The PCR conditions used with these primers were as described above for amplifying internal dcm sequences. All dcm-based PCR products were electrophoresed on a 2% agarose gel, which was then stained with ethidium bromide.
After agarose electrophoresis and transfer to nylon membranes (Roche), PCR products amplified with MET-F and UP-IS from selected C. perfringens isolates (see Fig. 6) were Southern blotted by conventional techniques (20). The blots were hybridized with a digoxigenin (DIG)-labeled PCR probe prepared (9) with a PCR DIG-labeling kit (Roche), KCpeFX28 template DNA, and MET-F and UP-IS.
FIG. 6.
PCR and Southern blotting analyses of putative cytosine methyltransferase gene sequences (dcm sequences) in other C. perfringens types. (A) PCR analyses with primers to internal dcm sequences; (C) Southern blotting results with a F4969 dcm-specific probe generated with the same primer pair and pKCpeFX28 template DNA. (A) This PCR assay generated an ∼0.4 kbp product (arrow) in C. perfringens types A to E which is similar in size to the PCR product this primer pair generates with most type A strains (Fig. 3); those PCR products also hybridized to a dcm-specific probe (C). (B) PCR analysis using primers based on dcm sequences and sequences ∼400 bp downstream from dcm sequences; (D) Southern blotting analysis with a dcm-specific probe. An ∼0.9-kbp product was detected in type A isolate F4969 (single arrow); otherwise, ∼2.0-kbp products were detected in type B, D, and E isolates (double arrow). Those products also hybridized with an F4969 dcm-specific probe. Numbers on the left indicate migration of molecular size markers.
Southern blotting analyses for the presence of cpe, dcm sequences, IS1470-like sequences, and IS1151-like sequences in C. perfringens isolates.
For additional Southern blotting analyses, four more DIG-labeled probes were also generated with the PCR DIG-labeling kit. A dcm sequence-specific probe was prepared in a PCR using the plasmid present in KCpeFX28 as the template DNA, along with MET-F and MET-R primers. A cpe-specific probe was prepared in a PCR using F4969 DNA and primers 2F (5′-GGTACCTTTAGCCAATCA-3′) and 5R (5′-TCCATCACCTAAGGACTG-3′), which are specific for internal cpe sequences. To prepare a probe for internal IS1470-like sequences, PCR was performed with the primers IS1470-likeF (5′-GATAATCACTATGGACTACC-3′) and IS1470-likeR (5′-CGTATTAGACCATTATGACG-3′), with KCpeFX28 as the template DNA. Finally, a probe for internal IS1151-like sequences was prepared from F4013 DNA by using the primers IS1151-F (5′-TCCGATATCTGTACGGCTCC-3′) and IS1151-R (5′-AGAGCCAATATAGATATCAGTAAG-3′), which are complementary to the previously reported (11) IS1151 sequence.
For Southern blotting analyses to detect the presence of cpe, dcm sequences, IS1470-like sequences, and IS1151-like sequences, plasmid preparations isolated from specified C. perfringens isolates were digested overnight with XbaI at 37°C. To detect the presence of dcm sequences in C. perfringens food poisoning isolates, total DNA was obtained from each isolate and digested overnight with XbaI at 37°C. Digested plasmid or total DNAs were electrophoresed on a 1% agarose gel (Bio-Rad) and then transferred with a vacuum blotter (Bio-Rad) onto a nylon membrane. The membranes were hybridized, as previously described (20), with one of the probes described above.
Comparative PCR analyses of the DNA region upstream of plasmid or chromosomal cpe genes in C. perfringens type A isolates.
Based upon the KCpeFX28 sequencing results, the DNA region between dcm sequences and IS1469 was explored with a PCR using the MET-F primer and IS1469R (5′-CCGCATACTTCTTTACTTGC-3′), a primer specific for IS1469 sequences. These primers (1 μM final concentration) were used in a PCR with template DNA (total DNA extracted from specified C. perfringens isolates), 0.2 mM deoxynucleotide triphosphates, and 1 μl of Advantage 2 Taq polymerase (Clontech). The reaction mixtures, in a total volume of 50 μl, were placed in a thermal cycler (Techne) under the following conditions: 1 cycle at 94°C for 5 min; 33 cycles at 94°C for 30 s, 50°C for 60 s, and 68°C for 180 s; and 1 cycle at 94°C for 90 s, 50°C for 90 s, and 68°C for 7 min. The products were run on a 2% agarose gel and stained with ethidium bromide.
A second PCR to amplify the DNA region between dcm sequences and the cpe gene was also performed with the primers MET-F and CPE-up, along with Advantage 2 Taq polymerase, under the following conditions: 1 cycle at 94°C for 2 min; 34 cycles at 94°C for 30 s; 60°C for 60 s, and 68°C for 180 s; and 1 cycle at 94°C for 90 s, 60°C for 90 s, and 68°C for 7 min.
Products of both the dcm-IS1469 and dcm-cpe PCRs were electrophoresed on a 2% agarose gel and then stained with ethidium bromide.
Comparative PCR analysis of the DNA region downstream of plasmid versus chromosomal cpe genes in C. perfringens type A isolates.
Primers complementary to sequences present in cpe (CPE-4.5F, 5′-CAGTCCTTAGGTGATGGA-3′) or the IS1470-like sequences of F4969 (IS1470-likeR) were used to investigate the DNA region downstream of the plasmid or chromosomal cpe gene in C. perfringens type A isolates. This PCR utilized total DNA extracted from specified type A isolates as the template and also included each primer (1 μM final concentration), 0.2 mM deoxynucleotide triphosphates (Promega), and 1 μl of Advantage 2 Taq polymerase (Clontech). The reaction mixtures, in a total volume of 50 μl, were placed in a thermal cycler (Techne) and amplified under the conditions described above for the dcm-cpe PCR. PCR products were run on a 2% agarose gel and stained with ethidium bromide.
A second PCR to amplify internal IS1470-like sequences was also performed with both the IS1470-likeF and -R primers. These primers were added (at a 1 μM final concentration) to a PCR utilizing template DNA (total DNA extracted from specified C. perfringens isolates), 0.1 mM deoxynucleotide triphosphates (Promega), 2.5 mM MgCl2, and 2.5 U of Taq polymerase. The reaction mixtures, in a total volume of 50 μl, were then placed in a thermal cycler (Techne) under the following conditions: 1 cycle at 94°C for 2 min; 38 cycles at 94°C for 30 s, 44°C for 60 s, and 72°C for 60 s; and 1 cycle at 94°C for 90 s, 44°C for 90 s, and 72°C for 10 min.
PCR analysis for IS1151 sequences.
To amplify internal IS1151 sequences, a PCR was performed with IS1151-F and -R primers, which are complementary to the previously reported (11) IS1151 sequence. These primers were used in a PCR under amplification conditions described earlier for detecting dcm sequences.
A final PCR assay to evaluate the location of IS1151 sequences relative to the cpe gene was performed using either CPE-4.5F or CPE-d1F (5′-CTAACTCATACCCTTGGACTC-3′), along with the IS1151-R primer, in a PCR run under conditions described above for amplifying internal IS1151 sequences.
Nucleotide sequence accession number.
The sequence of the ∼9-kbp XbaI fragment in KCpeFX28 was submitted to GenBank under accession number AF416450. The sequences obtained for the PCR products described below were submitted to GenBank under accession numbers AF506816 to -18 and AF511071.
RESULTS
Cloning and sequencing of ClaI and XbaI fragments carrying the plasmid cpe gene of F4969, a C. perfringens type A human sporadic-diarrhea isolate.
Initial Southern blotting results (data not shown) indicated that the plasmid cpe gene of type A isolate F4969 is present on an ∼5-kbp ClaI fragment and an ∼9-kbp XbaI fragment. Therefore, those two cpe-containing fragments were separately cloned into a pSK vector. The resultant plasmids were then used to transform E. coli, as described in Materials and Methods. Southern blotting with cpe probes (data not shown) confirmed the presence of cpe sequences in two transformants, named KCpeFC5-1 and KCpeFX28, which carry (respectively) an ∼5-kbp ClaI fragment and an ∼9-kbp XbaI fragment.
DNA sequencing of KCpeFC5-1 and KCpeFX28 confirmed a previous report (6) that the cpe open reading frame (ORF) of isolate F4969 is completely homologous to the chromosomal cpe ORF of type A food poisoning strain NCTC8239 (10). Sequences downstream or upstream of the cpe ORF had not yet been examined for any type A isolate carrying a cpe plasmid. However, our KCpeFC5-1 and KCpeFX28 sequencing data indicate that, with the exception of a few nucleotide differences, the putative stem-loop structure located ∼40 bp downstream of the chromosomal cpe stop codon in NCTC8239 DNA (10) is similarly present downstream of the plasmid cpe ORF of isolate F4969. As also noted in the chromosomal cpe locus of NCTC8239 (10), the putative stem-loop structure downstream of the F4969 cpe ORF is immediately followed by an oligo(T) tract, which is consistent with the idea that stem-loop-oligo(T) region functions as a rho-independent transcription terminator of cpe transcription in sporulating F4969 cells (10).
These sequencing analyses also revealed that, except for a few single-nucleotide differences, the downstream DNA of F4969 and NCTC8239 remain homologous for ∼1 kbp past their cpe ORF. Significant downstream sequence divergence occurs beyond that conserved region (Fig. 1), with the cpe plasmid of F4969 having a defective ORF that shares partial homology (Fig. 2) with an IS1470 element present, in about the same location, downstream of the NTCT8239 chromosomal cpe gene (5). However, relative to the IS1470 sequences downstream of the NCTC8239 cpe gene (10), the IS1470-like element of F4969 contains many nucleotide replacements (some introducing nonsense mutations), is oppositely oriented, and has a 84-bp deletion and a 88-bp insertion (Fig. 2).
FIG. 2.
Comparison of IS1470-like sequence in isolate F4969 and the downstream IS1470 sequence in the type A chromosomal-cpe food poisoning strain NTCT8239. #, putative start codon; +, putative stop codon. Underlined nucleotides are homologous between the IS1470 sequences downstream of the NCTC8239 cpe gene and the IS1470-like sequence of isolate F4969. ∗, missing nucleotide.
With respect to sequences upstream of the F4969 plasmid cpe ORF, our KCpeFC5-1 and KCpeFX28 sequencing results revealed that the Shine-Dalgarno sequence and four putative promoters of F4969 are identical, in location and sequence, to those identified previously in the NCTC8239 chromosomal cpe gene (3, 5, 17, 24). However, about 250 bp upstream of the F4969 cpe gene, a 45-bp insertion was detected. That insertion is absent from the chromosomal cpe locus of NCTC8239 but completely matches an insertion found, in the same location, upstream of the chromosomal cpe gene of another type A food poisoning strain, NCTC10240 (17).
Further (1.3 kbp) upstream of the F4969 plasmid cpe gene, IS1469 sequences were detected (Fig. 1). Except for two nucleotide changes, those insertion sequences were found to match IS1469 sequences located, in the same position and orientation, upstream of the NCTC8239 chromosomal cpe gene. The putative Shine-Dalgarno sequence for the F4969 IS1469 sequences was determined to be identical to that of NCTC8239 (3). Upstream of IS1469, the plasmid cpe locus of isolate F4969 was found to share homology, for another 200 bp, with the previously sequenced (3, 5) chromosomal cpe locus of strain NCTC8239. Beyond that conserved upstream region, significant sequence differences between these two cpe-positive type A isolates were detected. Notably, the IS1470 sequences present directly upstream of the IS1469 element of NCTC8239 were not apparent in the plasmid cpe locus of isolate F4969. However, a partial ORF was identified at the extreme 5′ end of the cpe-containing XbaI fragment cloned from F4969 (Fig. 1). That partial ORF, absent from the chromosomal cpe locus of NCTC8239 (3, 5), is predicted to encode the C-terminal half of a protein sharing partial amino acid homology with the cytosine methyltransferase encoded by Bacillus subtilis phage SPBc2.
Finally, our sequencing analyses also identified six sequences near the plasmid cpe gene of strain F4969 that resemble consensus binding sequences for the Hpr (hyperprotease-producing) regulatory protein of B. subtilis (3, 5). Five of those putative Hpr-binding sequences were found to share complete homology with the previously detected putative Hpr-binding sequences present near the chromosomal cpe gene of NCTC8239 (3, 5). However, the upstream Hpr-binding sequence nearest the F4969 cpe gene has a single nucleotide replacement (AATAGTGTT is present instead of AATAGTATT).
PCR evaluation of the association between the cpe gene and IS1469 in other type A strains.
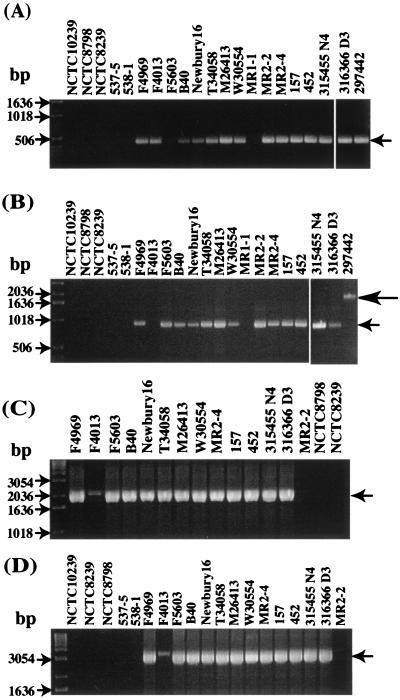
To further investigate previous suggestions (5, 8) that IS1469 sequences are consistently associated with the cpe gene of C. perfringens type A isolates, comparative PCR analyses were performed with a varied collection of cpe-positive, type A isolates (Table 1). Those analyses, using one primer specific for cpe ORF sequences and a second primer specific for IS1469 sequences, amplified an ∼1.7-kbp product using template DNA prepared from either F4969 or NCTC8239 (data not shown), which is fully consistent with expectations from KCpeFC5-1, KCpeFX28, and NCTC8239 sequencing results (3, 5; this study). A similar-sized PCR product was also amplified from all 16 other tested type A cpe-positive isolates (data not shown), regardless of their origin or cpe genotype (plasmid or chromosomal). However, no PCR product was amplified from the cpe-negative type A isolate MR2-2 (data not shown).
Detection of cytosine methyltransferase gene sequences (dcm sequences) in other C. perfringens strains.
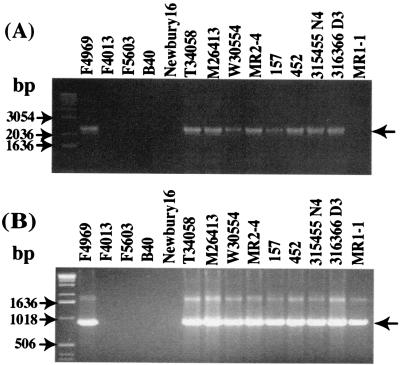
Following sequence analysis detection of putative dcm sequences near the F4969 cpe gene, two PCR studies were performed to evaluate whether similar dcm sequences might also be present in other C. perfringens type A isolates. As expected from KCpeFX28 sequencing results, a PCR assay using primers specific for internal F4969 dcm sequences amplified a single ∼0.4-kbp band from F4969 template DNA. A similar product was also observed when template DNA from 11 of 12 other type A isolates carrying a plasmid cpe gene was used (Fig. 3A).
FIG. 3.
PCR analyses of sequences upstream of the plasmid cpe gene of type A isolates. (A) Results of a dcm-specific PCR assay (expected ∼0.4-kbp product marked by arrow) with primers internal to F4969 dcm sequences. (B) Results of a PCR assay performed with primers designed to amplify a product from the DNA region between dcm sequences and sequences ∼400 bp downstream of them (based upon sequencing of the cpe-containing XbaI fragment of F4969). In this PCR assay, F4969 DNA yields an ∼0.9-kbp PCR product (small arrow), which is consistent with sequencing results for the F4969 plasmid cpe locus; note that one cpe-negative type A isolate, 297442, gave an ∼2.0-kbp product (large arrow). (C) PCR analysis of the association between dcm gene and IS1469 genes. An ∼2.1-kbp product (arrow) was detected in all type A isolates carrying a plasmid cpe gene. (D) PCR analysis of the association between dcm sequences and the cpe gene, which is consistent with sequencing results for the plasmid cpe locus of F4969. An ∼3.5-kbp product (arrow) was detected in all type A isolates carrying a plasmid cpe gene, which is consistent with sequencing results for the plasmid cpe locus of F4969. Numbers on the left indicate migration of molecular size markers.
To further confirm the presence of dcm sequences in many C. perfringens type A isolates carrying a cpe plasmid, Southern blotting analysis was performed with a probe specific for F4969 dcm sequences. This probe hybridized with XbaI-digested DNA from all 10 type A isolates carrying a plasmid cpe gene (including isolate F5603, which had tested negative by our dcm-specific PCR assay). In all cases, the dcm probe hybridized to an XbaI fragment similar in size to a fragment that hybridized with a cpe-specific probe (Fig. 4A and B). DNA from type A isolate 452, which carries a cpe plasmid, also had a second band hybridizing with the dcm-specific probe; however, this band did not hybridize with the cpe probe (Fig. 4A and B).
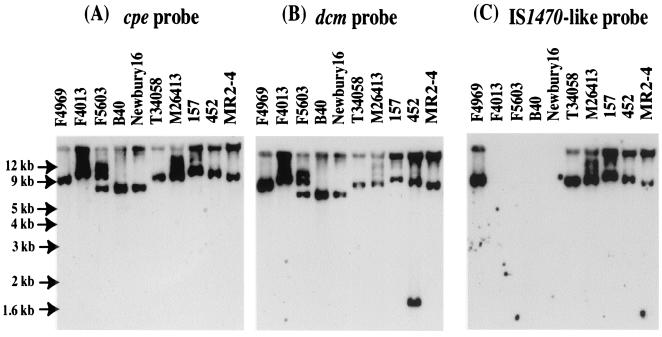
FIG. 4.
Southern blot analyses of cpe-positive C. perfringens type A isolates. Results shown are for type A isolates carrying a cpe plasmid hybridized with a cpe probe, a dcm probe, or an IS1470-like probe. The uppermost bands in panel C lie in the gel wells. The F5603 lane shows hybridization of the probes to incompletely digested DNA. Numbers on the left indicate migration of molecular size markers.
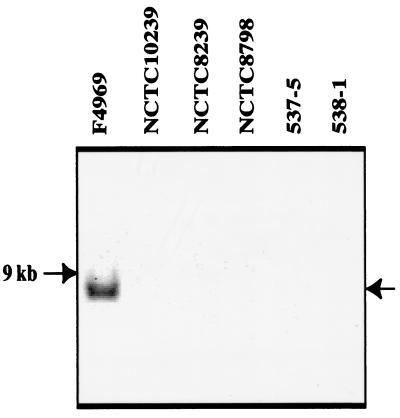
In contrast to the apparently widespread presence of dcm sequences in type A isolates carrying a plasmid cpe gene, template DNA from five type A food poisoning strains carrying a chromosomal cpe gene failed to amplify a product in our dcm-specific PCR assay (Fig. 3A). Moreover, Southern blot analysis detected no hybridization of a dcm probe to XbaI-digested DNA isolated from any of those five food poisoning isolates (Fig. 5). Finally, a PCR using dcm-specific primers and template DNA from three cpe-negative isolates (from both human and veterinary sources) amplified an ∼0.4-kbp product from two of three template DNAs of those type A isolates (Fig. 3A).
FIG. 5.
Southern blotting analysis of dcm sequences in C. perfringens type A food poisoning isolates. Note that the probe, generated by PCR with primers to F4969 dcm sequences and F4969 sequences ∼400 bp downstream of the dcm sequences, does not hybridize with DNA from any type A food poisoning isolates tested. However, this probe does react with DNA isolated from F4969.
A second PCR assay (using primers to F4969 dcm sequences and F4969 sequences ∼400 bp downstream of dcm) was then performed to further investigate the similarity of type A isolates carrying dcm sequences. As expected from KCpeFX28 sequencing results, this PCR amplified an ∼0.9-kbp product by using F4969 template DNA (Fig. 3B). A similar product was also amplified with template DNA from 11 of 12 other type A isolates carrying a plasmid cpe (Fig. 3B). Interestingly, template DNA from plasmid cpe type A isolate F5603, which had tested negative with the dcm-specific PCR but positive by Southern blotting with a dcm probe, did amplify the expected 0.9-kbp product of our second PCR. In contrast, template DNA from type A plasmid cpe strain F4013, which tested positive by dcm-specific PCR, gave no PCR product with that second PCR.
Our second PCR using one primer to F4969 dcm sequences and a second primer to F4969 sequences downstream of dcm also failed to amplify any product from five type A food poisoning strains carrying a chromosomal cpe gene or from MR1-1, a cpe-negative type A isolate (Fig. 3B). DNA from another cpe-negative type A isolate (canine diarrhea isolate 297442) did amplify a larger (∼2-kbp) product in this second PCR (Fig. 3B).
The possible presence of similar dcm sequences in other C. perfringens types was also investigated with representative (Table 1) type B, C, D, and E isolates. Template DNA prepared from isolates belonging to each C. perfringens type gave an ∼0.4-kbp PCR product using primers specific for F4969 dcm sequences (Fig. 6A). Our second PCR, using primers to the dcm sequences and adjacent downstream sequences of F4969 plasmid DNA, amplified an ∼2-kbp product with template DNA prepared from the representative type B, D, and E isolates (Fig. 6B). Both the ∼0.4 and ∼2-kbp PCR products from the type B, D, and E isolates also hybridized a dcm-specific probe (Fig. 6C and D).
The ∼2-kbp PCR products amplified by using template DNA from the tested type B and E isolates as well as from cpe-negative type A isolate 297442 were directly sequenced to determine the basis for these unexpectedly larger (∼2- versus 0.9-kbp) PCR products amplified with primers to dcm sequences and sequences downstream of dcm in F4969. The results confirmed the presence of similar dcm sequences in each of those isolates but also revealed the presence of an insertion containing an ∼0.7-kbp ORF that is predicted to encode a protein with partial homology to a hypothetical protein of Streptococcus pneumoniae bacteriophage MM1.
Comparative PCR analyses of DNA upstream of the plasmid cpe gene in type A isolates.
The relationship between dcm sequences and IS1469 in type A isolates was investigated with a PCR using one primer complementary to F4969 dcm sequences and a second primer specific for the IS1469 sequences of F4969 (Fig. 3C). Consistent with our KCpeFX28 sequencing results, this PCR amplified a product of ∼2.1 kbp using F4969 template DNA. A similar product was also amplified from all 12 other type A isolates carrying a plasmid cpe gene, although the amplification was weaker with F4013 template DNA. In contrast, two type A chromosomal-cpe isolates and one type A cpe-negative strain failed to amplify any PCR product in this reaction.
A second PCR was then performed using one primer complementary to dcm sequences and a second primer specific for cpe sequences (Fig. 3D). That PCR amplified a clear ∼3.5-kbp band using F4969 template DNA, consistent with KCpeFX28 sequencing results. A similar PCR product was also amplified with template DNA isolated from all 12 other type A isolates carrying a plasmid cpe gene, although the amplification was again weaker with template DNA from isolate F4013. However, five type A chromosomal-cpe strains and one type A cpe-negative strain failed to amplify any product in this PCR.
Comparative PCR and Southern blot analyses of the association between cpe and downstream IS1470-like sequences in C. perfringens type A isolates carrying a cpe plasmid.
To investigate possible similarity in the region downstream of the plasmid cpe gene among type A isolates, a PCR was performed with a primer pair specific for cpe sequences and the IS1470-like sequence present in F4969 (Fig. 7A). As expected from KCpeFX28 sequencing results, this PCR amplified a product of ∼2.2 kbp using F4969 template DNA. A similar product was also amplified with template DNA prepared from 8 of 12 other type A isolates carrying a plasmid cpe gene. By comparison, a chromosomal-cpe food poisoning isolate (not shown) and a type A cpe-negative isolate (Fig. 7A) both failed to amplify any product in this PCR.
FIG. 7.
PCR analyses of the region between the cpe gene and downstream IS1470-like sequences. (A) PCR product amplified from specified type A isolates using primers for cpe gene sequences and sequences present in IS1470-like sequences of F4969, which generate an ∼2.2-kbp product (arrow at right) with DNA from F4969. Numbers on the left indicate migration of molecular size markers. (B) PCR products amplified from specified type A isolates using primers specific for internal IS1470-like sequences. This reaction generated the expected 0.9-kbp product from F4969 DNA (arrow). Numbers on the left indicate migration of molecular size markers.
To further investigate the association between IS1470-like sequences and the cpe gene in type A isolates carrying a cpe plasmid, Southern blotting analyses were performed with a probe specific for the IS1470-like sequences of F4969. Those experiments (Fig. 4C) detected hybridization of the IS1470-like probe to XbaI-digested DNA isolated from five of nine type A isolates carrying a plasmid cpe gene. For each of those five isolates, a similar-sized XbaI fragment always hybridized to both the cpe and IS1470-like probes. The four type A isolates (F4013, F5603, B40, and NB16) carrying a plasmid cpe gene whose DNA failed to hybridize with the IS1470-like probe in the Southern blots (Fig. 4) were also unable to amplify a cpe-IS1470-like PCR product (Fig. 7A).
As a final assessment of the presence of IS1470-like sequences in type A isolates carrying a plasmid cpe gene, a second PCR was performed with primers to internal IS1470-like sequences (Fig. 7B). With those internal IS1470-like primers, template DNA from F4969 amplified the expected ∼0.9-kbp PCR product. The eight other type A plasmid cpe isolates whose DNA had amplified (Fig. 7A) a PCR product with cpe and IS1470-like primers also gave a similar ∼0.9-kbp PCR product with the internal IS1470-like primers. However, those same primers failed to support amplification of any PCR product (Fig. 7B) with template DNA from F4013, F5603, B40, or NB16, which is consistent with the fact that DNA from those four isolates failed to either hybridize an IS1470-like probe or amplify a cpe-IS1470-like PCR product (Fig. 4 and 7).
PCR detection of IS1151 sequences.
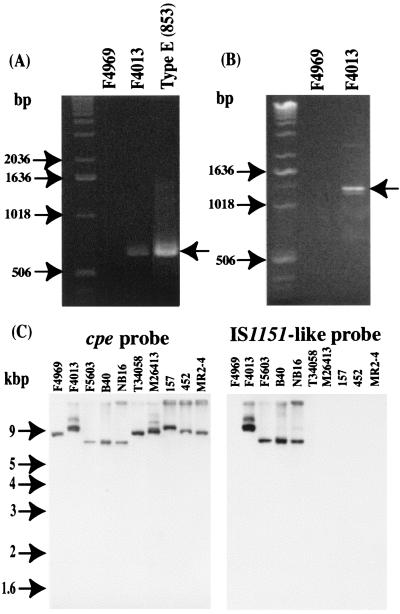
Since previous studies had detected IS1151 sequences near the plasmid cpe gene in two of three type A isolates (8), a PCR assay was performed with IS1151-specific primers and template DNAs prepared from our collection of type A isolates carrying a plasmid cpe gene. As shown in Fig. 8A, the internal IS1151 primers amplified a PCR product of ∼0.6 kbp using template DNA from type E isolate 853, consistent with previous sequencing results (1). A similar-sized PCR product was also detected when the same reaction was performed with template DNA prepared from type A plasmid cpe isolate F4013 (Fig. 8A). However, this PCR failed to amplify any product using template DNA from F4969 (Fig. 8A), the 11 other type A strains carrying a plasmid cpe gene that had also been tested in analyses whose results are shown in Fig. 7 (data not shown), or one type A cpe-negative strain (data not shown).
FIG. 8.
PCR detection of IS1151 sequences in C. perfringens type A isolates. (A) PCR results obtained with primers specific for IS1151 sequences which amplified (arrow at right) the expected ∼0.6-kbp product with total DNA from the previously sequenced IS1151 of type E isolate 853 (1). Numbers on the left indicate migration of molecular weight markers. (B) PCR results obtained with a primer specific for cpe sequences and a reverse primer specific for IS1151 sequences. Sequencing of the ∼1.2-kbp product obtained with F4013 DNA confirmed the inclusion of sequences spanning from cpe to IS1151. (C) Southern blot results obtained by hybridizing XbaI-digested DNA from type A isolates carrying a cpe plasmid with an internal IS1151-like probe.
To evaluate the proximity of the cpe gene to the putative IS1151 sequences of isolate F4013 and to further confirm the absence of IS1151 sequences in the other 12 type A isolates carrying a plasmid cpe gene examined by the internal IS1151 PCR, a second PCR was performed with one primer specific for cpe sequences and a second primer specific to IS1151 sequences. As shown in Fig. 8B, these primers amplified a PCR product using DNA from a single type A isolate carrying a cpe plasmid, i.e., F4013. No product was amplified from F4969 (Fig. 8B) or from the other 12 type A isolates also testing negative with the internal IS1151 PCR assay (data not shown).
Sequencing of the ∼1.1-kb cpe-IS1151 PCR product amplified from isolate F4013 DNA confirmed the presence of sequences resembling IS1151 ∼0.3 kbp downstream of the F4013 plasmid cpe gene (Fig. 1). The amplified F4013 sequences were determined to be ∼90% homologous to IS1151 but appear to be defective, as they contain several nonsense mutations. In addition, this sequencing analysis revealed that the region of F4013 DNA containing IS1151-like sequences also has an overlapping ORF (in a different reading frame from IS1151) sharing partial amino acid homology with transposase 11.
To further evaluate the presence of sequences with homology to IS1151 in the plasmid cpe locus of type A isolates, a Southern blot experiment was performed with a probe specific for internal IS1151-like sequences of isolate F4013. These analyses (Fig. 8C) demonstrated hybridization of the IS1151-like probe to a single XbaI DNA fragment from the type A plasmid cpe isolates F4013, F5603, B40, and NB16. For each of these isolates, a cpe probe also apparently hybridized to the XbaI fragment hybridizing to the IS1151-like probe. Coupling this observation with the apparent presence (Fig. 3) in these isolates of an F4969-like DNA region upstream of cpe (which should include the XbaI site of dcm sequences), the IS1151-like sequences of F5603, B40, and NB16 (like isolate F4013) appear to be located downstream of their cpe gene. No hybridization of the IS1151-like probe was observed with DNA prepared from any other type A isolates carrying a cpe plasmid (Fig. 8C), which all carry IS1470-like sequences downstream of their cpe gene (Fig. 7).
DISCUSSION
Recent studies (2, 3, 5-8, 23) established that most (or all) C. perfringens type A food poisoning isolates carry their enterotoxin gene (cpe) on the chromosome. Sequencing studies suggested that the chromosomal cpe gene of type A food poisoning isolate NCTC8239 (2, 3, 5, 10) may be present on Tn5565, a putative 6.3-kbp transposon with terminal IS1470 elements. Tn5565 has apparently inserted onto the C. perfringens chromosome between the purine permease (uapC) and quinolinate phosphorylribosyltransferase (nadC) genes (Fig. 1). Southern blot results suggest that other type A food poisoning strains have a similarly arranged chromosomal cpe locus (5, 8).
In contrast, most (if not all) enterotoxigenic C. perfringens type A isolates from non-food-borne human GI diseases or veterinary sources carry their cpe gene on a large plasmid. Prior to the present study, the organization of the plasmid cpe locus in type A isolates had not yet been thoroughly investigated, although an initial Southern blot study (8) suggested that the plasmid cpe locus of type A isolates substantially differs from the well-studied chromosomal cpe locus of NCTC8239. That study did not detect IS1470 sequences in the plasmid cpe locus of three tested type A isolates but instead found IS1151 sequences downstream of the cpe gene in two of those three isolates. Since those initial findings, the IS1151-containing cpe locus of F3686 (the only previously examined type A plasmid cpe isolate associated with human disease) has been considered representative of type A isolates carrying a cpe plasmid.
However, the apparent presence of IS1151 sequences in only two of the three initially examined isolates indicated that some variation must exist between the plasmid cpe loci of different type A isolates. To better evaluate the organization and diversity of the plasmid cpe locus in type A isolates, we first sequenced the plasmid cpe locus of F4969, a type A isolate from non-food-borne human GI disease used previously in both pathogenesis and genetic studies (4, 20). The arrangement determined for the plasmid cpe locus of F4969 (Fig. 1) was then compared, by PCR and Southern blotting, against the organization of the cpe locus in other type A strains carrying a cpe plasmid.
The results of this study are important for several reasons. First, the availability of sequence data for the F4969 plasmid cpe locus permits direct, detailed comparison of the plasmid cpe locus of a type A isolate against the previously sequenced chromosomal cpe locus of type A food poisoning strain NCTC8239 (3, 5, 10). Such comparisons indicate that the cpe loci of F4969 and NCTC8239 share nearly complete homology for ∼4 kbp. This conserved region extends ∼1.2 kbp downstream from the (∼1-kbp) cpe gene. Immediately beyond this common downstream region (Fig. 1), NCTC8239 carries IS1470 sequences, while F4969 has (in nearly the same position) IS1470-like sequences that are defective and present in the orientation opposite that of the NCTC8239 IS1470 sequences. Identification of IS1470-like sequences downstream of the plasmid cpe gene of F4969 contradicts previous studies that failed to find sequences with homology to IS1470 associated with the plasmid cpe locus of type A isolates (8).
With the exception of a 45-bp insert directly upstream of the F4969 cpe gene, F4969 and NCTC8239 were also found to share considerable homology for ∼1.8 kbp upstream of their cpe genes (Fig. 1). This result is consistent with previous proposals (3, 5, 8) that IS1469 sequences are located immediately upstream of the plasmid cpe gene in most (or all) type A isolates (see further discussion below). Approximately 250 bp beyond those shared IS1469 sequences, significant sequence divergence was detected between the cpe locus of F4969 and that of NCTC8239. In that divergent upstream region, F4969 lacks the IS1470 sequences present immediately upstream of the NCTC8239 IS1469 element (Fig. 1). Instead, sequences including the 3′ half of a putative cytosine methyltransferase gene (dcm) were identified ∼1.2 kbp upstream of the IS1469 element in isolate F4969 (Fig. 1).
This study also offers an improved assessment of plasmid cpe locus diversity among type A isolates. Using a collection of 13 type A isolates carrying a plasmid cpe gene from varied geographic and host or disease origins, it was determined that (as for all chromosomal-cpe isolates thus far examined [3, 5, 8; this study]) IS1469 sequences are present immediately upstream of the plasmid cpe gene in type A isolates. This finding further supports the already-mentioned proposals (3, 5, 8) of a consistent association between the cpe gene and IS1469. Additionally, dcm sequences (which are absent from chromosomal-cpe isolates) were localized to the same position upstream of the cpe gene in all 13 tested type A isolates carrying a plasmid cpe gene. Thus, results of this study strongly suggest that, for at least 3.5 kbp, sequences upstream of the cpe gene are conserved in most (or all) type A isolates carrying a cpe plasmid.
Our results also indicate that dcm sequences (when present) are not restricted to type A isolates carrying a cpe plasmid. Two of three cpe-negative type A strains from normal human and veterinary sources were also determined to carry dcm sequences, as were representative type B, C, D, and E isolates. Interestingly, a primer pair specific for dcm sequences and sequences located 400 bp downstream of dcm in strain F4969 DNA was able to amplify a larger (relative to the product generated with F4969 DNA) PCR product using template DNA from type A cpe-negative strain 297442 or representative type B, D, and E isolates, which all carry plasmids (21). Sequencing demonstrated that this larger PCR product has an insert containing an ORF with partial homology to a protein encoded by S. pneumoniae bacteriophage MM1. Since the dcm sequences of the F4969 cpe plasmid (and, by inference, other dcm-positive C. perfringens isolates) also share partial homology with a dcm gene found in B. subtilis phage SPBc2, future studies might explore whether some C. perfringens plasmids have a phage origin or have recombined with phage DNA. Further study is also warranted to examine whether dcm sequences are present on the virulence plasmids encoding beta, epsilon, and iota toxins in type B to E isolates. It also should be determined whether the detected dcm sequences actually express a functional cytosine methylase and, if so, whether that methylase plays any role in C. perfringens gene regulation.
The diversity of the DNA region downstream of the plasmid cpe gene of type A isolates was also addressed by the present study. When 13 randomly chosen type A plasmid cpe isolates from varied geographic and host or disease sources were surveyed, only 4 (∼30%) were found to carry IS1151-like sequences (which are defective) downstream of their cpe gene. Thus, contrary to initial indications (8), it now appears that organization of the plasmid cpe locus found in IS1151-like carrying type A isolates such as F3686 and F4013 is less representative than the IS1470-like containing plasmid cpe locus found in isolates such as F4969. The inability of our PCR assays to amplify a product from F5603, B40, or NB16 template DNA using primers specific for the IS1151-like sequences of F4013 suggests that diversity exists among the IS1151-like sequences of different type A isolates carrying a cpe plasmid. Finally, the failure of the present study to find more than two distinctly different types of organization in the plasmid cpe locus of type A isolates among 13 isolates suggests that that locus has only limited diversity.
The ever-increasing evidence for a close association between IS1469 sequences and the cpe gene in most (or all) type A isolates (3, 5, 8; this study) supports the possibility of chromosomal and plasmid cpe genes sharing a common origin. Since the NCTC8239 cpe gene is apparently (2, 5) located on Tn5565, a putative transposon with terminal IS1470 sequences that reportedly forms several circular intermediates (including an IS1469-cpe species without any IS1470 sequences), previous proposals (2, 5) that the plasmid cpe locus of type A isolates originated from integration of an IS1469-cpe genetic element into one or more C. perfringens plasmids remain viable. If so, the IS1470-like and IS1151-like sequences now present downstream of the plasmid cpe gene in type A isolates (8; this study) could reflect insertion events occurring after integration of the IS1469-cpe element onto plasmids.
Alternatively, discovery of IS1470-like sequences downstream of the plasmid cpe gene in F4969 and many other type A isolates opens the possibility of independent origins for the F4969-like and F4013-like plasmid cpe loci. A possible origin of the F4013-like plasmid cpe locus is discussed above, i.e., integration of an IS1469-cpe genetic element followed by a subsequent integration of an IS1151 element downstream of the cpe gene. In contrast, the F4969-like plasmid cpe locus could reflect integration of another cpe-containing genetic element (consisting of IS1469, cpe, and a single IS1470 sequence) onto a different C. perfringens plasmid. This hypothesis receives some support from previous PCR studies (2) purportedly identifying an IS1469-cpe-IS1470 circular intermediate in NCTC8239 DNA (2). However, for the F4969-like plasmid cpe locus to have this independent origin, some DNA rearrangements must have occurred during transposition to account for the reversed orientation of the IS1470-like sequences present in the F4969-like plasmid cpe locus. Finally, if IS1469-cpe and IS1469-cpe-IS1470 genetic elements have independently inserted onto different C. perfringens plasmids, the conserved nature of the upstream region of the plasmid cpe locus in most or all C. perfringens type A isolates, and the presence of dcm sequences in some cpe-negative isolates carrying plasmids, could suggest that plasmid DNA sequences downstream of dcm represent an insertional hot spot.
Regardless of how many different cpe-carrying genetic elements have integrated into C. perfringens plasmids, our results show that functional upstream and downstream IS1470 sequences are missing from the plasmid cpe locus of most (or all) type A isolates. If IS1470 insertion sequences are necessary for transposing the cpe gene of NCTC8239, as proposed elsewhere (2, 5), it is possible that the plasmid cpe gene of most type A isolates can no longer be mobilized from resident plasmids. This putative loss of function (i.e., mobilization of the cpe gene) could explain why large numbers of mutations, including nonsense mutations, are now present in the IS1470-like sequences of type A isolates carrying a plasmid with the F4969 type of plasmid cpe locus.
Finally, it is notable that an F4969-like plasmid cpe locus is also present in isolate 452, which was previously shown to lack a clonal relationship with F4969 (6). The presence of a similar plasmid cpe locus in two nonrelated type A isolates, from very different origins, is consistent with the possibility of these two type A isolates now sharing the same cpe plasmid. If true, this situation could reflect horizontal spread of a common plasmid carrying an F4969-like cpe locus to many other C. perfringens type A isolates, converting the recipients to enteric virulence. This hypothesis is consistent with recent studies demonstrating that the cpe plasmid of type A strain F4969 can transfer via conjugation (4). Studies are now under way to further assess the similarities between the cpe plasmids in different C. perfringens type A isolates and to evaluate if plasmids with the F4013 type of cpe locus organization can also transfer via conjugation.
Acknowledgments
This research was supported by Public Health Service grant AI19844 from the National Institute of Allergy and Infectious Diseases and by grant 2001-02517 from the Ensuring Food Safety Research Program of the U.S. Department of Agriculture.
We thank Shauna Sparks for technical assistance, Robert Carman for providing canine diarrhea isolates 315455N4, 316366D3, and 297442, and Julian Rood (Monash University, Melbourne, Australia) for helpful discussions.
Editor: D. L. Burns
REFERENCES
- 1.Billington, S. J., E. U. Wieckowski, M. R. Sarker, D. Bueschel, J. G. Songer, and B. A. McClane. 1998. Clostridium perfringens type E animal enteritis isolates with highly conserved, silent enterotoxin sequences. Infect. Immun. 66:4531-4536. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2.Brynestad, S., and P. E. Granum. 1999. Evidence that Tn5565, which includes the enterotoxin gene in Clostridium perfringens, can have a circular form which may be a transposition intermediate. FEMS Microbiol. Lett. 170:281-286. [DOI] [PubMed] [Google Scholar]
- 3.Brynestad, S., L. A. Iwanejko, G. S. A. B. Stewart, and P. E. Granum. 1994. A complex array of Hpr consensus DNA recognition sequences proximal to the enterotoxin gene in Clostridium perfringens type A. Microbiology 140:97-104. [DOI] [PubMed] [Google Scholar]
- 4.Brynestad, S., M. R. Sarker, B. A. McClane, P. E. Granum, and J. I. Rood. 2001. Enterotoxin plasmid from Clostridium perfringens is conjugative. Infect. Immun. 69:3483-3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brynestad, S., B. Synstad, and P. E. Granum. 1997. The Clostridium perfringens enterotoxin gene is on a transposable element in type A human food poisoning strains. Microbiology 143:2109-2115. [DOI] [PubMed] [Google Scholar]
- 6.Collie, R. E., J. F. Kokai-Kun, and B. A. McClane. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-foodborne human gastrointestinal diseases. Anaerobe 4:69-79. [DOI] [PubMed] [Google Scholar]
- 7.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornillot, E., B. Saint-Joanis, G. Daube, S. Katayama, P. E. Granum, B. Carnard, and S. T. Cole. 1995. The enterotoxin gene (cpe) of Clostridium perfringens can be chromosomal or plasmid-borne. Mol. Microbiol. 15:639-647. [DOI] [PubMed] [Google Scholar]
- 9.Czeczulin, J. R., R. E. Collie, and B. A. McClane. 1996. Regulated expression of Clostridium perfringens enterotoxin in naturally cpe-negative type A, B, and C isolates of C. perfringens. Infect. Immun. 64:3301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Czeczulin, J. R., P. C. Hanna, and B. A. McClane. 1993. Cloning, nucleotide sequencing, and expression of the Clostridium perfringens enterotoxin gene in Escherichia coli. Infect. Immun. 61:3429-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daube, G., P. Simon, and A. Kaeckenbeeck. 1993. IS1151, an IS-like element of Clostridium perfringens. Nucleic Acids Res. 21:352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daube, G., P. Simon, B. Limbourg, C. Manteca, J. Mainil, and A. Kaeckenbeeck. 1996. Hybridization of 2,659 Clostridium perfringens isolates with gene probes for seven toxins (α, β, ɛ, ι, θ, μ and enterotoxin) and for sialidase. Am. J. Vet. Res. 57:496-501. [PubMed] [Google Scholar]
- 13.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClane, B. A. 2001. Clostridium perfringens, p. 351-372. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers, 2nd ed. ASM Press, Washington, D.C.
- 15.McClane, B. A., D. M. Lyerly, J. S. Moncrief, and T. D. Wilkins. 2000. Enterotoxic clostridia: Clostridium perfringens type A and Clostridium difficile, p. 551-562. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 16.McClane, B. A., and J. I. Rood. 2001. Clostridial toxins involved in human enteric and histotoxic infections, p. 169-209. In H. Bahl and P. Dürre (ed.), Clostridia: biotechnology and medical applications. Wiley-VCH, Weinheim, Germany.
- 17.Melville, S. B., R. Labbe, and A. L. Sonenshein. 1994. Expression from the Clostridium perfringens cpe promoter in C. perfringens and Bacillus subtilis. Infect. Immun. 62:5550-5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts, I., W. M. Holmes, and P. B. Hylemon. 1986. Modified plasmid isolation method for Clostridium perfringens and Clostridium absonum. Appl. Environ. Microbiol. 52:197-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sambrook, J. E., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Sarker, M. R., R. J. Carman, and B. A. McClane. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946-958. [DOI] [PubMed] [Google Scholar]
- 21.Songer, J. G. 1996. Clostridial enteric diseases of domestic animals. Clin. Microbiol. Rev. 9:216-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Songer, J. G., and R. M. Meer. 1996. Genotyping of Clostridium perfringens by polymerase chain reaction is a useful adjunct to diagnosis of clostridial enteric disease in animals. Anaerobe 2:197-203. [Google Scholar]
- 23.Sparks, S. G., R. J. Carman, M. R. Sarker, and B. A. McClane. 2001. Genotyping of enterotoxigenic Clostridium perfringens isolates associated with gastrointestinal disease in North America. J. Clin. Microbiol. 39:883-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]