Abstract
Survival of macrophage microbicidal activity is a prerequisite for invasive disease caused by the enteric pathogen Salmonella enterica serovar Typhimurium. Flavohemoglobins, such as those of Escherichia coli, Salmonella, and yeast, play vital roles in protection of these microorganisms in vitro from nitric oxide (NO) and nitrosative stress. A Salmonella hmp mutant defective in flavohemoglobin (Hmp) synthesis exhibits growth that is hypersensitive to nitrosating agents. We found that respiration of this mutant exhibited increased inhibition by NO, whereas wild-type cells pregrown with sodium nitroprusside or S-nitrosoglutathione showed enhanced tolerance of NO. Most significantly, hmp mutants internalized by primary human peripheral monocyte-derived macrophages survived phagocytosis relatively poorly compared with similarly bound and internalized wild-type cells. That the enhanced sensitivity to macrophage microbicidal activity is due primarily to the failure of Salmonella to detoxify NO was suggested by the ability of l-NG-monomethyl arginine—an inhibitor of NO synthase—to eliminate the difference in killing between wild-type and hmp mutant Salmonella cells. These observations suggest that Salmonella Hmp contributes to protection from NO-mediated inhibition by human macrophages.
Salmonella enterica serovar Typhimurium causes human invasive disease, particularly in frail and immunocompromised individuals. In animal models of Salmonella disease, invasion of M cells or CD18-expressing mononuclear phagocytes within ileal mucosa is an initial step in the pathogenesis (38). Subsequent survival within macrophages is a key feature of Salmonella virulence in vivo. Salmonella actively resists killing by macrophages (12), a property that confers invasiveness on the microorganism and increases the likelihood of disease in the challenged host (31, 38). Mutants incapable of intracellular survival are nonvirulent in experimental infections (11). Macrophages phagocytize invading bacteria and kill them by a number of mechanisms, including production of antimicrobial peptides, lysosomal enzymes, reactive oxygen species (ROS), and reactive nitrogen species (RNS).
Macrophages generate nitric oxide (NO) in response to a variety of stimuli (43), and subsequent modification of NO (to produce NO2, N2O3, and S-nitrosothiols) and reactions with oxygen species (for example, with O2·− to form ONOO−) may result in enhanced bactericidal activity. There is continuing controversy over the role of NO and its products in phagocytic killing by human macrophages in contrast to murine macrophages, in which NO has an undisputed role (34, 39). In mice, failure to generate NO leads to susceptibility to infection with Salmonella serovar Typhimurium (22, 39). Both nitrosative and oxidative stress are exerted by murine macrophages via production of ROS and RNS during intracellular killing of Salmonella serovar Typhimurium (34, 39), though ROS are considered to have the dominant role, particularly within the period immediately following internalization (39).
Salmonella is adapted to the intracellular environment and has some characteristics which confer resistance to RNS. These include DNA repair systems (e.g., umuC and recBC), small thiol molecules (e.g., homocysteine), and detoxifying enzymes (e.g., copper zinc superoxide dismutase) (6, 7, 19). In addition, there is a family of bacterial hemoglobins (29) possessed by many bacteria, including Salmonella. Bacterial hemoglobins may be classified into three broad groups, namely, the small single-domain proteins widely distributed but of uncertain function (36, 41), the truncated globins that are 20 to 40 amino acid residues shorter than vertebrate hemoglobins (26), and the flavohemoglobins, which are now the best understood (29). The first gene for a microbial flavohemoglobin, hmp, was discovered in Escherichia coli (37) and encodes a monomeric, chimeric flavohemoprotein, comprising a single polypeptide of about 44 kDa. The C-terminal domain of Hmp resembles ferredoxin reductase, binds NAD(P)H, and transfers electrons to the heme in the globin domain via flavin adenine dinucleotide (1, 17). The N-terminal domain is homologous to animal and plant globins and contains a single heme B (37).
Evidence that flavohemoglobins are involved in resisting NO came first from the demonstration that the E. coli hmp gene is markedly up-regulated by addition of NO to cultures (28). The Salmonella serovar Typhimurium hmp gene is also up-regulated by spermine NONOate (3). Subsequently, mutants of both Salmonella serovar Typhimurium (4) and E. coli (24) defective in Hmp synthesis were shown to exhibit growth defects in the presence of NO or upon nitrosative stress and were more prone to killing by these agents. It is now recognized that E. coli Hmp detoxifies NO aerobically (29), via an oxygenase (14, 15) or denitrosylase (16) reaction producing nitrate, or anaerobically, by reducing NO to N2O (18). In E. coli, hmp transcription is also increased upon exposure to the redox cycling agent and superoxide generator methyl viologen (paraquat) (2, 23, 27), and hmp mutant cells are hypersensitive to killing by paraquat (24), but the physiological significance of this is not understood. However, a Salmonella hmp mutant is not more sensitive (4) than the isogenic wild-type strain to paraquat and H2O2; the effects of oxidative stress on hmp expression in Salmonella have not been reported.
In the present study, we tested the hypothesis that expression of Salmonella serovar Typhimurium Hmp contributes to intracellular survival following phagocytosis. Here we describe the novel finding that Hmp protects Salmonella from killing within human macrophages and attribute this to the NO-detoxifying activity of the protein.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
All studies were performed using wild-type Salmonella serovar Typhimurium ATCC 14028s or its isogenic derivative carrying an antibiotic resistance cassette inserted in the hmp gene (14028 hmp) (MCS2A) (4). Bacteria were cultured in TY broth (32) containing kanamycin (final concentration, 50 μg/ml). Where indicated, S-nitrosoglutathione (GSNO) or sodium nitroprusside (SNP) (both 100 μM) was added to liquid cultures. The optical density of the culture was measured spectrophotometrically at 600 nm, in cells with a pathlength of 1 cm. Salmonella serovar Typhimurium cells were transferred directly from nutrient agar plates (3 to 4 large colonies) into TY broth and incubated at 37°C with shaking until an optical density at 600 nm of 0.25 was achieved (mid-exponential phase), unless otherwise stated. Prior to experiments, bacteria were harvested and washed three times in phosphate-buffered saline (PBS). MICs and minimum bactericidal concentrations were determined for both strains by conventional methods.
Determination of respiration rates and the effect of NO on respiration of Salmonella serovar Typhimurium.
Bacteria were grown to stationary phase at 37°C with shaking (200 rpm), harvested by centrifugation, and washed once with HEPES buffer. Respiration rates were measured with a Clark type polarographic oxygen electrode (Rank Bros., Bottisham, Cambridge, United Kingdom) comprising a water-jacketed (37°C) Perspex chamber stirred magnetically (35). The suspension (2 ml) was supplemented with glucose (final concentration, 10 μM), and respiration rates were measured in the closed system. Additions of anoxic NO-saturated solutions (28) were made with a Hamilton microsyringe in the same way. The electrode was calibrated with air-saturated water (assumed to contain 220 μM O2). Anoxia was achieved by adding a few grains of sodium dithionite. The period of inhibition of respiration is calculated as the period between addition of the NO solution, when respiration becomes inhibited, and the point where oxygen uptake is reinitiated (35).
Human macrophages.
Primary human peripheral monocyte-derived macrophages (MDM) were isolated from healthy volunteers by density centrifugation of heparinized blood using standard techniques (30). Macrophages were cultured for 10 to 12 days in 24-well flat-bottom plates (106 cells/well), in RPMI 1640 medium (Gibco BRL) supplemented with 2 mM l-glutamine and 10% fetal calf serum, at 37°C in 95% air-5% CO2. Wells were incubated with RPMI 1640 containing 4% (vol/vol) bovine serum albumin for 15 min and then washed with RPMI 1640 immediately prior to experiments.
Macrophage phagocytosis of Salmonella serovar Typhimurium.
Suspensions of bacteria were declumped by vortexing twice for 30 s each time, resuspended to produce an inoculum of 2 × 107 CFU in 250 μl of RPMI 1640, and then added to wells containing macrophages. In some experiments, there was initial cultivation for 60 min at 4°C in order to allow binding but not internalization of bacteria. For microscopy, cells were fixed with 4% paraformaldehyde in PBS at pH 7.4 and were stained with the nucleic acid stain 4′,6′-diamidino-2-phenylindole (DAPI). Coverslips were removed, dried, mounted in Vectashield (Molecular Probes), and viewed at a magnification of ×1,000 by using a DMRB 1000 fluorescence microscope (Leica, Wetzlar, Germany). To measure internalization during warm incubation, cells were fixed with paraformaldehyde and then irrigated with PBS containing a 1:20-diluted fluorescein isothiocyanate-conjugated rabbit anti-Salmonella O antibody (Bacto) (to localize extracellular salmonellae), followed by secondary irrigation with a 1:20-diluted fluorescein isothiocyanate-conjugated goat anti-rabbit antibody (Sigma) and a DAPI counterstain (which identifies nucleic acid within eukaryotic and prokaryotic DNA). Internalization of bacteria was determined by subtracting the number of extracellular bacteria, identified by their colocalization with the fluorescein isothiocyanate-conjugated antibody, from the total number of bacteria exhibiting the DAPI stain, as previously described (30).
Assay of intracellular Salmonella viability.
Bacteria and macrophages were incubated at 4°C for 60 min, then washed and reincubated at 37°C as above, for 90 min (period of maximal internalization). The supernatant fluid was then aspirated, and wells were washed twice with PBS. Extracellular bacteria were killed by incubation with RPMI 1640 containing gentamicin at a concentration of 200 μg ml−1 at 37°C for 30 min. (The minimum gentamicin concentration required for total kill of a suspension of 2 × 107 CFU of bacteria in RPMI 1640 over 30 min [150 ml−1] was identical for the two strains of Salmonella used in these experiments.) Cells were washed twice in PBS and then incubated for a further 120 min at 37°C in RPMI 1640. The numbers of surviving intracellular bacteria were estimated by measuring viable counts from wells before and after lysis of cells with saponin (1%) by using a standard dilution technique. This was done both immediately prior to gentamicin treatment and 30, 60, 90, or 120 min after addition of gentamicin. To determine the effect of inhibiting the production of NO by NO synthase (NOS), macrophages were exposed to 1 mM l-NG-monomethyl arginine (L-NMMA) for 48 h prior to infection of macrophages with bacteria.
Assay of NO produced by MDMs.
Macrophage production of NO was measured by an assay of nitrite accumulation in the supernatant liquid over 6 h of incubation at 37°C. Nitrite was first reduced to NO gas in a mixture of sulfuric acid and potassium iodide and then quantified by chemiluminescence (Model Mk2B; Glaxo-SmithKline, Beckenham, United Kingdom). Nitrite concentrations were calculated from the integral of the detected signal over time and were compared to those of a series of standards. The assay was linear up to 500 μM nitrite, with a limit of detection of 0.2 μM nitrite per 100-μl sample. Data were recorded at the rate of 1 Hz by using Biopac data acquisition software (MP100; Biopac Systems, Goleta, Calif.) (8).
Statistical analysis.
Values are given as means ± standard errors of the means (SEM) unless stated otherwise. Differences between means of continuous values were evaluated for statistical significance by the Student t test or by means of a multivariate analysis of variance (ANOVA), as appropriate. Nonparametric data were analyzed by the Wilcoxon sign rank test, the Mann-Whitney U test, or the Freedman two way ANOVA, as appropriate. Statistical significance was established at a P value of <0.05.
RESULTS
Expression of hmp protects Salmonella from NO inhibition of respiration.
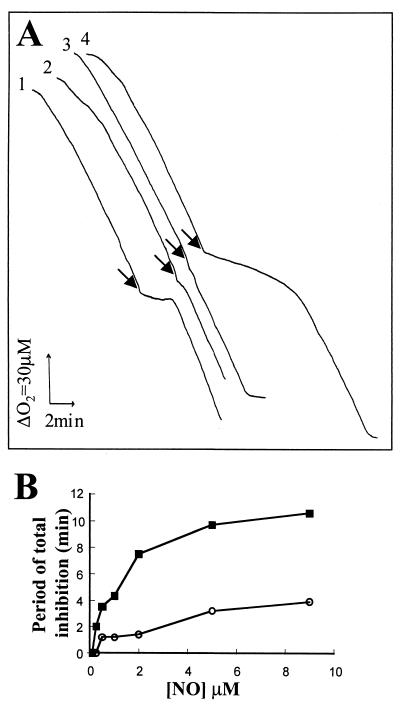
There was no significant difference between the rates of respiration of the wild-type and hmp mutant strains prior to NO addition (Fig. 1A; compare traces 1 and 4). However, the mutant was more sensitive to NO inhibition than the wild type, as judged by the period of inhibition after NO addition, as defined by Stevanin et al. (35) (Fig. 1B). Moreover, the rate of respiration of the wild type returned to its uninhibited level 2 min after addition of NO, while respiration of the hmp mutant recovered slowly and, at lower oxygen concentrations, failed to recover to preinhibition levels (data not shown). At a fixed concentration of O2 (approximately 60 μM) the period of inhibition by NO was dose dependent (Fig. 1B), and the mutant was considerably more sensitive. Thus, at 2 μM NO, the mutant was about five times more sensitive than the wild-type strain. Growing cells in the presence of the nitrosating agent SNP or GSNO significantly protected Salmonella serovar Typhimurium from subsequent inhibition of respiration with 9 μM NO (Fig. 1A; compare traces 2 and 3 with trace 1). Only when NO concentrations exceeded 25 μM could some inhibition be observed in cells preconditioned with these agents (data not shown). Thus, as in E. coli (35), Hmp provides inducible protection from transient inhibition of respiration.
FIG. 1.
Sensitivity of the Salmonella serovar Typhimurium hmp mutant to NO inhibition of oxygen consumption. Respiration rates of washed cell suspensions of strains 14028s (wild type) and 14028 hmp (MCS2A) were measured in an oxygen electrode apparatus. (A) A saturated anoxic solution of NO was added at intervals to produce (at each point indicated by arrows) a final concentration of 9 μM, when oxygen concentration was approximately 60 μM. Alongside the traces is a scale of the respiration rate expressed as a 30 μM change in O2 concentration and an elapse of 2 min of recording time. Traces (which have been offset for clarity) are as follows: trace 1, wild type; trace 2, wild type grown in the presence of 100 μM SNP; trace 3, wild type grown in the presence of 100 μM GSNO; trace 4, hmp mutant. (B) Relationship between NO concentration and inhibition of respiration of the wild type (open circles) and the hmp mutant (filled squares). The uninhibited rate of respiration was 60 nmol of O2/min/mg of cell protein in each case. Similar results were obtained in three separate experiments.
Binding and internalization of Salmonella serovar Typhimurium by human macrophages.
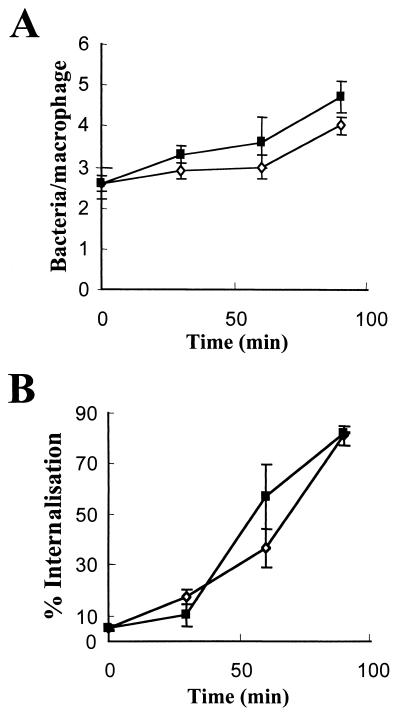
Both strains bound to the surfaces of cells at a density of 2.6 bacteria per macrophage (±0.4 for the hmp mutant and ±0.2 for the wild-type strain) (Fig. 2A). After further incubation of macrophages for 90 min at 37°C, the number of bacteria associated with macrophages increased for both strains (Fig. 2A). Maximal internalization of macrophage-associated hmp mutant bacteria (approximately 80% of those that were macrophage associated) was achieved after 90 min of incubation at 37°C. Although the hmp mutant exhibited slightly greater adherence and internalization than the wild type, this difference was not significant (Fig. 2B).
FIG. 2.
Binding and internalization of Salmonella serovar Typhimurium by human macrophages. Bacteria were incubated at 4°C for 60 min with macrophages, allowing binding but not internalization. Wells were fixed with 4% paraformaldehyde to determine bound bacteria. Thereafter, the trays were transferred to 37°C (time zero) to initiate internalization. Wells containing wild-type (open circles) or hmp mutant (solid squares) bacteria were scored after 30, 60, and 90 min of incubation at 37°C. The number of bound bacteria associated with each macrophage (A) was assessed microscopically. Internalization (B) is expressed as the percentage of total macrophage-associated bacteria that was internalized as observed by immunofluorescence, compared to the total number of bacterial cells stained with the nucleic acid stain DAPI. Data are means ± SEM from no fewer than five separate experiments performed in duplicate by using macrophages from different human donors.
Sensitivity of hmp mutants to macrophage microbicidal activity.
At the point of maximal internalization (90 min of warm incubation [see Fig. 2B]), saponin lysis of macrophages within wells yielded approximately equivalent numbers of viable bacteria of each strain: 24 × 105 ± 6 × 105 CFU (wild type) and 20 × 105 ± 5 × 105 (hmp) organisms (P > 0.05) (Fig. 3). No bacteria were recovered from similarly washed control wells that did not contain macrophages. Macrophages were then treated with gentamicin for 30 min, and at 30-min intervals over a subsequent 120-min chase, the number of viable hmp mutant bacteria collected after saponin lysis (Fig. 3) was consistently two to threefold lower than the number of viable wild-type bacteria recovered (P < 0.05).
FIG. 3.
Killing of Salmonella serovar Typhimurium by human macrophages. MDM infected with wild-type (solid bars) or hmp mutant (shaded bars) bacteria were incubated for 90 min at 37°C to allow internalization of bacteria. External bacteria were then killed with gentamicin for 30 min at 37°C. Samples were taken immediately before gentamicin treatment (0 h) and every 30 min thereafter for viable counts following saponin lysis. Results, expressed as CFU per milliliter, are means ± SEM from no fewer than six separate experiments performed in duplicate by using macrophages from different human donors. ∗, P < 0.05.
Effect of inhibition of NOS.
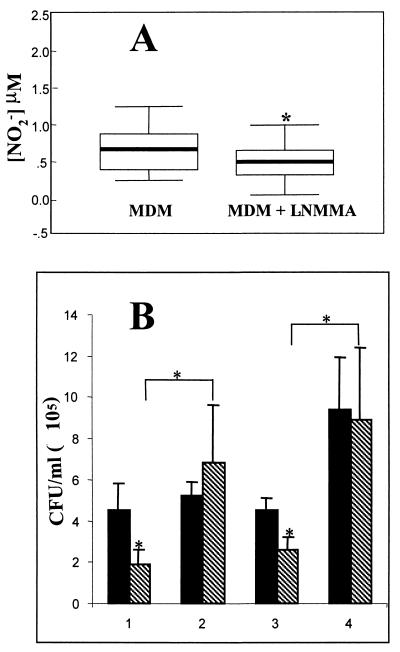
Treatment with L-NMMA (1 mM) for 48 h reduced release of NO by resting macrophages, as estimated by measurement of accumulated nitrite levels (Fig. 4A). Viable counts of bacteria recovered from macrophage lysates obtained at 30 and 120 min after gentamicin treatment are shown in Fig. 4B. In the absence of L-NMMA, the wild-type strain was, again, significantly more resistant to macrophage bactericidal activity at both 30 and 120 min after gentamicin treatment. Treatment with L-NMMA (Fig. 4B, data sets 2 and 4) significantly increased the resistance to killing of the hmp mutant and eliminated the difference in macrophage killing of wild-type and hmp mutant strains. Although there was a trend toward increased survival of wild-type Salmonella serovar Typhimurium in the presence of L-NMMA, this was not statistically significant.
FIG. 4.
Effect of the NOS inhibitor L-NMMA on intracellular survival of Salmonella serovar Typhimurium. (A) Effect of a 48-h pretreatment with L-NMMA on resting NO production, measured as the accumulation of NO2− in the supernatants of 12-day adherent MDM cultures over 6 h of incubation in fresh medium at 37°C (n = 8; P < 0.05). (B) Viable counts of Salmonella serovar Typhimurium within macrophage lysates infected with wild-type (solid bars) or hmp mutant (hatched bars) strains and incubated for 30 min (data sets 1 and 2) or 120 min (data sets 3 and 4) at 37°C, after addition of gentamicin to wells. Data sets 1 and 3 represent cultures not treated with L-NMMA, whereas data sets 2 and 4 represent cultures treated with 1 mM L-NMMA prior to infection of MDM with Salmonella. Results are means ± SEM from no fewer than six experiments performed in duplicate with macrophages from different donors. ∗, P < 0.05.
DISCUSSION
Studies with experimental animals suggest an important role for inducible NOS (iNOS) in host defense against Salmonella infections (34, 39). iNOS knockout mice show increased mortality during Salmonella serovar Typhimurium challenge (22, 34). Furthermore, macrophages derived from these animals display impaired killing of Salmonella serovar Typhimurium even during the early (<4 h) phase of intracellular processing of the organism (39). During this phase, NO provides microbicidal activity in concert with the relatively potent phagocyte respiratory burst. The role of NO in human host defense against Salmonella serovar Typhimurium is far less clear, but elevated production of NO has been detected in patients with bacterial gastroenteritis (10). Although most investigators have demonstrated NO production by human macrophages (43), the role of NO in the microbicidal activity of human macrophages remains controversial. Weinberg (43) has reviewed more than 100 examples of iNOS mRNA expression, iNOS protein expression, and NO production or NO-related antimicrobial activity of human mononuclear phagocytes. Humans with phox mutations resulting in chronic granulomatous disease suffer multiple infections as a result of the impaired phagocyte respiratory burst (25). Although no natural human deficiency of iNOS has been described, a point mutation in the promoter region of human NOS2 is associated with higher NOS activity and relative protection against malaria (20), suggesting a role for NO in natural protection of humans against infection. Some investigators have failed to demonstrate a role for NO in human macrophage microbicidal activity. One possible explanation for the successful demonstration of NO production and NO-related antimicrobial activity of human MDM in the present study is the prolonged in vitro cultivation (10 to 12 days) prior to experimentation. Martin and Edwards (21) showed that the role of reactive nitrogen intermediates in macrophage cytotoxic activity is much increased relative to that in immature cells.
The present study has demonstrated that Salmonella serovar Typhimurium Hmp protects the organism against NO-induced inhibition of respiration in vitro. We have not extended these experiments to precondition the wild-type and mutant strains with nitrosating agents or performed complementation studies of the hmp mutant with a plasmid expressing hmp, but these experiments will be done in the future. We have also demonstrated that Salmonella serovar Typhimurium protects the organism against killing by human macrophages during the first few hours following nonopsonic phagocytosis. That the latter effect was intracellular was suggested by equivalent internalization of the wild-type and mutant strains, and the absence of any difference in viability immediately prior to the gentamicin exclusion assay. Furthermore, treatment of macrophages with the iNOS inhibitor L-NMMA reduced resting NO production by adherent cultured macrophages and removed the survival advantage of the wild type over the hmp mutant following subsequent challenge with the organisms. Treatment with L-NMMA did not result in differential internalization of the two strains (data not shown). That L-NMMA resulted in incomplete blockade of nitrite production (Fig. 4) suggests that some nitrite may have arisen from unknown, non-NOS-derived pathways.
The microbicidal activity of NO against Salmonella is relatively weak compared to that of other reactive species such as peroxynitrite (5), but our data clearly show that at least some protection against intracellular killing is afforded by Hmp, presumably because of its ability to detoxify NO (29). That this effect was small (less than a threefold difference) may be a reflection of the relatively minor microbicidal activities of NO compared with those of the RNS, against which Salmonella might employ other mechanisms of defense. The additional significance of this study is that it supports an overall role for NO in intracellular killing of Salmonella serovar Typhimurium by human macrophages. A number of studies, reviewed by Vazquez-Torres and Fang (40), have failed to demonstrate a role for NO in macrophage killing of Salmonella, but this may be due partly to the greater magnitude of the microbicidal activity of phox mechanisms during the early phase of intracellular killing. It is significant that in Salmonella, mutation of the hmp gene leads to hypersensitivity to nitrosative stress generated by GSNO or S-nitrosylated N-acetylcysteine but not to oxidative stress generated by paraquat or H2O2 (4). Sensitivity to SIN-1 (3-morpholinosydnonimine hydrochloride), an in vitro generator of peroxynitrite, was also unaffected in the hmp mutant (4). These results indicate that the physiological role of Hmp in this pathogen is protection from nitrosative stress, and not from ROS or peroxynitrite, a product of the reaction of the superoxide anion and NO. These strains of Salmonella are therefore useful for examination of the contribution of NO to macrophage killing.
There is clear evidence that murine macrophage iNOS is activated and contributes to Salmonella killing very early on during phagocytosis (42), but Vazquez-Torres et al. (39) demonstrated that the NO-related bactericidal activity of murine macrophages during the first few hours of intracellular processing of Salmonella serovar Typhimurium is of relatively minor importance compared with their bacteriostatic activity 24 h after internalization. We have not examined the performance of the hmp mutant during extended cocultivation with human macrophages, because we find, like others (31), that Salmonella serovar Typhimurium is cytotoxic to human macrophages over 24-h cocultivation; our present data reveal significant differences in survival over the first few hours.
The experiments described here were performed without opsonization of Salmonella. Despite this, the organisms bound to the surfaces of macrophages in adequate numbers and were internalized presumably by pathogen-directed mechanisms. Although there is evidence for low-level production of complement proteins in colonic mucosa (13), we reasoned that in vivo, bacteria taken up into M cells may not be exposed to high concentrations of opsonins prior to contact with the epithelial surface and that a nonopsonic experimental procedure would most likely mimic the physiological interaction between Salmonella and host macrophages. Likewise, we did not activate macrophages with IFN-γ or IFN-α, which has been suggested to be a more potent inducer of iNOS in human macrophages (33), as Salmonella is likely to encounter resting macrophages during the early phase of invasion. This probably explains the relatively low concentrations of accumulated nitrite that we measured in the supernatants of cultured macrophages; it is noteworthy that an effect of Hmp on resistance to killing was observed despite this.
Our experimental methodology to compare intracellular survival relies heavily on the gentamicin exclusion assay to estimate numbers of viable intracellular organisms. This antibiotic penetrates cells relatively poorly, but it can enter by pinocytosis (9), and this is clearly possible at the concentration used in our experiments. The microbicidal activity of gentamicin is greatly reduced below pH 5.5 to 6.0, and internalized bacteria entering phagosomes will be exposed rapidly to pH values lower than this. It is unlikely that there was differential killing of wild-type and mutant bacteria by intracellular gentamicin, since their sensitivity to this antibiotic is identical, and although the difference in the numbers of viable intracellular bacteria observed could be explained by differences in compartmentalization, or some inhibitory effect upon phagosome acidification exhibited by the mutant but not the wild-type strains, the probabilities of these confounders seem remote. The modest reduction of CFU yielded from lysates of macrophages infected with the Salmonella hmp mutant may imply that NO may be bacteriostatic rather than bactericidal under the experimental conditions we used, which resulted in relatively low NO concentrations.
Flavohemoglobins have now been identified in a wide range of bacteria and yeasts, and a role in resisting nitrosative stress has been clearly demonstrated for the proteins from E. coli (15, 24), Ralstonia eutropha, and Saccharomyces cerevisiae (29). It is highly probable that other, related flavohemoglobins, particularly those that are up-regulated by nitrosative stress, have similar roles. This study has confirmed that Salmonella Hmp confers protection against nitrosative stress, and this may contribute to the distinctive pathogenic potential of this human pathogen.
Acknowledgments
We thank the Biotechnology and Biological Sciences Research Council (BBSRC) for Research Grant PRS12199 (to R.K.P. and Martin N. Hughes) and a Committee Studentship (to R.K.P. for T.M.S.). R.C.R. receives support from the Wellcome Trust for related work.
We are indebted to Martin N. Hughes for a gift of GSNO and advice on NO chemistry, to Margaret Lee for technical support, and to Pauline Whitaker for administrative assistance.
Editor: A. D. O'Brien
REFERENCES
- 1.Andrews, S. C., D. Shipley, J. N. Keen, J. B. Findlay, P. M. Harrison, and J. R. Guest. 1992. The haemoglobin-like protein (HMP) of Escherichia coli has ferrisiderophore reductase activity and its C-terminal domain shares homology with ferredoxin NADP+ reductases. FEBS Lett. 302:247-252. [DOI] [PubMed] [Google Scholar]
- 2.Anjum, M. F., N. Ioannidis, and R. K. Poole. 1998. Response of the NAD(P)H-oxidising flavohaemoglobin (Hmp) to prolonged oxidative stress and implications for its physiological role in Escherichia coli. FEMS Microbiol. Lett. 166:219-223. [DOI] [PubMed] [Google Scholar]
- 3.Crawford, M. J., and D. E. Goldberg. 1998. Regulation of the Salmonella typhimurium flavohemoglobin gene. A new pathway for bacterial gene expression in response to nitric oxide. J. Biol. Chem. 273:34028-34032. [DOI] [PubMed] [Google Scholar]
- 4.Crawford, M. J., and D. E. Goldberg. 1998. Role for the Salmonella flavohemoglobin in protection from nitric oxide. J. Biol. Chem. 273:12543-12547. [DOI] [PubMed] [Google Scholar]
- 5.De Groote, M. A., D. Granger, Y. Xu, G. Campbell, R. Prince, and F. C. Fang. 1995. Genetic and redox determinants of nitric oxide cytotoxicity in a Salmonella typhimurium model. Proc. Natl. Acad. Sci. USA 92:6399-6403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Groote, M. A., T. Testerman, Y. Xu, G. Stauffer, and F. C. Fang. 1996. Homocysteine antagonism of nitric oxide-related cytostasis in Salmonella typhimurium. Science 272:414-417. [DOI] [PubMed] [Google Scholar]
- 7.De Groote, M. A., U. A. Ochsner, M. U. Shiloh, C. Nathan, J. M. McCord, M. C. Dinauer, S. J. Libby, T. Vazquez, Y. Xu, and F. C. Fang. 1997. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 94:13997-14001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Demoncheaux, E. A. G., T. W. Higenbottam, P. J. Foster, C. D. R. Borland, A. P. L. Smith, H. Marriott, D. Bee, S. Akamine, and M. B. Davies. 2002. Circulating nitrite anions are a directly acting vasodilator and are donors for nitric oxide. Clin. Sci. (London) 102:77-83. [PubMed] [Google Scholar]
- 9.Drevets, D. A., B. P. Canono, P. J. Leenen, and P. A. Campbell. 1994. Gentamicin kills intracellular Listeria monocytogenes. Infect. Immun. 62:2222-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dykhuizen, R. S., J. Masson, G. McKnight, A. N. Mowat, C. C. Smith, L. M. Smith, and N. Benjamin. 1996. Plasma nitrate concentration in infective gastroenteritis and inflammatory bowel disease. Gut 39:393-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fields, P. I., R. V. Swanson, C. G. Haidaris, and F. Heffron. 1986. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 83:5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fields, P. I., E. A. Groisman, and F. Heffron. 1989. A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243:1059-1062. [DOI] [PubMed] [Google Scholar]
- 13.Fujiyama, A. A., H. Sakumoto, H. Uchihara, T. Kimura, S. Koyama, and T. Bamba. 1998. Detection of complement C3 and factor B gene expression in normal colorectal mucosa, adenomas and carcinoma. Clin. Exp. Immunol. 111:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gardner, P. R., A. M. Gardner, L. A. Martin, and A. L. Salzman. 1998. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc. Natl. Acad. Sci. USA 95:10378-10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner, P. R., A. M. Gardner, L. A. Martin, Y. Dou, T. Li, J. S. Olson, H. Zhu, and A. F. Riggs. 2000. Nitric-oxide dioxygenase activity and function of flavohemoglobins. Sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 275:31581-31587. [DOI] [PubMed] [Google Scholar]
- 16.Hausladen, A., A. Gow, and J. S. Stamler. 2001. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc. Natl. Acad. Sci. USA 98:10108-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ioannidis, N., C. E. Cooper, and R. K. Poole. 1992. Spectroscopic studies on an oxygen-binding haemoglobin-like flavohaemoprotein from Escherichia coli. Biochem. J. 288:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S. O., Y. Orii, D. Lloyd, M. N. Hughes, and R. K. Poole. 1999. Anoxic function for the Escherichia coli flavohaemoglobin (Hmp): reversible binding of nitric oxide and reduction to nitrous oxide. FEBS Lett. 445:389-394. [DOI] [PubMed] [Google Scholar]
- 19.Kosaka, H., Y. Oda, and M. Uozumi. 1985. Induction of umuC gene expression by nitrogen dioxide in Salmonella typhimurium. Mutat. Res. 142:99-102. [DOI] [PubMed] [Google Scholar]
- 20.Kun, J. F., B. Mordmüller, D. J. Perkins, J. May, D. Bercereau-Puijalan, M. Alpers, J. B. Weinberg, P. G. Kremsner, and Z. Lambarénè. 2001. Nitric oxide synthase (G-954C), increased nitric oxide production, and protection against malaria. J. Infect. Dis. 184:330-336. [DOI] [PubMed] [Google Scholar]
- 21.Martin, J. H. J., and S. W. Edwards. 1993. Changes in mechanisms of monocyte/macrophage-mediated cytotoxicity during culture. J. Immunol. 150:3478-3486. [PubMed] [Google Scholar]
- 22.Mastroeni, P., T. Vazquez, F. C. Fang, Y. Xu, S. Khan, C. E. Hormaeche, and G. Dougan. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. II. Effects on microbial proliferation and host survival in vivo. J. Exp. Med. 192:237-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Membrillo-Hernández, J., S. O. Kim, G. M. Cook, and R. K. Poole. 1997. Paraquat regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12 is SoxRS independent but modulated by σs. J. Bacteriol. 179:3164-3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Membrillo-Hernández, J., M. D. Coopamah, M. F. Anjum, T. M. Stevanin, A. Kelly, M. N. Hughes, and R. K. Poole. 1999. The flavohemoglobin of Escherichia coli confers resistance to a nitrosating agent, a “nitric oxide releaser,” and paraquat and is essential for transcriptional responses to oxidative stress. J. Biol. Chem. 274:748-754. [DOI] [PubMed] [Google Scholar]
- 25.Mouy, R., A. Fischer, E. Vilmer, R. Seger, and C. Griscelli. 1989. Incidence, severity, and prevention of infections in chronic granulomatous disease. J. Pediatr. 114:555-560. [DOI] [PubMed] [Google Scholar]
- 26.Pesce, A., M. Couture, S. Dewilde, M. Guertin, K. Yamauchi, P. Ascenzi, L. Moens, and M. Bolognesi. 2000. A novel two-over-two alpha-helical sandwich fold is characteristic of the truncated hemoglobin family. EMBO J. 19:2424-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pomposiello, P. J., M. H. J. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poole, R. K., M. F. Anjum, H. Membrillo, S. O. Kim, M. N. Hughes, and V. Stewart. 1996. Nitric oxide, nitrite, and Fnr regulation of hmp (flavohemoglobin) gene expression in Escherichia coli K-12. J. Bacteriol. 178:5487-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poole, R. K., and M. N. Hughes. 2000. New functions for the ancient globin family: bacterial responses to nitric oxide and nitrosative stress. Mol. Microbiol 36:775-783. [DOI] [PubMed] [Google Scholar]
- 30.Read, R. C., S. Zimmerli, C. Broaddus, D. A. Sanan, D. S. Stephens, and J. D. Ernst. 1996. The (α2→8)-linked polysialic acid capsule of group B Neisseria meningitidis modifies multiple steps during interaction with human macrophages. Infect. Immun. 64:3210-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richter, D., A. M. Buchan, and B. B. Finlay. 1997. Murine salmonellosis studied by confocal microscopy: Salmonella typhimurium resides intracellularly inside macrophages and exerts a cytotoxic effect on phagocytes in vivo. J. Exp. Med. 186:569-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Sharara, A. I., D. J. Perkins, M. A. Misukonis, S. U. Chan, J. A. Dominitz, and J. B. Weinberg. 1997. Interferon (IFN)-α activation of human blood mononuclear cells in vitro and in vivo for nitric oxide synthase expression: possible relationship of induced NOS2 to the anti-hepatitis C effects of IFN-α in vivo. J. Exp. Med. 186:1495-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiloh, M. U., J. D. MacMicking, S. Nicholson, J. E. Brause, S. Potter, M. Marino, F. Fang, M. Dinauer, and C. Nathan. 1999. Phenotype of mice and macrophages deficient in both phagocyte oxidase and inducible nitric oxide synthase. Immunity 10:29-38. [DOI] [PubMed] [Google Scholar]
- 35.Stevanin, T. M., N. Ioannidis, C. E. Mills, S. O. Kim, M. N. Hughes, and R. K. Poole. 2000. Flavohemoglobin Hmp affords inducible protection for Escherichia coli respiration, catalyzed by cytochromes bo′ or bd, from nitric oxide. J. Biol. Chem. 275:35868-35875. [DOI] [PubMed] [Google Scholar]
- 36.Tarricone, C., A. Galizzi, A. Coda, P. Ascenzi, and M. Bolognesi. 1997. Unusual structure of the oxygen-binding site in the dimeric bacterial hemoglobin from Vitreoscilla sp. Structure 5:497-507. [DOI] [PubMed] [Google Scholar]
- 37.Vasudevan, S. G., W. L. Armarego, D. C. Shaw, P. E. Lilley, N. E. Dixon, and R. K. Poole. 1991. Isolation and nucleotide sequence of the hmp gene that encodes a haemoglobin-like protein in Escherichia coli K-12. Mol. Gen. Genet. 226:49-58. [DOI] [PubMed] [Google Scholar]
- 38.Vazquez-Torres, A., C. Jones, A. J. Baumler, S. Falkow, R. Valdivia, W. Brown, M. Le, R. Berggren, W. T. Parks, and F. C. Fang. 1999. Extraintestinal dissemination of Salmonella by CD18-expressing phagocytes. Nature 401:804-808. [DOI] [PubMed] [Google Scholar]
- 39.Vazquez-Torres, A., J. Jones-Carson, P. Mastroeni, H. Ischiropoulos, and F. C. Fang. 2000. Antimicrobial actions of the NADPH phagocyte oxidase and inducible nitric oxide synthase in experimental salmonellosis. I. Effects on microbial killing by activated peritoneal macrophages in vitro. J. Exp. Med. 192:227-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vazquez-Torres, A., and F. C. Fang. 2001. Oxygen-dependent anti-Salmonella activity of macrophages. Trends Microbiol. 9:29-33. [DOI] [PubMed] [Google Scholar]
- 41.Wakabayashi, S., H. Matsubara, and D. A. Webster. 1986. Primary sequence of a dimeric bacterial haemoglobin from Vitreoscilla. Nature 322:481-483. [DOI] [PubMed] [Google Scholar]
- 42.Webb, J. L., N. W. Harvey, D. W. Holden, and T. J. Evans. 2001. Macrophage nitric oxide synthase associates with cortical actin but is not recruited to phagosomes. Infect. Immun. 69:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weinberg, J. B. 1999. Human mononuclear phagocyte nitric oxide production and inducible nitric oxide synthase expression, p. 95-150. In F. C. Fang (ed.), Nitric oxide and infection. Academic Press, New York, N.Y.