Abstract
In Listeria monocytogenes the acid tolerance response (ATR) takes place through a programmed molecular response which ensures cell survival under unfavorable conditions. Much evidence links ATR with virulence, but the molecular determinants involved in the reactivity to low pHs and the behavior of acid-exposed bacteria within host cells are still poorly understood. We have investigated the effect of acid adaptation on the fate of L. monocytogenes in human macrophages. Expression of genes encoding determinants for cell invasion and intracellular survival was tested for acid-exposed bacteria, and invasive behavior in the human myelomonocytic cell line THP-1 activated with gamma interferon was assessed. Functional approaches demonstrated that preexposure to an acidic pH enhances the survival of L. monocytogenes in activated human macrophages and that this effect is associated with an altered pattern of expression of genes involved in acid resistance and cell invasion. Significantly decreased transcription of the plcA gene, encoding a phospholipase C involved in vacuolar escape and cell-to-cell spread, was observed in acid-adapted bacteria. This effect was due to a reduction in the quantity of the bicistronic plcA-prfA transcript, concomitant with an increase in the level(s) of the monocistronic prfA mRNA(s). The transcriptional shift from distal to proximal prfA promoters resulted in equal levels of the prfA transcript (and, as a consequence, of the inlA, hly, and actA transcripts) under neutral and acidic conditions. In contrast, the sodC and gad genes, encoding a cytoplasmic superoxide dismutase and the glutamate-based acid resistance system, respectively, were positively regulated at a low pH. Morphological approaches confirmed the increased intracellular survival and growth of acid-adapted L. monocytogenes cells both in vacuoles and in the cytoplasm of interferon gamma-activated THP-1 macrophages. Our data indicate that preexposure to a low pH has a positive impact on subsequent challenge of L. monocytogenes with macrophagic cells.
Listeria monocytogenes is a widespread gram-positive bacterium (25) associated with severe food-borne disease (listeriosis) in humans (37, 41). To establish infection, orally ingested listeriae must reach the intestinal tract and invade the cells lining the intestinal epithelium (62). Along the route to target tissues, L. monocytogenes is faced with different stresses, including the low (<2.5) pH of the stomach (16), the deleterious effects of volatile fatty acids produced in the gut by sugar fermentation (28), and attack by macrophages (19). Like other gastrointestinal bacteria, L. monocytogenes has evolved different strategies for survival at low pHs, including the stationary-phase-dependent acid shock response and the logarithmic-phase-dependent acid tolerance response (ATR) (17, 29, 35).
The ATR is a complex phenomenon involving multiple systems for maintenance of intracellular pH homeostasis (16). The ATR has been correlated to the induction or down-regulation of as many as 53 proteins (43, 45) and to a modification of the fatty acid composition of the lipid layers of the bacterial membrane (60). As in other bacteria, the ATR in L. monocytogenes has been shown to require a two-component signal transduction system, which is essential for sensing different environmental stresses (15). Both the acid shock response and the ATR have been suggested to promote L. monocytogenes survival in acidic foods (9) and in gastric fluids (16), thereby contributing to the ability of this bacterium to cause infection (49).
The recently published complete genome sequence of L. monocytogenes EGD-e disclosed the existence of multiple genes involved in the homeostatic response to acidic stress (31). Among these, the most extensively characterized is the gene system coding for glutamic acid decarboxylases, which is structurally and functionally related to those present in other enteric bacteria (54) and whose expression is enhanced at low pHs (16). L. monocytogenes has evolved triplicate genes encoding putative isoforms of glutamic acid decarboxylase, here designated gadA, gadB, and gadB′, mapping at nucleotides (nt) 2503804 (lmo2434), 2433224 (lmo2363), and 481250 (lmo447), respectively (16, 31). As in Lactococcus lactis, gadB is preceded by and cotranscribed with gadC (lmo2362), encoding a putative glutamate:γ-aminobutyric acid (GABA) antiporter (16, 50). An inverted gene arrangement is observed for the gadB′ paralog, which is followed by the gadC′ antiporter gene (lmo448), resembling the gad (xas) system configuration of Escherichia coli and Shigella flexneri (18, 64). Evidence has emerged that L. monocytogenes strains differ in their levels of intrinsic acid tolerance (24) and that strain-dependent variations in Gad expression correlate with survival in the hyperacidic environment of the gastric juices (16).
While robust evidence linking the ATR with virulence has been obtained (39, 43, 49), the underlying molecular circuitry and the behavior of acid-stressed bacteria within host cells are not yet well known. Following penetration of the gut epithelium, the multiplication of L. monocytogenes in macrophages contributes to its spread to various target tissues (14, 21, 22, 38). In the case of murine macrophages there is evidence for simultaneous killing and survival of intracellular bacteria (19, 23). It has been shown that some macrophages kill L. monocytogenes while others do not, depending on intracellular iron levels in macrophages and the types of receptors expressed (26). In human THP-1 macrophages, which are normally not listericidal, bacteria are restricted to phagosomes and intracellular growth is prevented by preexposure to gamma interferon (IFN-γ) (44). Intracellular killing occurs through early challenge with the acidic pH in the phagolysosomal environment and activation of oxygen-dependent and -independent mechanisms, including the production of hydrogen peroxide and superoxide radicals (28).
In a previous report we have demonstrated that both invasion of human enterocyte-like cells and survival in lipopolysaccharide-activated murine macrophages were significantly enhanced for acid-adapted L. monocytogenes (13). The mechanisms involved in the entry and survival of L. monocytogenes in macrophages have been studied in the murine macrophage-like cell line J774 (19) and the human myelomonocytic cell line THP-1 (51). Phagocytosed L. monocytogenes cells dissolve the phagosomal and endosomal membrane and escape into the macrophagic cytoplasm. Then the bacteria move through polymerization of F-actin and coating with actin filaments (57). Several genes play a key role and are coordinately expressed during the infection process (reviewed in references 20 and 62): inlA, encoding internalin, a protein required for invasion of epithelial cells; hly, plcA, and plcB, encoding listeriolysin O (LLO) and two phospholipases, respectively, mediating in combination vacuolar escape and cell-to-cell spread; and actA, encoding a protein essential for actin-based intracellular mobility of the bacterium. Moreover, a zinc-metalloprotease, encoded by mpl, is involved in processing of PlcB by proteolytic cleavage in secondary vacuoles. Successful survival and multiplication in host cells may also depend on the expression of listerial stress proteins, including the sodC gene product, a manganese cytoplasmic superoxide dismutase (61), and the ClpC ATPase, encoded by the clpC gene (48). Remarkably, most genes which are essential for survival in phagocytic cells, namely, prfA, plcA, hly, mpl, actA, and plcB, are clustered in a single 10-kb chromosomal region (36).
Expression of the virulence gene cluster is under the control of the global regulatory gene prfA (8). The PrfA protein belongs to the Crp-Fnr family of transcriptional activators and recognizes 14-bp dyad-symmetry sites (PfrA boxes) within target promoters (52). Regulation of prfA is complex; the prfA gene is located in a dicistronic operon downstream of plcA and is transcribed from three differentially regulated promoters (42). The two prfA proximal promoters (P1prfA and P2prfA) generate overlapping monocistronic transcripts of 0.8 and 0.9 kb. Autogenous regulation of the system is ensured by the positive activity of PfrA at the upstream plcA promoter (PplcA), resulting in transcription of the plcA-prfA dicistronic messenger of 2.1 kb (27). Regulation of prfA expression via multiple promoters allows a sophisticated control of PrfA levels during various stages of intracellular parasitism (reviewed in reference 34), and this correlates, at least in part, with the observation that numerous PrfA-controlled genes (plcA, hly, inlC, actA, and plcB) are differentially expressed in murine macrophagic cells (5, 7, 33). An additional level of complexity of PrfA regulation is inferred by the observation that this protein may undergo strain-dependent sequence variation and posttranslational control, since its activity is influenced by numerous environmental cues as well as by host factors (3, 46, 63).
In the present work we report on the effect of acid adaptation on the fate of L. monocytogenes in human macrophages activated with IFN-γ. Expression of genes encoding determinants for cell invasion and intracellular survival was compared in acid-exposed and nonexposed bacteria, and their invasive behavior was assessed in a human myelomonocytic cell line (THP-1 cells) activated with IFN-γ. Induction of gad and sodC genes was observed in acid-adapted bacteria, concomitant with decreased transcription of the plcA gene. Functional and morphological approaches demonstrate increased intracellular survival and growth of acid-adapted L. monocytogenes cells both in vacuoles and in the cytoplasm of IFN-γ-activated human macrophages.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are L. monocytogenes LM2, previously isolated in our laboratory from the spinal fluid of a newborn with listeriosis, and reference strains ATCC 7644 and EGD (kindly provided by P. Cossart). All these L. monocytogenes strains belong to serotype 1. LM2 is a hemolytic wild-type strain capable of invading and multiplying in Caco-2 cells similarly to L. monocytogenes ATCC 7644 (10).
Bacteria were routinely grown in brain heart infusion broth (BHI; Oxoid), pH 7.2, and maintained by serial passages onto tryptone soy agar (TSA; Oxoid).
Acid adaptation of L. monocytogenes and ATR measurements.
Fourteen clinical isolates of L. monocytogenes, including strain LM2, and the two reference strains ATCC 7644 and EGD were preliminarily screened for acid tolerance (pH 3.5). Overnight cultures of L. monocytogenes in BHI were diluted (1:300) into fresh medium, and bacteria were grown at 37°C until the A600 of the culture reached approximately 0.15 (early log phase). Duplicate samples were centrifuged, and the pellets were resuspended in an equal volume of BHI, adjusted to pH 5.1 with 1 M lactic acid (acid-adapted bacteria) or to pH 7.2 (nonadapted bacteria), and incubated for 1 h at 37°C. These culture conditions were taken as the standard for further adhesion, invasion, and gene expression assays. To determine the ATR, cells were harvested by centrifugation and resuspended in BHI acidified to pH 3.5 with 3 M lactic acid. The acidified cultures were incubated for up to 2 h at 37°C. At given times (0, 1, and 2 h), samples were serially diluted in phosphate-buffered saline, and survival was determined by performing plate counts of viable cells (CFU) on TSA plates.
Transcriptional analysis of virulence genes in acid-adapted and nonadapted L. monocytogenes.
Total cellular RNA was extracted from acid-adapted and nonadapted cells of L. monocytogenes. A 1-μl sample from each culture was centrifuged at 10,000 × g for 5 min at room temperature. Cells were washed with 0.5 ml SET buffer (50 mM NaCl, 30 mM Tris-HCl, 5 mM EDTA [pH 8]) and harvested by centrifugation at 10,000 × g for 5 min at room temperature. The supernatant was discarded, and the bacterial pellet was suspended in 0.5 ml of ice-cold acetone and kept at 0°C for 10 min. Acetone was then completely decanted, and cells were resuspended in 0.5 ml of lysis buffer containing 10 U of M1-mutanolysin (Sigma Chemical Co., St. Louis, Mo.), 50 mM Tris-HCl (pH 6.5), and 10 mM vanadyl-ribonucleoside complex (Gibco BRL/Life Technologies). The final mixture was incubated for 30 min at 37°C and centrifuged at 15,000 × g for 15 min at 4°C to remove cell debris. The supernatant was extracted twice with phenol and once with chloroform. Nucleic acids were precipitated with 3 volumes of absolute ethanol in the presence of 0.3 M sodium acetate, resuspended in water, and treated with 20 U each of RNase-free DNase I and RNasin (Boehringer) in a volume of 50 μl. RNA was subsequently extracted twice with a phenol-chloroform mixture (1:1, vol/vol), ethanol precipitated, redissolved in water, and stored at −80°C.
For slot blot hybridization, 10-μl samples containing 5 μg of total RNA were mixed with 3 volumes of denaturing solution [1× MOPS solution, comprising 0.4 M 3-(N-morpholino)-propanesulfonic acid, 10 mM sodium acetate, 1 mM EDTA, 50% (vol/vol) formamide, 2.2 M formaldehyde], incubated at 65°C for 5 min, and poured on ice. Samples were mixed with 15 volumes of 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), pH 8, and vacuum blotted onto Hybond-C extra nitrocellulose membranes (Amersham Corp.) previously soaked in the same solution. For Northern blot hybridization, 10-μg samples of total RNA were denatured at 65°C for 15 min in the presence of 2 M formaldehyde and 50% formamide and then electrophoresed for 4 h at 4 V/cm on a 1% agarose gel containing 2 M formaldehyde in MOPS buffer (18). RNA was cross-linked to the membrane by baking at 80°C for 2 h. The total amount of RNA present in each slot was checked by probing the membranes with a 16S ribosomal DNA (rDNA) probe from L. monocytogenes LM2. Membrane hybridization and washings were carried out as previously described (12). Transcription of the actA, clpC, gadA, gadBC(B′C′), hly, inlA, plcA, prfA, and sodC genes was monitored by using intragenic probes generated by PCR with the oligonucleotide pairs listed in Table 1. For generation of the gadA/B probe, degenerated primers annealing to internal regions of both gadA and gadB were used. Because both gadA and gadB contain extended regions of homology with gadB′ (>90% over 243 nt and >70% overall), the gadA/B probe was expected to recognize gadB′-specific transcripts also. The 16S rDNA probe was obtained with eubacterial primers as described elsewhere (58).
TABLE 1.
List of oligonucleotide primers used in PCR amplification
| Primer paira | Accession no. | Sequence (nt position) | Product size (bp) |
|---|---|---|---|
| actA-F | AF281897 | 5′-GTGATAAAATCGACGAAAATCC-3′ (679-701) | 400 |
| actA-R | 5′-CTTGTAAAACTAGAATCTAGCG-3′ (1057-1079) | ||
| clpC-F | U40604 | 5′-TAGGGCTTGTAAGAGAAG-3′ (2350-2367) | 654 |
| clpC-R | 5′-CCACGATATTTTGTACCTG-3′ (3004-2986) | ||
| gadA/B-F | AF309076 | 5′-TCTAGTGAKGCGTGTATGCTTGG-3′ (445-467 for gadA; 1937-1959 for gadB) | 771 |
| gadA/B-R | AF309077 | 5′-TARTTTRTARCAKACGATTGGTAAG-3′ (1215-1191 for gadA; 2707-2683 for gadB) | |
| hly-F | M24199 | 5′-CGGAGGTTCCGCAAAAGATG-3′ (2532-2551) | 234 |
| hly-R | 5′-CCTCCAGAGTGATCGATGTT-3′ (2765-2746) | ||
| inlA-F | M67471 | 5′-GCCAACCTGTCACTATTGG-3′ (2952-2970) | 549 |
| inlA-R | 5′-CTTGATAGTCTACTGCTTG-3′ (3499-3481) | ||
| plcA-F | U25453 | 5′-TTGTTTTCACACTCGGACCA-3′ (166-185) | 491 |
| plcA-R | 5′-TAACGGAGACATGACGTGGA-3′ (656-637) | ||
| prfA-F | M55160 | 5′-GAAGTCATTAGCGAGCAGGCTACC-3′ (667-690) | 305 |
| prfA-R | 5′-CTAACAGCTGAGCTATGTGC-3′ (971-951) | ||
| sodC-F | M80526 | 5′-TGGCAATTTAAAAGCAGCAA-3′ (431-450) | 306 |
| sodC-R | 5′-TGCGTCAAAGCGTTTGTTAG-3′ (737-717) |
Except for the gadA/B primer pair, accession numbers are for both forward and reverse primers.
For RNA slot blot hybridizations, DNA probes were labeled by PCR with the PCR DIG Probe Synthesis kit (Boehringer), using digoxigenin (DIG)-11-dUTP as directed by the manufacturer. The annealing temperatures were 55 to 60°C for all primer pairs. Amplification reactions were carried out in 50 μl containing 1× PCR buffer (Perkin-Elmer), 1.5 μM MgCl2, 200 μM each deoxynucleoside triphosphate, 10 μM each primer, 2.5 U of Taq polymerase (Perkin-Elmer), and 1 ng of total DNA from L. monocytogenes LM2. A total of 35 cycles were performed; each cycle comprised 30 s at 95°C, 1 min at the annealing temperature, and 45 s at 74°C. Filters were developed with a commercial chemiluminescence-based detection kit (Boehringer), and signals were detected by exposure to a Kodak XAR film. Comparative estimation of transcript levels was performed by membrane analysis with a Molecular Analyst-Imaging Densitometer (model GS670; Bio-Rad), and results were normalized according to the level of 16S RNA.
For Northern blot analysis, PCR-generated fragments internal to the coding regions of the gadA/B, plcA, and prfA genes were labeled with [α-32P]dATP by use of a commercial random priming kit (Boehringer) and were used for hybridization at ≈104 cpm/cm2 of filter. After washings, individual membranes were first exposed to Kodak XAR film and then analyzed in a Storm electronic autoradiographer (Molecular Dynamics) to quantify the total amount of bound probe for each RNA sample and the signal intensities of individual bands. Results were adjusted by subtracting the background level of the filter.
Computational DNA and RNA analysis.
The presence of putative prokaryotic promoter elements upstream of gad genes was predicted by the Neural Network Promoter Prediction (NNPP) software, available at http://www.fruitfly.org/seq_tools/promoter.html. The NNPP program is a time-delay neural network which allows recognition of about 50% prokaryotic promoters with 0.3% false positivity using a score threshold of 0.9. The prediction was then confirmed by using the HBR software, available at http://genomic.sanger.ac.uk/gf/gf.shtml. Putative promoter elements were considered when the score was ≥0.9. RNA secondary-structure predictions were performed with the mfold software (version 3.1), available at http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi (40). Potential stem-loop-like elements involved in the generation of the 3′ termini of transcripts were considered when their ΔG° was ≤−25 kcal/mol.
THP-1 cells.
THP-1 cells, a myelomonocytic cell line derived from the blood of a 1-year-old boy with acute monocytic leukemia (59), were maintained in RPMI 1640 medium supplemented with 10% fetal calf serum and 2 mM glutamine in an atmosphere of 95% air and 5% CO2. Cells which grow spontaneously in loose suspension under these conditions were subcultured every third day by gentle shaking followed by pelleting and reseeding at a density of approximately 106 cells per ml. THP-1 cells were differentiated by incubation with phorbol myristate acetate (0.16 μM; Sigma Chemical Co.) for 48 h at 37°C in 24-well Nunc plates by following the procedure described by Scorneaux et al. (51). Cells that had become adherent were activated by exposure to IFN-γ (100 U/ml; specific activity, 2 × 107 U/mg of protein) for 24 h at 37°C, according to the work of Ouadrhiri et al. (44). Human recombinant IFN-γ was purchased from Roche Diagnostics (Mannheim, Germany) and stored in aliquots at −20°C.
Adhesion and invasion assays.
Infection was performed in nonactivated and IFN-γ-activated adherent THP-1 cell monolayers. Adhesion of L. monocytogenes was determined by adding logarithmically grown nonadapted and acid-adapted bacteria at a multiplicity of infection (MOI) of approximately 100 bacteria per cell, followed by incubation at 4°C for 1 h. Then cells were carefully washed five times in RPMI medium to remove unattached bacteria and were lysed by addition of ice-cold 0.1% Triton X-100. Invasion assays were performed by incubating logarithmically grown nonadapted and acid-adapted bacteria for 1 h at 37°C at an MOI of approximately 10 bacteria per cell. Then the macrophages were washed five times in RPMI medium and incubated at 37°C with the same medium, supplemented with 5 μg of gentamicin/ml to kill extracellular bacteria. One, three, and five hours after infection, monolayers were lysed by addition of ice-cold 0.1% Triton X-100.
Viable adherent or intracellular bacteria were determined by plate counts. Adhesion was expressed as the percentage of inoculated bacteria which adhered to THP-1 cells. Invasion was expressed as the percentage of the initial inoculum of bacteria that was gentamicin resistant at 1 and 5 h postinfection. Intracellular growth was also expressed as the replication index (RI), corresponding to the number of CFU at a given time (3 or 5 h postinfection) divided by the number of CFU at time zero (1 h postinfection).
Transmission electron microscopy.
Macrophages infected with acid-adapted or nonadapted L. monocytogenes LM2 (100 bacteria per cell) were fixed at 5 h after addition of gentamicin-containing medium in 2.5% cacodylate-buffered (0.1 M; pH 7.2) glutaraldehyde for 1 h at room temperature and were postfixed in 1% OsO4 for 1 h. Fixed specimens were dehydrated through a graded series of ethanol solutions. They were then scraped off the surfaces of culture dishes and embedded in Agar 100 (Agar AIDS, Cambridge, Essex, United Kingdom). Serial ultrathin sections were collected on 200-mesh grids and then counterstained with uranyl acetate and lead citrate. Sections were observed on a Zeiss 902 transmission electron microscope at 80 kV.
Statistical analysis.
Values were expressed as means ± standard deviations (SD). The statistical significance of differences between different experimental conditions was determined by the paired Student t test.
RESULTS
Survival of L. monocytogenes at an acidic pH.
Among different L. monocytogenes strains that were preliminarily screened for low pH tolerance, the clinical isolate LM2 was selected for its strong ATR. As an example, a comparison of the acid-resistant phenotypes of L. monocytogenes LM2 and ATCC 7644 is shown in Fig. 1. Exponentially grown nonadapted cells of both strains were rapidly killed upon exposure to pH 3.5 (14- to 33-fold reduction in viable counts within 2 h). When LM2 cells were preexposed to a moderate, nonlethal acidic pH (pH 5.1) for 1 h at 37°C before challenge at pH 3.5, they were able to mount a strong ATR which resulted in elevated survival (87% recovery) after 1 h at pH 3.5. In contrast, a substantial reduction in cell viability was observed for acid-adapted ATCC 7644 cells (11% recovery). In the same experimental setting, an even more pronounced reduction in cell viability (4% recovery) was observed for L. monocytogenes EGD (data not shown), consistent with the previously reported acid-sensitive phenotype of this prototypic strain (16).
FIG. 1.
ATR in wild-type L. monocytogenes LM2 and ATCC 7644. Survival rates are reported for acid (pH 5.1)-adapted (•) and nonadapted (○) strain LM2 and for acid-adapted (▪) and nonadapted (□) strain ATCC 7644. Bacterial counts were performed at different times of incubation in BHI acidified at pH 3.5. Error bars, SD for five independent experiments.
Transcriptional response of L. monocytogenes virulence-related genes to acid adaptation.
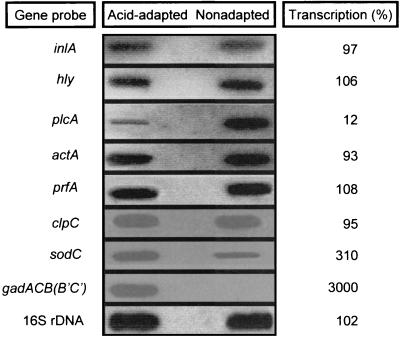
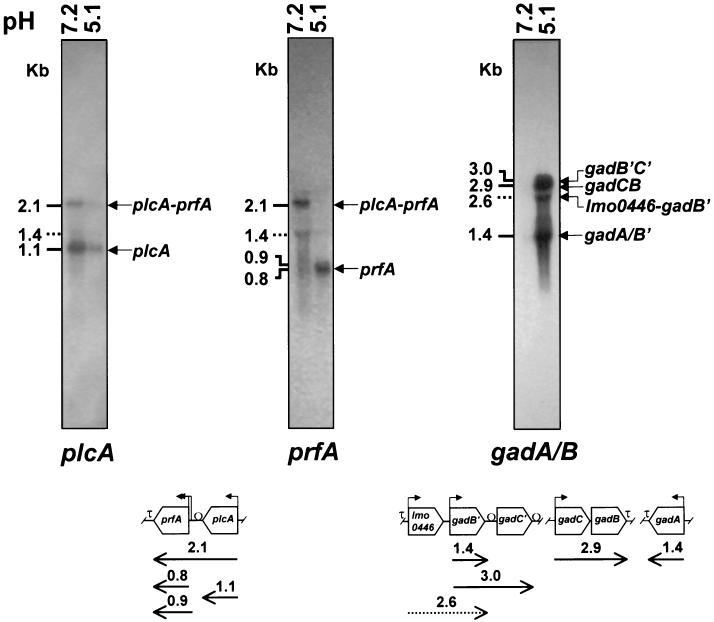
To investigate the effect of acid adaptation on the expression of L. monocytogenes virulence-related genes, RNA slot blot hybridization assays were preliminarily performed with total bacterial RNA extracted from acid-adapted and nonadapted LM2 cells (Fig. 2). Internal fragments of genes implicated in the invasive pathway (inlA, hly, plcA, and actA), in the regulation of virulence (prfA), and in the stress response (clpC, gadB, and sodC) were used as probes. The hybridization profiles of total RNA with the prfA, inlA, hly, actA, and clpC probes appeared similar in exponential-phase cultures of L. monocytogenes LM2 at pH 5.1 and 7.2. A dramatic up-regulation (>50-fold increase) of the gad gene system was observed in acid-exposed bacteria, while transcription of sodC was moderately enhanced (3-fold increase). Interestingly, transcription of the plcA gene was strongly decreased during growth at pH 5.1 (eightfold reduction), though transcription of the downstream gene prfA was unchanged. This finding suggests that acid sensing could differently affect transcription starting from the tandemly arranged prfA and plcA promoters. To address this point, the levels of prfA- and plcA-specific transcripts in acid-exposed and nonexposed cells were analyzed by quantitative Northern blot assays. For an in-depth transcriptional analysis of the acid-inducible gad gene system, duplicate RNA preparations were also hybridized with the gadB probe. The results shown in Fig. 3 provide an explanation for the pH-dependent differential control of plcA and prfA transcription. Comparative hybridization analysis with the plcA probe of total RNAs extracted from neutral (pH 7.2) and acidic (pH 5.1) cultures shows that the levels of both the dicistronic plcA-prfA transcript and the more abundant monocistronic plcA transcript are cumulatively fivefold reduced in acidic cultures, with negligible differences in reduction rates of individual mRNA species. Accordingly, RNA hybridization analysis with the prfA probe barely detected the plcA-prfA transcript in acid-exposed cells, while the dicistronic mRNA represented the major prfA transcript at the neutral pH. Despite the fact that overall levels of prfA-specific messengers were not significantly affected by pH, the pattern of prfA transcription was clearly pH dependent. Thus, the 2.1-kb plcA-prfA transcript was prevalent at the neutral pH, while the 0.8-kb prfA transcript represented the main species at the acidic pH. Under this condition, the dramatic decrease in the quantity of the dicistronic plcA-prfA transcript was fully compensated for by a comparable increase in the quantity of the monocistronic prfA messenger. A similar transcriptional shift from distal to proximal prfA promoters was observed also in acid-exposed L. monocytogenes EGD (data not shown). Interestingly, in L. monocytogenes LM2 there was a substantially larger amount of prfA transcript starting from the downstream P2 promoter (0.8-kb mRNA) than from the upstream P1 promoter (0.9-kb mRNA), irrespective of pH.
FIG. 2.
RNA slot blot analysis of actA, clpC, gad, hly, inlA, plcA, prfA, and sodC transcripts in nonadapted and acid-adapted L. monocytogenes LM2. Total RNAs were extracted from exponential cultures (A600 ≅ 0.4) of L. monocytogenes LM2 in BHI at pH 5.1 and 7.2. The amount of bound probe for each RNA slot was normalized by the amount of bound 16S rRNA probe. The percentage of transcription is relative to that for the non-acid-adapted condition.
FIG. 3.
Transcriptional analysis of gad, plcA, and prfA genes in nonadapted and acid-adapted L. monocytogenes LM2. (Top) Gels show the Northern blot hybridization of total RNAs extracted from exponential cultures (A600 ≅ 0.4) of L. monocytogenes LM2 in BHI at pH 7.2 and 5.1, as indicated above each lane. Aliquots (10 μg) of total RNA were subjected to electrophoresis, transferred onto nitrocellulose filters, and hybridized with [α-32P]dATP-labeled probes, as indicated at the bottom of each gel. RNA standards of 4.06, 2.19, 1.70, 1.54, 0.55, and 0.12 kb (not shown) were used for size estimation of transcripts. The positions of predicted transcripts are indicated by arrows on the right, and their sizes are shown on the left. Transcripts of uncertain assignment are marked with dotted lines indicating their size. (Bottom) Schematic representation (not drawn to scale) of the L. monocytogenes chromosomal regions containing the plcA-prfA, lmo0446-gadB′-gadC′, gadC-gadB, and gadA genes. Predicted or experimentally confirmed promoter elements are indicated by bent arrows. Gene orientations and positions of transcriptional terminators (τ) are given according to the published genome sequence (31). The Ω symbols map stem-loop-like structures likely involved in generation of the 3′ termini of transcripts, either by RNA processing or by protection from exonucleolytic cleavage.
The Northern blot results shown in Fig. 3 also confirmed that gad genes are not transcribed at a neutral pH (in BHI at pH 7.2), while dramatic up-regulation of the whole gad gene system (27-fold overall induction) occurs during exponential growth in mildly acidic medium (pH 5.1). A complex transcriptional profile was observed, due to the generation of four different-sized mRNA species. The longer transcripts of 3.0 and 2.9 kb can be predicted to be the dicistronic gadB′C′ and gadCB mRNAs, respectively, since their sizes are consistent with the lengths of the duplicate GABA antiporter/glutamic acid decarboxylase gene systems of L. monocytogenes (approximately 1.4 and 1.5 kb for gadB/B′ and gadC/C′, respectively, plus an 84-bp gadB′C′ intergenic spacer that is 12 nt in gadCB [see Fig. 3]). The shorter transcript of 1.4 kb matches the expected size of the triplicate paralogs encoding the three putative isoforms of glutamic acid decarboxylase (gadA, gadB, and gadB′) of L. monocytogenes. Based on sequence analysis (Fig. 3), the gadCB transcript is predicted to lack intergenic processing sites, implying that gadB should not contribute to the generation of these mRNA species. Interestingly, the levels of the dicistronic (3.0- and 2.9-kb) and monocistronic (1.4-kb) transcripts were similarly increased following acid exposure (14- and 15-fold, respectively), though the extent of induction was lower than that determined by whole-lane comparison of Northern blots and by RNA slot blot hybridization. This can be ascribed to a substantial fraction of smeared RNA hybridizing with the gadA/B probe and to the presence, in acid-exposed cells, of a 2.6-kb transcript accounting for 4% of the total β-emission of the RNA sample. Based on length estimation, this minor transcript could originate either from processing of the larger mRNA species at intragenic sites or, more likely, from readthrough transcription starting from gene lmo446 (1 kb), encoding a putative conjugated bile acid hydrolase, and ending at a stem-loop-like structure (ΔG° = −27.4 kcal/mol) within the intergenic spacer downstream of gadB′ (lmo447). A similar pattern of gad transcripts was observed in L. monocytogenes EGD, but the quantity of individual transcripts was approximately sixfold lower in this acid-sensitive strain than in LM2 (data not shown).
Survival and intracellular growth of acid-adapted L. monocytogenes in nonactivated and IFN-γ-activated THP-1 macrophages.
To evaluate the effect of the ATR on the early interactions between L. monocytogenes and macrophage surfaces, assays of adherence to nonactivated or IFN-γ-activated THP-1 macrophages were performed with nonadapted and acid-adapted L.monocytogenes LM2. Table 2 shows that the adhesion abilities of bacteria (at an MOI of approximately 100 bacteria per cell) after a 1-h incubation at 4°C were similar irrespective of whether they had been preexposed to mild acidic conditions (pH 5.1) or not.
TABLE 2.
Adhesion and intracellular viability of acid-adapted and nonadapted L. monocytogenes LM2 cells with regard to nonactivated and IFN-γ-activated THP-1 macrophages
| THP-1 macrophages | % Adherent bacteriaa ± SD
|
% Intracellular bacteriab ± SD at the indicated time after infection
|
||||
|---|---|---|---|---|---|---|
| Nonadaptedc | Acid adaptedd | Nonadapted
|
Acid adapted
|
|||
| 1 h | 5 h | 1 h | 5 h | |||
| Nonactivated | 4.6 ± 0.5 | 5.0 ± 0.6 | 10.1 ± 2.1 | 33.0 ± 3.1 | 8.0 ± 1.9 | 31.9 ± 3.2 |
| IFN-γ activated | 4.7 ± 0.6 | 5.2 ± 0.7 | 6.5 ± 1.5 | 7.1 ± 2.0 | 6.5 ± 1.6 | 31.0 ± 2.7 |
Adherence is expressed as the percentage of the initial inoculum of bacteria that adhered to macrophages 1 h after infection at 4°C. Data are means ± standard deviations from at least three experiments.
Intracellular growth is expressed as the percentage of the initial inoculum of bacteria that were gentamicin resistant at 1 and 5 h postinfection. Data are means ± standard deviations from at least six experiments.
Acid adapted, pH 5.1.
Nonadapted, pH 7.2.
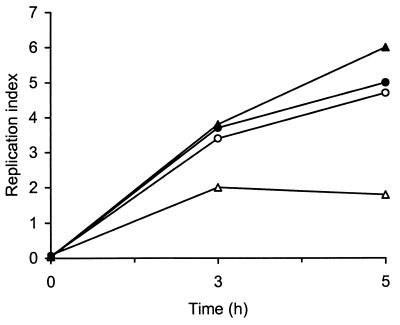
To evaluate the ability of acid-adapted L. monocytogenes LM2 to enter and proliferate within phagocytic cells, invasion experiments were carried out by infecting THP-1 cells for 1 h at 37°C at an MOI of approximately 10 bacteria per cell. Table 2 shows the entry and growth of LM2 in nonactivated and IFN-γ-activated macrophages at 1 and 5 h postinfection. Both acid-adapted and nonadapted bacteria were phagocytosed and multiplied efficiently in nonactivated THP-1 cells. When THP-1 macrophages were preexposed to IFN-γ, they became nonpermissive to growth of non-acid-adapted bacteria. However, when activated macrophages were infected with acid-adapted bacteria, the invasion rate was not affected but the intracellular growth rate was greatly enhanced, as shown by the increase in the number of intracellular bacteria 5 h after infection. As shown in Fig. 4, this effect was more evident when intracellular growth was expressed as the RI (the number of CFU determined 3 or 5 h postinfection divided by the number of CFU at time zero).
FIG. 4.
RI of L. monocytogenes LM2 in THP-1 macrophages. Circles, bacteria in nonactivated cells; triangles, bacteria in IFN-γ-activated cells. Solid symbols, acid-adapted bacteria; open symbols, nonadapted bacteria. Each data point is the mean intracellular L. monocytogenes RI from one experiment representative of six performed.
Transmission electron microscopy of L. monocytogenes-infected THP-1 macrophages.
Invasion of macrophages by acid-adapted and nonadapted listerial cells was followed by electron microscopy. These experiments were carried out in both nonactivated and IFN-γ-activated THP-1 macrophages. No appreciable differences between acid-adapted and nonadapted bacteria were observed in control (nonactivated) macrophages. In these cells, intact bacteria were seen in phagosomes, from which they escaped to reach the cytosol (data not shown).
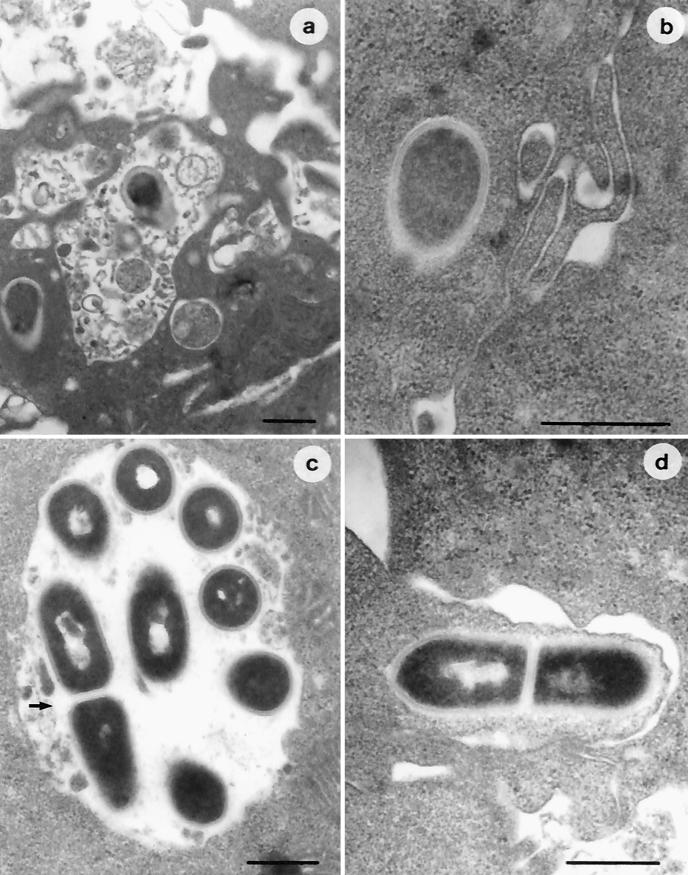
When ultrastructural investigations were carried out in macrophages pretreated with IFN-γ, different behaviors were observed. Infection of IFN-γ-activated THP-1 cells with nonadapted strain LM2 resulted in numerous bacteria enclosed in the phagosomes, a number of which showed extensive structural damage (Fig. 5a), whereas only a few bacterial cells were observed to spread into the cytoplasm (Fig. 5b). In contrast, IFN-γ-activated macrophages infected with acid-adapted L. monocytogenes contained a greater number of bacteria, often intact and almost all free in the cytoplasm (Fig. 5c and d). Acid-adapted LM2 cells appeared to be surrounded by a network of actin filaments, and bacteria with actin tails were frequently observed.
FIG. 5.
Electron micrographs of IFN-γ-activated THP-1 macrophages infected with nonadapted (a and b) and acid-adapted (c and d) L. monocytogenes LM2. A number of nonadapted bacterial cells show dramatic structural damage due to phagosomal digestion (a), whereas only a few cells can be observed free in the cytoplasm (b). Acid-adapted L. monocytogenes cells appear either intact and in active multiplication (arrow) in the phagosome (c) or free in the cytoplasm, with actin tails (d). Bars, 0.5 μm.
DISCUSSION
A wealth of evidence indicates that the response to different environmental stressors can modulate the expression of the virulent phenotype of L. monocytogenes (4, 56). We previously demonstrated that the expression of virulence determinants of L. monocytogenes can be influenced by low temperature and iron deficiency (10, 11, 12). Sensing an acidic environment, as that encountered by the bacterium during passage through the gastrointestinal tract, can be considered an additional stimulus triggering expression of the virulent phenotype, and this is consistent with the evidence that induction of the ATR enhances the survival of L. monocytogenes both in vitro and in vivo (9, 16, 39, 43, 49). Acid-tolerant variants have increased lethality for mice when inoculated by the intraperitoneal route (43). Acid adaptation also correlates with increased translocation rates of bacteria to mesenteric lymph nodes in mice infected by intragastric inoculation (49). Recently, we showed that acid-tolerant L. monocytogenes strains invade enterocyte-like cells more efficiently and survive longer in murine macrophage-like cells (13). In this report we present novel evidence of the effect of pH, a major environmental signal, on the expression of listerial genes relevant to host invasion and intracellular parasitism. We demonstrated that preexposure to an acidic pH enhances the survival of L. monocytogenes in IFN-γ-activated human macrophages, and we correlated this effect with an altered pattern of expression of genes involved in acid resistance and intracellular survival.
Among prfA-controlled genes, only plcA was significantly down-regulated at a low pH; its transcription was >8-fold decreased in acid-exposed bacteria. This effect appeared to be specific, in so far as transcription of the inlA, hly, and actA genes was unchanged in acid-adapted and nonadapted bacteria. Although PlcA was initially regarded as a major virulence determinant of L. monocytogenes (6, 7), subsequent studies with an in-frame plcA deletion mutant revealed only a minor defect in virulence and escape from primary vacuoles (55). This secondary role of PlcA in virulence can be related to our observation that, despite poor plcA expression at low pHs, acid-adapted bacteria survived and multiplied in IFN-γ-activated THP-1 macrophages, as demonstrated by both intracellular growth assays and morphological analysis.
We noticed that the reduced transcription of plcA at pH 5.1 was not paralleled by decreased transcription of the pfrA gene, encoding the pleiotropic activator of the virulence regulon. Under this condition, the pattern of transcription of the plcA-pfrA operon was altered, likely reflecting down-regulation of the upstream PplcA promoter concomitant with up-regulation of the downstream P1prfA and P2prfA promoters. This transition ensured that the prfA gene was transcribed at similar levels in neutral and acidic environments. The effect of low pH on pfrA expression is therefore reminiscent of that caused by the stationary phase (42, 53) or by addition of the disaccharide cellobiose (2). In particular, cellobiose has been observed to repress the expression of the PrfA regulon and change the pattern of prfA transcription without affecting the intracellular levels of the PrfA protein (47). Because cellobiose-dependent repression of PrfA takes place at the posttranslational level, an indirect effect of the sugar on cofactors required for PrfA activity was hypothesized. In contrast to cellobiose, however, low pH negatively affected the expression of a single PrfA-controlled gene (plcA), leaving the transcription of several other genes of the prfA regulon (inlA, hly, and actA) unchanged. This observation indicates that PrfA is equally active under neutral and acidic conditions on at least three genes of its regulon, and it suggests that the reduced levels of plcA transcripts at low pHs derive either from mRNA instability or from specific repression of PrfA activity at the PplcA promoter.
If our in vitro observations reflect the intracellular behavior of L. monocytogenes, then the differential regulation of hly and plcA would constitute a strategy for fine tuning of genes during the intraphagosomal phase, preventing expression of plcA under conditions in which its product could be detrimental to the parasite. LLO is a pore-forming cytolysin that requires cholesterol as the membrane receptor (30), while PlcA hydrolyzes phosphatidylinositol residues, which are prevalent in the cytoplasmic leaflet of the phagosomal membrane (32). Both LLO and PlcA are expressed in the phagosome and contribute to vacuolar escape (5, 33), but the pH ranges for their enzymatic activities are different (30, 32); LLO requires a more acidic pH (4.5 to 6.5) than PlcA (5.5 to 7.5). A plausible interpretation of our results is that, during the intraphagosomal life of L. monocytogenes, transcription of hly and plcA is differentially regulated depending on the vacuolar pH, thereby ensuring that LLO and PlcA will be produced in the environments most favorable for their activities. Early after cell invasion, the acidic environment of the phagosome could prevent plcA expression, while enabling LLO to be accumulated in an enzymatically active state. Pore formation by LLO would then raise the phagosomal pH to neutral values, thereby increasing PlcA expression. Perturbation of membrane integrity would also facilitate PlcA access to the cytoplasmic leaflet of the phagosomal membrane and cleavage of phosphatidylinositol-containing substrates (6). In line with this hypothesis, activation of the plcA promoter inside J774 macrophages has been shown to persists for 5 h after infection (33), though acidification of macrophage phagosomes and perforation by LLO occur shortly after internalization of bacteria (1).
In this report we also demonstrate that exposure of L monocytogenes to a low pH causes up-regulation of the gad and sodC genes, involved in the responses to acid and oxidative stresses, respectively. From this point of view, acidic pH should be considered an overall stressor which influences the survival strategies of L. monocytogenes by triggering the expression of at least some genes required for successful in vivo multiplication. This is consistent with the notion that acid adaptation provides cross-protection against a variety of secondary stresses and promotes virulence in the mouse model (43). It is conceivable that increased expression of superoxide dismutase at low pHs might help an intracellular pathogen, such as L. monocytogenes, to cope with the oxygen-dependent bactericidal activity of phagocytic cells (61). Plausibly, it would allow the infecting bacteria to produce an oxygen radical-detoxifying enzyme in the acidic intestinal environment, thereby preventing damage upon subsequent challenge with an oxidative stress in the macrophagic phagolysosome.
The increased gad transcription at low pHs is in good agreement with the function of the glutamate decarboxylase system in the maintenance of intracellular pH homeostasis (54). At an initial stage of this study, expression of gad genes was used as a reporter system to probe the effect of low pHs on gene expression, and the dramatic induction of gad messengers at pH 5.1 served as the positive control for RNA slot blot experiments aimed at investigating the transcriptional response to acidic stress of virulence-related genes. However, only after publication of the complete genome sequence of L. monocytogenes (31) did the complex transcriptional profile of gad genes become interpretable. We conducted an in silico analysis of the three gad loci of L. monocytogenes and predicted, with good confidence, the existence of multiple promoters and putative transcription termination sites. Sequence-based predictions were supported by size estimations of gad-specific transcripts, also showing that all paralogs encoding the multiple isoforms of glutamate decarboxylase and cognate antiporters, namely, gadA, gadCB, and (lmo446)gadB′C′, were coordinately transcribed at low pHs. The presence of duplicate gene clusters for cycling of glutamic acid and GABA, both expressed in acid-exposed L. monocytogenes cells, can also explain why ΔgadAB and ΔgadC mutants were found to retain residual resistance to acid killing (16). Remarkably, the amount of gad transcripts, but not their quality, varied in a strain-dependent fashion, likely reflecting differences in acid tolerance. In fact, gad transcripts were significantly more abundant in the acid-resistant strain LM2 than in the acid-susceptible strain EGD, in accordance with the extremely low levels of glutamic acid decarboxylase activity previously evidenced in L. monocytogenes EGD (16). Induction of gad genes was observed during exponential growth in acidic medium and did not require addition of exogenous glutamate, indicating that acid sensing constitutes the specific stimulus for induction of the glutamate-based acid resistance system. Such a strict acid dependence could make gad promoters ideal tools for future studies aimed at monitoring pH changes during the infectious cycle of L. monocytogenes.
The overall resistance to environmental stressors of acid-adapted L. monocytogenes was documented by invasion assays of IFN-γ-activated-THP-1 human macrophages combined with morphological investigations. It has been reported that in nonactivated THP-1 cells L. monocytogenes can easily escape from phagosomes and multiply in the cytoplasm, whereas in IFN-γ-activated macrophages listerial growth is greatly inhibited and bacteria are rapidly killed (44). In our experimental setup, when infection of activated THP-1 cells was performed with non-acid-adapted bacteria, the results obtained were concordant with those of Ouadrhiri et al. (44), as intracellular growth of L. monocytogenes was restricted. A different behavior was observed for acid-adapted bacteria; after being efficiently phagocytosed by IFN-γ-activated cells, they were able to survive and proliferate intracellularly to a significantly higher extent than nonadapted bacteria. The RIs of acid-adapted bacteria were similar in IFN-γ-activated and nonactivated THP-1 cells, demonstrating a generally increased resistance of L. monocytogenes to the hostile macrophagic environment following ATR induction. The different fate of acid-exposed bacterial cells after phagocytosis was also confirmed at the ultrastructural level: electron microscopy showed that unadapted L. monocytogenes cells were digested into the phagosome, and only a few bacterial cells reached the cytoplasm. In contrast, acid-adapted bacteria appeared either intact and in active multiplication within the phagosome or free in the cytoplasm, thus confirming their ability to resist intraphagosomal killing and to replicate both in vacuoles and in the intracytoplasmic compartment.
The survival of acid-adapted bacteria in THP-1 macrophages could therefore be related to the synergistic effect of different factors, including modification of the protein and fatty acid composition of the bacterial surface (43, 45, 60), the pH-neutralizing activity of the glutamic acid decarboxylase system (16), and the superoxide dismutase-dependent escape from oxidative attack (19). In perspective, a more detailed molecular and functional dissection of the listerial response to low pHs could help in understanding the mechanism by which this bacterium survives in the harsh intestinal environment and endures the stress associated with exposure to phagocytic cells.
Acknowledgments
We thank Lamberto Camilli for photographic work.
This work was supported by ENEA (Programmi Fondi Strutturali) and MURST grants to L. Seganti and by grants from MURST and the Ministry of Health (Targeted Projects) to P. Visca.
Editor: A. D. O'Brien
REFERENCES
- 1.Beauregard, K. E., K. D. Lee, R. J. Collier, and J. A. Swanson. 1997. pH-dependent perforation of macrophage phagosomes by listeriolysin O from Listeria monocytogenes. J. Exp. Med. 186:1159-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behari, J., and P. Youngman. 1998. Regulation of hly expression in Listeria monocytogenes by carbon sources and pH occurs through separate mechanisms mediated by PrfA. Infect. Immun. 66:3635-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bohne, J., H. Kestler, C. Uebele, Z. Sokolovic, and W. Goebel. 1996. Differential regulation of the virulence genes of Listeria monocytogenes. Mol. Microbiol. 20:1189-1198. [DOI] [PubMed] [Google Scholar]
- 4.Brehm, K., J. Kreft, M. T. Ripio, and J. A. Vazquez-Boland. 1996. Regulation of virulence gene expression in pathogenic Listeria. Microbiologia 12:219-236. [PubMed] [Google Scholar]
- 5.Bubert, A., Z. Sokolovic, S.-K. Chun, L. Papatheodorou, A. Simm, and W. Goebel. 1999. Differential expression of Listeria monocytogenes virulence genes in mammalian host cells. Mol. Gen. Genet. 261:323-336. [DOI] [PubMed] [Google Scholar]
- 6.Camilli, A., H. Goldfine, and D. A. Portnoy. 1991. Listeria monocytogenes mutants lacking phosphatidylinositol-specific phospholipase C are avirulent. J. Exp. Med. 173:751-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camilli, A., L. G. Tilney, and D. A. Portnoy. 1993. Dual roles of plcA in Listeria monocytogenes pathogenesis. Mol. Microbiol. 8:143-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, M. B., M. V. Jones, and C. Holyoak. 1990. The effect of pH, salt concentration and temperature on the survival and growth of Listeria monocytogenes. J. Appl. Bacteriol. 69:63-72. [DOI] [PubMed] [Google Scholar]
- 10.Conte, M. P., C. Longhi, G. Petrone, M. Polidoro, P. Valenti, and L. Seganti. 1994. L. monocytogenes infection of Caco-2 cells: role of growth temperature. Res. Microbiol. 145:677-682. [DOI] [PubMed] [Google Scholar]
- 11.Conte, M. P., C. Longhi, G. Petrone, M. Polidoro, P. Valenti, and L. Seganti. 2000. Iron modulates actA gene expression in Listeria monocytogenes. J. Med. Microbiol. 49:681-683. [DOI] [PubMed] [Google Scholar]
- 12.Conte, M. P., C. Longhi, M. Polidoro, G. Petrone, V. Buonfiglio, S. Di Santo, E. Papi, L. Seganti, P. Visca, and P. Valenti. 1996. Iron availability affects entry of Listeria monocytogenes into the enterocyte-like cell line Caco-2. Infect. Immun. 64:3925-3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conte, M. P., G. Petrone, A. M. Di Biase, M. G. Ammendolia, F. Superti, and L. Seganti. 2000. Acid tolerance in Listeria monocytogenes influences invasiveness of enterocyte-like cells and macrophage-like cells. Microb. Pathog. 29:137-144. [DOI] [PubMed] [Google Scholar]
- 14.Cossart, P., and J. Mengaud. 1989. Listeria monocytogenes: a model system for the molecular study of intracellular parasitism. Mol. Biol. Med. 6:463-474. [PubMed] [Google Scholar]
- 15.Cotter, P. D., N. Emerson, C. G. Gahan, and C. Hill. 1999. Identification and disruption of lisRK, a genetic locus encoding a two-component signal transduction system involved in stress tolerance and virulence in Listeria monocytogenes. J. Bacteriol. 181:6840-6843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cotter, P. D., C. G. Gahan, and C. Hill. 2001. A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40:465-475. [DOI] [PubMed] [Google Scholar]
- 17.Davis, M. J., P. J. Coote, and C. P. O'Byrne. 1996. Acid tolerance in Listeria monocytogenes: the adaptive acid tolerance response (ATR) and growth-phase-dependent acid resistance. Microbiology 142:2975-2982. [DOI] [PubMed] [Google Scholar]
- 18.De Biase, D., A. Tramonti, F. Bossa, and P. Visca. 1999. The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol. Microbiol. 32:1198-1211. [DOI] [PubMed] [Google Scholar]
- 19.de Chastellier, C., and P. Berche. 1994. Fate of Listeria monocytogenes in murine macrophages: evidence for simultaneous killing and survival of intracellular bacteria. Infect. Immun. 62:543-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dramsi, S., M. Lebrun, and P. Cossart. 1996. Molecular and genetic determinants involved in invasion of mammalian cells by Listeria monocytogenes. Curr. Top. Microbiol. Immunol. 209:61-68. [DOI] [PubMed] [Google Scholar]
- 21.Dramsi, S., S. Levi, A. Triller, and P. Cossart. 1998. Entry of Listeria monocytogenes into neurons occurs by cell-to-cell spread: an in vitro study. Infect. Immun. 66:4461-4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drevets, D. A. 1999. Dissemination of Listeria monocytogenes by infected macrophages. Infect. Immun. 67:3512-3517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Drevets, D. A., B. P. Canono, and P. A. Campbell. 1992. Listericidal and nonlistericidal mouse macrophages differ in complement receptor type 3-mediated phagocytosis of L. monocytogenes and in preventing escape of the bacteria into the cytoplasm. J. Leukoc. Biol. 52:70-79. [DOI] [PubMed] [Google Scholar]
- 24.Dykes, G. A., and S. M. Moorhead. 2000. Survival of osmotic and acid stress by Listeria monocytogenes strains of clinical or meat origin. Int. J. Food Microbiol. 56:161-166. [DOI] [PubMed] [Google Scholar]
- 25.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fleming, S. D., and P. A. Campbell. 1997. Some macrophages kill Listeria monocytogenes while others do not. Immunol. Rev. 158:69-77. [DOI] [PubMed] [Google Scholar]
- 27.Freitag, N. E., and D. A. Portnoy. 1994. Dual promoters of the Listeria monocytogenes prfA transcriptional activator appear essential in vitro but are redundant in vivo. Mol. Microbiol. 12:845-853. [DOI] [PubMed] [Google Scholar]
- 28.Gahan, C. G., and C. Hill. 1999. The relationship between acid stress responses and virulence in Salmonella typhimurium and Listeria monocytogenes. Int. J. Food Microbiol. 50:93-100. [DOI] [PubMed] [Google Scholar]
- 29.Gahan, C. G., B. O'Driscoll, and C. Hill. 1996. Acid adaptation of Listeria monocytogenes can enhance survival in acidic foods and during milk fermentation. Appl. Environ. Microbiol. 62:3128-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geoffroy, C., J. L. Gaillard, J. E. Alouf, and P. Berche. 1987. Purification, characterization, and toxicity of the sulfhydryl-activated hemolysin listeriolysin O from Listeria monocytogenes. Infect. Immun. 55:1641-1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 32.Goldfine, H., and C. Knob. 1992. Purification and characterization of Listeria monocytogenes phosphatidylinositol-specific phospholipase C. Infect. Immun. 60:4059-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klarsfeld, A. D., P. L. Goossens, and P. Cossart. 1994. Five Listeria monocytogenes genes preferentially expressed in infected mammalian cells: plcA, purH, purD, pyrE and an arginine ABC transporter gene, arpJ. Mol. Microbiol. 13:585-597. [DOI] [PubMed] [Google Scholar]
- 34.Kreft, J., J. Bohne, R. Gross, H. Kestler, Z. Sokolovic, and W. Goebel. 1995. Control of Listeria monocytogenes virulence by the transcriptional regulator PrfA, p. 129-142. In R. Rappuoli, V. Scarlato, and B. Arico (ed.), Signal tranduction and bacterial virulence. R. G. Landes Company, Austin, Tex.
- 35.Kroll, R. G., and R. A. Patchett. 1992. Induced acid tolerance in Listeria monocytogenes. Lett. Appl. Microbiol. 14:224-227. [Google Scholar]
- 36.Kuhn, M., and W. Goebel. 1995. Molecular studies on the virulence of Listeria monocytogenes. Genet. Eng. (New York) 17:31-51. [PubMed] [Google Scholar]
- 37.Lorber, B. 1996. Listeriosis. Clin. Infect. Dis. 24:1-11. [DOI] [PubMed] [Google Scholar]
- 38.Mackaness, G. B. 1962. Cellular resistance to infection. J. Exp. Med. 116:381-406. [PubMed] [Google Scholar]
- 39.Marron, L., N. Emerson, C. G. Gahan, and C. Hill. 1997. A mutant of Listeria monocytogenes LO28 unable to induce an acid tolerance response displays diminished virulence in a murine model. Appl. Environ. Microbiol. 63:4945-4947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 41.McLauchlin, J. 1996. The relationship between Listeria and listeriosis. Food Control 7:187-193. [Google Scholar]
- 42.Mengaud, J., S. Dramsi, E. Gouin, J. A. Vazquez-Boland, G. Milon, and P. Cossart. 1991. Pleiotropic control of Listeria monocytogenes virulence factors by a gene that is autoregulated. Mol. Microbiol. 5:2273-2283. [DOI] [PubMed] [Google Scholar]
- 43.O'Driscoll, B., C. G. Gahan, and C. Hill. 1996. Adaptive acid tolerance response in Listeria monocytogenes: isolation of an acid-tolerant mutant which demonstrates increased virulence. Appl. Environ. Microbiol. 62:1693-1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ouadrhiri, Y., B. Scorneaux, Y. Sibille, and P. M. Tulkens. 1999. Mechanism of the intracellular killing and modulation of antibiotic susceptibility of Listeria monocytogenes in THP-1 macrophages activated by gamma interferon. Antimicrob. Agents Chemother. 43:1242-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Phan-Thanh, L., F. Mahouin, and S. Aligé. 2000. Acid responses in Listeria monocytogenes. Int. J. Food Microbiol. 55:121-126. [DOI] [PubMed] [Google Scholar]
- 46.Renzoni, A., P. Cossart, and S. Dramsi. 1999. PrfA, the transcriptional activator of virulence genes, is upregulated during interaction of Listeria monocytogenes with mammalian cells and in eukaryotic cell extracts. Mol. Microbiol. 34:552-561. [DOI] [PubMed] [Google Scholar]
- 47.Renzoni, A., A. Klarsfeld, S. Dramsi, and P. Cossart. 1997. Evidence that PrfA, the pleiotropic activator of virulence genes in Listeria monocytogenes, can be present but inactive. Infect. Immun. 65:1515-1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouquette, C., M. T. Ripio, E. Pellegrini, J. M. Bolla, R. I. Tascon, J. A. Vazquez-Boland, and P. Berche. 1996. Identification of a ClpC ATPase required for stress tolerance and in vivo survival of Listeria monocytogenes. Mol. Microbiol. 21:977-987. [DOI] [PubMed] [Google Scholar]
- 49.Saklani-Jusforgues, H., E. Fontan, and P. L. Goossens. 2000. Effect of acid adaptation on Listeria monocytogenes survival and translocation in a murine intragastric infection model. FEMS Microbiol. Lett. 193:155-159. [DOI] [PubMed] [Google Scholar]
- 50.Sanders, J. W., K. Leenhouts, J. Burghoorn, J. R. Brands, G. Venema, and J. A. Kok. 1998. Chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27:299-310. [DOI] [PubMed] [Google Scholar]
- 51.Scorneaux, B., Y. Ouadrhiri, G. Anzalone, and P. M. Tulkens. 1996. Effect of recombinant human gamma interferon on intracellular activities of antibiotics against Listeria monocytogenes in the human macrophage cell line THP-1. Antimicrob. Agents Chemother. 40:1225-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sheehan, B., A. Klarsfeld, R. Ebright, and P. Cossart. 1996. A single substitution in the putative helix-turn-helix motif of the pleiotropic activator PrfA attenuates Listeria monocytogenes virulence. Mol. Microbiol. 20:785-797. [DOI] [PubMed] [Google Scholar]
- 53.Sheehan, B., A. Klarsfeld, T. Msadek, and P. Cossart. 1995. Differential activation of virulence gene expression by PrfA, the Listeria monocytogenes virulence regulator. J. Bacteriol. 177:6469-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Small, P. L., and S. R. Waterman. 1998. Acid stress, anaerobiosis and gadCB: lessons from Lactococcus lactis and Escherichia coli. Trends Microbiol. 6:214-216. [DOI] [PubMed] [Google Scholar]
- 55.Smith, G. A., H. Marquis, S. Jones, N. C. Johnston, D. A. Portnoy, and H. Goldfine. 1995. The two distinct phospholipases C of Listeria monocytogenes have overlapping roles in escape from a vacuole and cell-to-cell spread. Infect. Immun. 63:4231-4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sokolovic, Z., J. Riedel, M. Wuenscher, and W. Goebel. 1993. Surface-associated, PrfA-regulated proteins of Listeria monocytogenes synthesized under stress conditions. Mol. Microbiol. 8:219-227. [DOI] [PubMed] [Google Scholar]
- 57.Tilney, L. G., and D. A. Portnoy. 1989. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite Listeria monocytogenes. J. Cell Biol. 109:1597-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiveljung, A., J. Backstrom, U. Forsum, and H. J. Monstein. 1995. Broad-range PCR amplification and DNA sequence analysis reveals variable motifs in 16S rRNA genes of Mobiluncus species. AMPIS 103:755-763. [DOI] [PubMed] [Google Scholar]
- 59.Tsuchiya, S., M. Yamabe, Y. Yamaguchi, Y. Kobayashi, T. Konno, and K. Tada. 1980. Establishment and characterisation of a human acute monocytic leukemia cell line (THP-1). Int. J. Cancer 26:171-176. [DOI] [PubMed] [Google Scholar]
- 60.van Schaik, W., C. G. Gahan, and C. Hill. 1999. Acid-adapted Listeria monocytogenes displays enhanced tolerance against the antibiotics nisin and lacticin 3147. J. Food Prot. 62:536-539. [DOI] [PubMed] [Google Scholar]
- 61.Vasconcelos, J. A., and H. G. Deneer. 1994. Expression of superoxide dismutase in Listeria monocytogenes. Appl. Environ. Microbiol. 60:2360-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vega, Y., C. Dickneite, M. T. Ripio, R. Böckmann, B. González-Zorn, S. Novella, G. Domínguez-Bernal, W. Goebel, and J. A. Vázquez-Boland. 1998. Functional similarities between the Listeria monocytogenes virulence regulator PrfA and cyclic AMP receptor protein: the PrfA∗ (Gly145Ser) mutation increases binding affinity for target DNA. J. Bacteriol. 180:6655-6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Waterman, S. R., and P. L. Small. 1996. Identification of sigma S-dependent genes associated with the stationary-phase acid-resistance phenotype of Shigella flexneri. Mol. Microbiol. 21:925-940. [DOI] [PubMed] [Google Scholar]