Abstract
Sixteen of 22 omp-1 paralogs encoding 28-kDa-range immunodominant outer membrane proteins of Ehrlichia chaffeensis were transcribed in blood monocytes of dogs throughout a 56-day infection period. Only one paralog was transcribed by E. chaffeensis in three developmental stages of Amblyomma americanum ticks before or after E. chaffeensis transmission to naïve dogs.
Ehrlichia chaffeensis, an obligatory intramonocytic bacterium, causes human monocytic ehrlichiosis (HME), an emerging tick-borne zoonosis (1, 5, 19). E. chaffeensis has been detected in field-collected Amblyomma americanum ticks, white-tailed deer, and dogs (2-4, 6, 9-17, 20, 21, 24). Although A. americanum has been shown to transmit E. chaffeensis among deer (7), development of an easily accessible tick transmission model using the dog would facilitate the analysis of molecular mechanisms of ehrlichial transmission.
To date, only a few E. chaffeensis genes have been characterized. We recently characterized the omp-1 multigene families encoding outer membrane protein 1 (OMP-1)-immunodominant major OMPs of E. chaffeensis (18). A total of 22 paralogs are clustered in the 27-kb locus of E. chaffeensis. There has been no report of any protein gene transcription by E. chaffeensis in mammals or ticks. In the present study, (i) we analyzed the transcription of the entire family of 22 omp-1 multigenes in experimentally infected dogs and A. americanum ticks and (ii), since E. chaffeensis transmission from ticks to dogs has never been demonstrated, we examined whether E. chaffeensis can be transmitted from A. americanum to dogs.
Eight pathogen-free female dogs (1 to 2 years old) were used. All dogs were free of E. chaffeensis infection as determined by indirect fluorescent antibody and PCR tests of their blood specimens. Dogs 133 and 146 were each intravenously inoculated with 5 × 106 DH82 cells infected with E. chaffeensis Arkansas (low-passage 1993 stock). E. chaffeensis 16S rRNA was detected in the peripheral blood mononuclear cells (PBMCs) of both dogs by reverse transcriptase PCR (RT-PCR) starting on day 7 and continuing through day 56 postinoculation (p.i.) (8). A. americanum ticks at three developmental stages were attached to both dogs and removed as previously described (8). The remaining six dogs were used to reattach these infected ticks. Indirect fluorescent antibody tests, isolation of PBMCs, dissection of ticks, DNA and RNA extraction, RT-PCR, and PCR based on the 16S rRNA gene were described previously (8, 23).
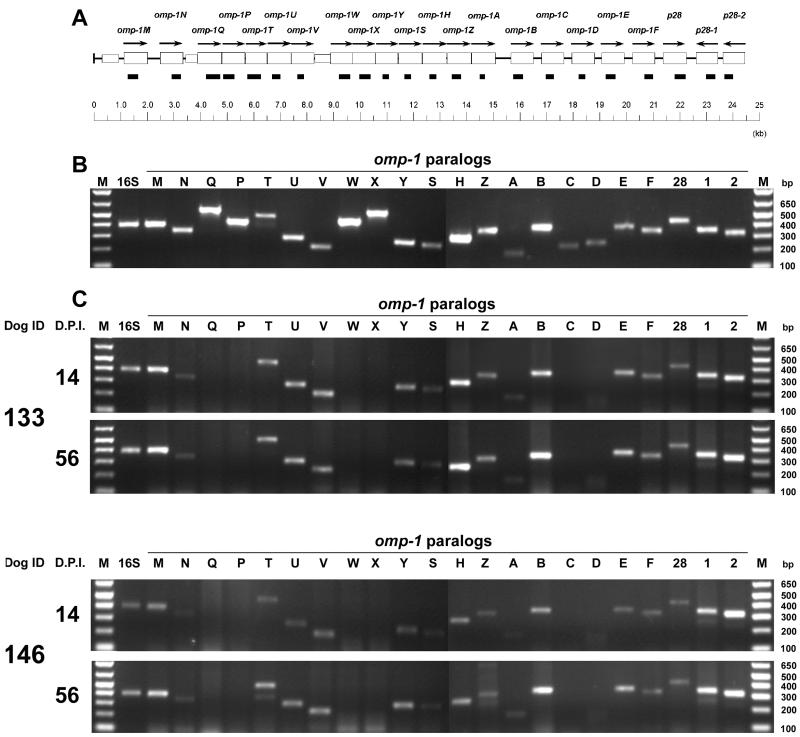
The 22 pairs of primers that amplify the regions shown in Table 1 were shown to be specific for each omp-1 by PCR using 0.5 ng of purified E. chaffeensis DNA as template (Fig. 1A and B). By RT-PCR using these omp-1-specific primers, 16 omp-1 paralogs were found to be transcribed by E. chaffeensis in PBMCs from dogs 133 and 146 throughout the 56 days p.i., and transcripts of the remaining six paralogs were undetectable in either dog (Fig. 1C). To normalize levels of ehrlichial RNA present in the PBMCs at every time point in each dog, constitutively expressed E. chaffeensis 16S rRNA was amplified by RT-PCR in the linear range (27 cycles) using primer HE1-HE3 (2). E. chaffeensis 16S rRNA levels were slightly increased in both dogs from day 14 to day 56 p.i., and so was the level of expression of omp-1 paralogs (Fig. 1C). Without addition of RT, none of the RNA specimens was positive in the RT-PCR, indicating the absence of DNA contamination (data not shown). Ehrlichia canis, the agent of canine monocytic ehrlichiosis, is phylogenetically and biologically closely related to E. chaffeensis. This finding is similar to that with a p30-immunodominant major OMP multigene family of E. canis that has a gene structure and arrangement similar to those of the omp-1 gene family (18). Like omp-1 genes, the same set of p30 genes is expressed by E. canis in PBMCs regardless of the individual dog or infection time period (23).
TABLE 1.
Gene-specific oligodeoxynucleotides used in PCR and RT-PCR
| Target gene | Regions for RT-PCR
|
|
|---|---|---|
| Nucleotide positiona | Amplicon size (bp) | |
| omp-1M | 1275-1666 | 392 |
| omp-1N | 2932-3261 | 330 |
| omp-1Q | 4198-4749 | 552 |
| omp-1P | 4861-5285 | 425 |
| omp-1T | 5789-6269 | 481 |
| omp-1U | 6726-7007 | 282 |
| omp-1V | 7670-7892 | 223 |
| omp-1W | 9211-9659 | 449 |
| omp-1X | 9953-10500 | 548 |
| omp-1Y | 10811-11072 | 262 |
| omp-1S | 11685-11932 | 248 |
| omp-1H | 12596-12878 | 283 |
| omp-1Z | 13469-13807 | 339 |
| omp-1A | 14687-14868 | 182 |
| omp-1B | 15774-16138 | 365 |
| omp-1C | 17053-17282 | 230 |
| omp-1D | 18200-18445 | 246 |
| omp-1E | 19229-19594 | 366 |
| omp-1F | 20680-21015 | 331 |
| p28 | 21826-22248 | 423 |
| p28-1 | 23029-23373 | 345 |
| p28-2 | 23708-24028 | 321 |
Nucleotide position is based on the 27-kb omp locus of E. chaffeensis (18).
FIG. 1.
(A) RT-PCR region in the omp-1 multigene cluster of E. chaffeensis. The open boxes and arrows show respective omp-1 paralogs and their orientation. Closed boxes indicate amplified RT-PCR region as indicated in Table 1. (B) PCR utilizing purified E. chaffeensis DNA (0.5 ng) as template and gene-specific primer pairs showing the strength and specificity of each reaction. The amplified products were resolved on agarose gels containing ethidium bromide. omp-1 genes were identified on the top in the order of their genomic localization. M, molecular size markers (1-kb plus DNA marker [Life Technologies]). Nucleotide base pair sizes of the marker are indicated on the left. (C) Expression of mRNA of omp-1 paralogs and 16S rRNA of E. chaffeensis in the PBMCs of two dogs infected by intravenous inoculation of E. chaffeensis. Total RNA was extracted and subjected to RT-PCR. The amplified products were resolved on agarose gels containing ethidium bromide. omp-1 genes were identified on the top in the order of their genomic localization. M, molecular size markers (1-kb plus DNA marker [Life Technologies]); D.P.I., days p.i. Nucleotide base pair sizes of the marker are indicated on the right.
E. chaffeensis 16S rRNA was detected in all 16 different groups of tick tissues (salivary glands, midgut, or whole body of ticks at three different developmental stages) using 16S rRNA prior to (8) and after attachment to six naïve dogs (Fig. 2), indicating that the tick infection was stable. Of 22 omp-1 paralogs examined by RT-PCR, omp-1B was the only omp-1 transcript detected in all 16 groups of tick tissues (Fig. 2). Without RT, all of these tick tissues were negative by RT-PCR using 16S rRNA or omp-1 paralog primers, indicating the absence of DNA contamination (data not shown). No ehrlichial 16S rRNA or transcripts of omp-1 paralogs were detected in control uninfected tick tissues. It was previously demonstrated that only one p30 paralog, p30-10, the ortholog of omp-1B, is expressed by E. canis in Rhipicephalus sanguineus ticks and that, of all 22 p30 paralogs examined, only p30-10 expression is up-regulated in DH82 cells at 25°C in culture compared to its expression at 37°C (23). Present results support our speculation that expression of p30-10 and omp-1B, in E. canis and E. chaffeensis, respectively, is induced in ticks, since the temperature is lower in ticks than in mammals.
FIG. 2.
Transcriptional profiles of the omp-1 paralogs by RT-PCR in 16 different specimens of ticks. Total tick RNA was extracted and subjected to RT-PCR. The panels show ethidium bromide-stained RT-PCR products of E. chaffeensis 16S rRNA and omp-1B. M, molecular size markers (1-kb plus DNA marker [Life Technologies]); Pos, positive control utilizing E. chaffeensis DNA as template; lanes 1 to 16, different specimens of ticks; and NT, no template. Nucleotide base pair sizes of amplified product are indicated on the right.
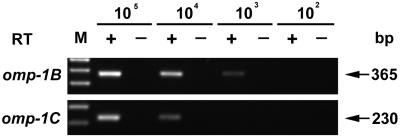
To estimate the detection limit of the RT-PCR, omp-1-specific transcripts were generated in vitro. omp-1B and omp-1C were selected as representatives, because omp-1B was universally expressed in both dogs and ticks (Fig. 1 and 2) and because omp-1C was the weakest amplicon detected by the gene-specific primers (Fig. 1B). The template for the transcription was prepared by PCR using the following primer pairs: for omp-1B, forward primer 5′-TAATACGACTCACTATAGGGAACGACAGCAGAGAAGGC-3′ and reverse primer 5′-GCGGAAACTTCTGGTGTG-3′, which was 220 bp downstream of the 3′ end (nucleotide 16138) of the RT-PCR region; for omp-1C, forward primer 5′-TAATACGACTCACTATAGGGCTTCAAGTCATGCTGATGC-3′ and reverse primer 5′-ATGATGGTGTAGCAAACGC-3′, which was 301 bp downstream from the 3′ end (nucleotide 17282) of the RT-PCR region. The T7 binding site sequences of the above primers are underlined. Specific transcripts of omp-1B and omp-1C generated in vitro as previously described (23) were 10-fold serially diluted and used in RT-PCR against a background of the total RNA from 2.5 × 106 uninfected dog PBMCs to mimic the experimental conditions (Fig. 3). Under our standard RT-PCR conditions, 365- and 230-bp cDNA fragments of omp-1B and omp-1C, respectively, were detected to levels of 103 and 104 transcripts, respectively. The detection limit per reaction of nested PCR based on the 16S rRNA gene was 48 fg of DNA from E. chaffeensis-infected DH82 cells (8). This amount of DNA corresponds to 250 E. chaffeensis genomes in 2.5 × 106 PBMCs. All dog and tick specimens examined in the present study were positive for 16S rRNA gene-based nested PCR. This means that at least 250 E. chaffeensis genomes were present in each specimen. Therefore, when the omp-1 paralogs were not detectable in these specimens by RT-PCR, the transcript number was less than 4 to 40 per E. chaffeensis genome.
FIG. 3.
Estimation of RT-PCR sensitivity in detecting the transcripts of omp-1 paralogs. +, RT-PCR analysis was performed using decreasing amounts of in vitro generated transcripts as template; −, shown is the result of an identical reaction without the addition of RT as control for DNA contamination. The transcript numbers are shown at the top. omp-1 genes are identified on the left. M, molecular size markers (1-kb plus DNA marker [Life Technologies]). Nucleotide base pair sizes of amplified product are indicated on the right.
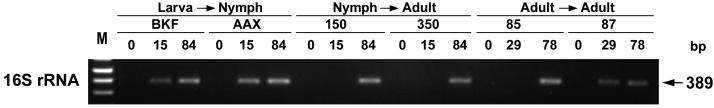
To test E. chaffeensis transmission from ticks to dogs, approximately 150 nymphs infected as larvae were placed on dogs BKF and AAX, respectively; 50 and 60 adult ticks infected as nymphs were placed on dogs 150 and 350, respectively; and 50 and 70 male ticks infected as adults were placed on dogs 85 and 87, respectively. Ticks were allowed to feed for 7 days and were then removed. E. chaffeensis 16S rRNA was detected by RT-PCR in the PBMCs, starting from days 15 to 36 through 153 (dogs 85 and 87) or through day 159 (dogs 150, 350, BKF, and AAX) postattachment (only representative time points are shown in Fig. 4). In addition, groEL transcripts of E. chaffeensis were detected using primers described previously (22) on day 35 or 36 in the 16S rRNA-positive samples (data not shown), indicating that E. chaffeensis was transmitted among dogs by A. americanum ticks.
FIG. 4.
Expression of 16S rRNA of E. chaffeensis in the PBMCs of six dogs infected by tick attachment. Total RNA was extracted and subjected to RT-PCR. The amplified products were resolved on agarose gels containing ethidium bromide. Stages of ticks fed on naïve dogs, dog identifications, and days postattachment are indicated in this sequential order at the top. Larva→Nymph, nymphs infected as larvae; Nymph→Adult, adults infected as nymphs; Adult→Adult, adult males infected as adults; and M, molecular size markers (1-kb plus DNA marker [Life Technologies]). Nucleotide base pair size of amplified product is indicated on the right.
Acknowledgments
This research is supported by grants RO1AI40934 and RO1AI47407 from the National Institutes of Health.
We thank Robert Hamlin, Debra Grover, and Nelson Orellana for providing dogs from Batelle, helping in the technical aspects of tick attachment, and helping in the blood collection, respectively.
Editor: J. T. Barbieri
REFERENCES
- 1.Anderson, B. E., J. E. Dawson, D. C. Jones, and K. H. Wilson. 1991. Ehrlichia chaffeensis, a new species associated with human ehrlichiosis. J. Clin. Microbiol. 29:2838-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, B. E., J. W. Sumner, J. E. Dawson, T. Tzianabos, C. R. Greene, J. G. Olson, D. B. Fishbein, M. Olsen-Rasmussen, B. P. Holloway, E. H. George, and A. F. Azad. 1992. Detection of the etiologic agent of human ehrlichiosis by polymerase chain reaction. J. Clin. Microbiol. 30:775-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, B. E., K. G. Sims, J. G. Olson, J. E. Childs, J. F. Piesman, C. M. Happ, G. O. Maupin, and B. J. Johnson. 1993. Amblyomma americanum: a potential vector of human ehrlichiosis. Am. J. Trop. Med. Hyg. 49:239-244. [DOI] [PubMed] [Google Scholar]
- 4.Breitschwerdt, E. B., B. C. Hegarty, and S. I. Hancock. 1998. Sequential evaluation of dogs naturally infected with Ehrlichia canis, Ehrlichia chaffeensis, Ehrlichia equi, Ehrlichia ewingii, or Bartonella vinsonii. J. Clin. Microbiol. 36:2645-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawson, J. E., B. E. Anderson, D. B. Fishbein, J. L. Sanchez, C. S. Goldsmith, K. H. Wilson, and C. W. Duntley. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J. Clin. Microbiol. 29:2741-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dawson, J. E., K. L. Biggie, C. K. Warner, K. Cookson, S. Jenkins, J. F. Levine, and J. G. Olson. 1996. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human ehrlichiosis, in dogs from southeast Virginia. Am. J. Vet. Res. 57:1175-1179. [PubMed] [Google Scholar]
- 7.Ewing, S. A., J. E. Dawson, A. A. Kocan, R. W. Barker, C. K. Warner, R. J. Panciera, J. C. Fox, and K. M. Kocan. 1995. Experimental transmission of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) among white-tailed deer by Amblyomma americanum (Acari: Ixodidae). J. Med. Entomol. 32:368-374. [DOI] [PubMed] [Google Scholar]
- 8.Felek, S., A. Unver, R. W. Stich, and Y. Rikihisa. 2001. Sensitive detection of Ehrlichia chaffeensis in cultured cells, blood, and ticks by reverse transcription-PCR. J. Clin. Microbiol. 39:460-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ijdo, J. W., C. Wu, L. A. Magnarelli, K. C. Stafford III, J. F. Anderson, and E. Fikrig. 2000. Detection of Ehrlichia chaffeensis DNA in Amblyomma americanum ticks in Connecticut and Rhode Island. J. Clin. Microbiol. 38:4655-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Irving, R. P., R. R. Pinger, C. N. Vann, J. B. Olesen, and F. E. Steiner. 2000. Distribution of Ehrlichia chaffeensis (Rickettsiales: Rickettsiaeceae) in Amblyomma americanum in southern Indiana and prevalence of E. chaffeensis-reactive antibodies in white-tailed deer in Indiana and Ohio in 1998. J. Med. Entomol. 37:595-600. [DOI] [PubMed] [Google Scholar]
- 11.Kordick, S. K., E. B. Breitschwerdt, B. C. Hegarty, K. L. Southwick, C. M. Colitz, S. I. Hancock, J. M. Bradley, R. Rumbough, J. T. McPherson, and J. N. MacCormack. 1999. Coinfection with multiple tick-borne pathogens in a Walker Hound kennel in North Carolina. J. Clin. Microbiol. 37:2631-2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Little, S. E., D. E. Stallknecht, J. M. Lockhart, J. E. Dawson, and W. R. Davidson. 1998. Natural coinfection of a white-tailed deer (Odocoileus virginianus) population with three Ehrlichia spp. J. Parasitol. 84:897-901. [PubMed] [Google Scholar]
- 13.Lockhart, J. M., W. R. Davidson, D. E. Stallknecht, and J. E. Dawson. 1996. Site-specific geographic association between Amblyomma americanum (Acari: Ixodidae) infestations and Ehrlichia chaffeensis-reactive (Rickettsiales: Ehrlichieae) antibodies in white-tailed deer. J. Med. Entomol. 33:153-158. [DOI] [PubMed] [Google Scholar]
- 14.Lockhart, J. M., W. R. Davidson, D. E. Stallknecht, J. E. Dawson, and E. W. Howerth. 1997. Isolation of Ehrlichia chaffeensis from wild white-tailed deer (Odocoileus virginianus) confirms their role as natural reservoir. J. Clin. Microbiol. 35:1681-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lockhart, J. M., W. R. Davidson, D. E. Stallknecht, J. E. Dawson, and S. E. Little. 1997. Natural history of Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) in the piedmont physiographic province of Georgia. J. Parasitol. 83:887-894. [PubMed] [Google Scholar]
- 16.Mueller-Anneling, L., M. J. Gilchrist, and P. S. Thorne. 2000. Ehrlichia chaffeensis antibodies in white-tailed deer, Iowa, 1994 and 1996. Emerg. Infect. Dis. 6:397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy, G. L., S. A. Ewing, L. C. Whitworth, J. C. Fox, and A. A. Kocan. 1998. A molecular and serologic survey of Ehrlichia canis, E. chaffeensis, and E. ewingii in dogs and ticks from Oklahoma. Vet. Parasitol. 79:325-339. [DOI] [PubMed] [Google Scholar]
- 18.Ohashi, N., Y. Rikihisa, and A. Unver. 2001. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect. Immun. 69:2083-2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rikihisa, Y. 1999. Clinical and biological aspects of infection caused by Ehrlichia chaffeensis. Microbes Infect. 1:367-376. [DOI] [PubMed] [Google Scholar]
- 20.Roland, W. E., E. D. Everett, T. L. Cyr, S. Z. Hasan, C. B. Dommaraju, and G. A. McDonald. 1998. Ehrlichia chaffeensis in Missouri ticks. Am. J. Trop. Med. Hyg. 59:641-643. [DOI] [PubMed] [Google Scholar]
- 21.Steiner, F. E., R. R. Pinger, and C. N. Vann. 1999. Infection rates of Amblyomma americanum (Acari: Ixodidae) by Ehrlichia chaffeensis (Rickettsiales: Ehrlichieae) and prevalence of E. chaffeensis-reactive antibodies in white-tailed deer in southern Indiana, 1997. J. Med. Entomol. 36:715-719. [DOI] [PubMed] [Google Scholar]
- 22.Sumner, J. W., K. G. Sims, D. C. Jones, and B. E. Anderson. 1993. Ehrlichia chaffeensis expresses an immunoreactive protein homologous to the Escherichia coli GroEL protein. Infect. Immun. 61:3536-3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unver, A., N. Ohashi, T. Tajima, R. W. Stich, D. Grover, and Y. Rikihisa. 2001. Transcriptional analysis of p30 major outer membrane multigene family of Ehrlichia canis in dogs, ticks, and cell culture at different temperatures. Infect. Immun. 69:6172-6178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Whitlock, J. E., Q. Q. Fang, L. A. Durden, and J. H. Oliver, Jr. 2000. Prevalence of Ehrlichia chaffeensis (Rickettsiales: Rickettsiaceae) in Amblyomma americanum (Acari: Ixodidae) from the Georgia coast and Barrier Islands. J. Med. Entomol. 37:276-280. [DOI] [PubMed] [Google Scholar]