Abstract
Several autotransporter proteins have previously been identified in Neisseria meningitidis. Using molecular features common to most members of the autotransporter family of proteins, we have identified an additional novel ca. 112-kDa autotransporter protein in the meningococcal genomic sequence data. This protein, designated autotransported serine protease A (AspA), has significant N-terminal homology to the secreted serine proteases (subtilases) from several organisms and contains a serine protease catalytic triad. The amino acid sequence of AspA is well-conserved in serogroup A, B, and C meningococci. In Neisseria gonorrhoeae, the AspA homologue appears to be a pseudogene. The gene encoding AspA was cloned and expressed from meningococcal strain MC58 (B15:P1.16b). Anti-AspA antibodies were detected in patients' convalescent-phase sera, suggesting that AspA is expressed in vivo during infection and is immunogenic and cross-reactive. Rabbit polyclonal monospecific anti-AspA serum was used to probe whole-cell proteins from a panel of wild-type meningococcal strains and two AspA mutant strains. Expression of the ca. 112-kDa precursor polypeptide was detected in 12 of 20 wild-type meningococcal strains examined, suggesting that AspA expression is phase variable. Immunogold electron microscopy and cellular fractionation studies showed that the AspA precursor is transported to the outer membrane and remains surface exposed. Western blot experiments confirmed that smaller, ca. 68- or 70-kDa components of AspA (AspA68 and AspA70, respectively) are then secreted into the meningococcal culture supernatant. Site-directed mutagenesis of S426 abolished secretion of both rAspA68 and rAspA70 in Escherichia coli, confirming that AspA is an autocleaved autotransporter protein. In conclusion, we characterized a novel, surface-exposed and secreted, immunogenic, meningococcal autotransporter protein.
Recently, we described two independent approaches, each combining genetic and immunological protocols, based on expression cloning, that led to the identification of autotransporter A (AutA), autotransporter B (AutB), and adhesion and penetration (App) proteins (1, 2). AutA and App are potent B- and T-cell immunogens, and belong to the autotransporter (type V secretion) family of proteins (1, 2). AutB is probably a pseudogene (2). The autotransporter mechanism of protein transportation and secretion was first described for the immunoglobulin A1 (IgA1) proteases of pathogenic Neisseria (26) and is a rapidly expanding family of proteins (14, 15). The characteristic structure of autotransporter proteins and the secretion mechanism have been reviewed elsewhere (15, 17). Autotransporter proteins, possessing a diverse array of N-terminal “functional” domains, have been reported in many gram-negative organisms (1, 2, 5, 6, 9, 10, 12, 13, 23-25, 29-31). Typically, these proteins exhibit virulence-associated functions, such as adhesion, cytotoxicity, serum resistance, and proteolysis (14). Several organisms (e.g., Bordetella pertussis, Escherichia coli, and Haemophilus influenzae) are now recognized to produce an array of autotransporter proteins. After the discovery by Ait-Tahar et al. (2) and others that the meningococcus also possesses multiple autotransporter proteins, we performed an in silico search for additional, unidentified meningococcal autotransporter proteins. This led to the identification of a novel putative autotransporter protein, which was designated autotransported serine protease A (AspA). The aim of the work presented here was to determine whether AspA is: (i) a naturally expressed protein in meningococci, (ii) an autotransporter protein, (iii) surface-exposed and secreted, (iv) expressed in vivo and immunogenic, and (v) conserved and cross-reactive.
MATERIALS AND METHODS
Screening of the meningococcal Z2491 genome for autotransporter proteins.
The predicted coding sequences (CDSs) of the Neisseria meningitidis strain Z2491 (Sanger Centre, Wellcome; http://www.sanger.ac.uk/Projects/N_meningitidis/blast_server.shtml) were searched by using the amino acid sequences of previously reported autotransporter proteins from N. meningitidis (IgA1 protease, AutA, and NhhA), H. influenzae (Hap, Hia, and Hsf), E. coli (Aida-1, EspC, Pet, and Pic), and Serratia marcescens (PrtT, Ssp-H1, and Ssp-H2). Homologous peptides were examined for the typical characteristics of autotransporter proteins and used to perform searches of nonredundant protein databases (using the BLAST program available at http://www.ncbi.nlm.gov/blast.index.html), the N. meningitidis strain MC58 genome (The Institute for Genomic Research [TIGR]; http://www.tigr.org/tdb/tgi.shtml) and the Neisseria gonorrhoeae strain FA1090 genome (University of Oklahoma; http://dna1.chem.ou.edu/gono/html). Putative peptides and their encoding DNA sequences were analyzed by using the DNAMAN programs (Lynnon Biosoft). The promoter prediction by neural network program (http://www.fruitfly.org/seq_tools/promoter.html) and GeneMark (http://genemark.biology.gatech.edu/GeneMark/hmmchoice.html) were used to examine DNA segments for promoter sequences and the presence of ribosome-binding sites. The SignalP (http://www.cbs.dtu.dk/services//SignalP/) program was used to predict the presence of a signal sequence and the Prosite program (http://www.expasy.ch/prosite/) was used to search for conserved amino acid motifs.
Bacterial strains, growth conditions, and media.
E. coli strains JM109 (Promega), TOP10F′ and BL21(DE3)pLysS (Invitrogen) were grown at 37°C in Luria-Bertani (LB) broth or on LB agar supplemented where appropriate, with ampicillin (50 to 100 μg ml−1) or chloramphenicol (34 μg ml−1). N. meningitidis isolates (Tables 1 and 2) were grown at 37°C on chocolate agar or on Mueller-Hinton agar supplemented with Vitox (MHA/V) at the concentration suggested by the manufacturer (Oxoid) in an atmosphere of 5% CO2, or in Mueller-Hinton broth supplemented with Vitox (MHB/V). Where appropriate, streptomycin (50 to 100 μg ml−1) and spectinomycin (50 to 100 μg ml−1) were added to the MHA/V or MHB/V for selection of meningococcal mutants. All meningococcal strains were clinical isolates belonging to different serogroups and types (Tables 1 and 2) and included representative isolates of recognized hypervirulent lineages. The latter were kindly provided by D. Caugant. These were fully characterized by multilocus enzyme sequence typing (22).
TABLE 1.
Phenotypically characterized strains of N. meningitidis used in this studya
| Straina | Serological identity | AspA expression |
|---|---|---|
| Z2491 | A:4,21:P1.7b,13a | + |
| AS | A:4:P1.9 | + |
| BT | A:4:P1.7 | − |
| H44/76 | B:15:P1.7,16 | + |
| SD | B:15:P1.16 | − |
| MC58 | B:15:P1.16b | + |
| OR | C:2a:P1.10 | − |
| GLD | C:2a:P1.5 | − |
| GN | C:NT | − |
| C3 1.1 | C | − |
Strains of different serological phenotypes.
TABLE 2.
Strains of hypervirulent lineages of N. meningitidis used in this study
| Straina | Serogroup | Clonal subgroup | Origin | AspA expression |
|---|---|---|---|---|
| Z1035 | A | I | Pakistan | + |
| Z5010 | A | II | Djibouti | + |
| Z1503 | A | III | China | + |
| Z1269 | A | IV-1 | Burkina Faso | − |
| Z1001 | A | IV-2 | United States | − |
| Z5035 | A | V | China | + |
| Z4696 | B | ET-5 | Norway | + |
| Z6424 | B | Lineage 3 | Netherlands | + |
| Z4181 | C | ET-37 | Mali | + |
| Z6414 | C | Cluster A4 | New Zealand | + |
MLEE/MLST typed strains of hypervirulent lineages.
Patient convalescent-phase sera.
Serum samples were obtained from patients who were convalescing from invasive meningococcal infection. In this study, acute and convalescent-phase (obtained 5 weeks after discharge from hospital) serum samples from six patients were used (Table 3). Human sera were used in Western blots at a dilution of 1:50.
TABLE 3.
Patients from whom convalescent serum was obtained
| Patient no. | Isolate | Code | Age (yr) | Sex | Clinical syndrome |
|---|---|---|---|---|---|
| 1 | B:NT:P1.15 | CC | 28 | M | Meningitis |
| 2 | B:NT:P1.4 | CG | 20 | M | Meningitis |
| 3 | Ba | TJ | 17 | F | Meningitis |
| 4 | B:NT:NST | JB | 50 | M | Meningitis |
| 5 | Ba | JS | 19 | F | Meningitis |
| 6 | C:2a:NT | LW | 15 | M | Septicemia |
Serotype and subtype data were not available.
PCR amplification, cloning, and expression of AspA.
Genomic DNA was isolated from meningococcal strains SD and MC58 by using the method described by Chen and Kuo (7). Plasmid DNA was isolated by using the Qiaprep spin miniprep kit (Qiagen). The aspA gene was amplified by PCR from the first ATG initiation codon (aspA) by using genomic DNA of strain SD and MC58 and from base 301 (tr-aspA) by using the genomic DNA of strain MC58. The sequences of the primers used—AspAFor, tr-AspAFor, and AspARev—and their location are shown in Table 4 and Fig. 1, respectively. Taq polymerase used in PCR was purchased from Boehringer Mannheim. PCRs were performed according to the manufacturer's recommendations. Plasmid pGEMAspA(SD) was constructed by ligating gel-purified, undigested aspA PCR product into the T-vector pGEM-T Easy according to the manufactures' instructions (Promega). pGEMAspA(SD) was digested with BamHI and SacI and the aspA DNA fragment was gel purified. Plasmid pQEAspA(SD) was constructed by ligating the BamHI/SacI-digested aspA DNA fragment to the expression vector pQE30 (Qiagen) cut with the same enzymes. Plasmid pQEAspA(SD)2 was constructed by ligating BamHI/SacI-digested aspA PCR product amplified from meningococcal strain SD directly into the expression vector pQE30 cut with the same enzymes. SacI was purchased from Boehringer Mannheim. Restriction endonuclease digestions were performed according to the manufacturer's instructions or at room temperature overnight to cut sites close to the ends of PCR products. The resulting ligations were transformed into E. coli JM109 (Promega) by heat shock according to the manufacturer's instructions.
TABLE 4.
Primers used in the cloning and mutation of AspA
| Primer name | DNA sequence |
|---|---|
| AspAFor | CGC GGA TCC ATG CGA ACG ACC CCA ACC TTC C |
| tr-AspAFor | CGC GGA TCC CTG CAT ACC GGA GAC TTT CC |
| AspARev | CGC GAG CTC TCA GAA CCG GTA GCC TAC GCC |
| AspAfl-1 | CGC GGA TCC CGC GGT TAT TTC TTC GAG CCG |
| AspAfl-2 | CGC GTC GAC GGG CTG AGC TTG TGC GTC ATT G |
| T3Kpn1 | CGC GGT ACC AAT TAA CCC TCA CTA AAG GG |
| T7Kpn1 | CGC GGT ACC GTA ATA CGA CTC ACT ATA GGG C |
| AspAS426A-1.2 | GGG TGC GGA AAA GGC TGT TCC GG |
| AspAS426A-2.0 | CCG GAA CAG CCT TTT CCG CAC CC |
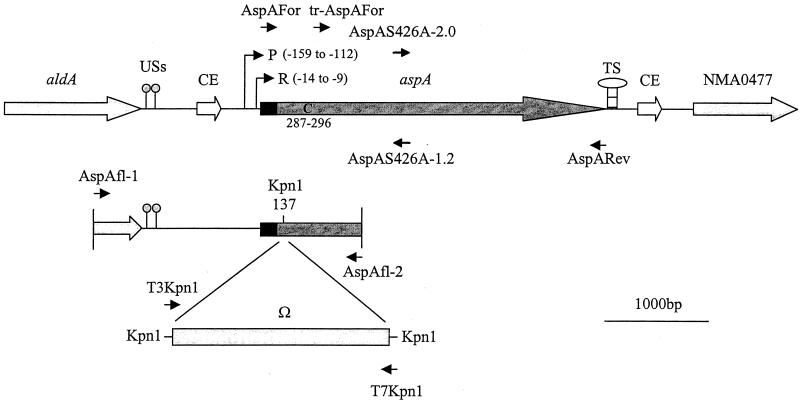
FIG. 1.
aspA locus and mutagenesis of aspA. The aspA gene is flanked by two open reading frames in the same orientation as aspA. Correia elements (CE) are present between these open reading frames and the aspA gene. Two uptake sequences (USs), a putative promoter sequence (P) and a ribosome-binding site (R) are indicated upstream of aspA. A putative transcription termination signal (TS) is indicated downstream of aspA. The predicted signal sequence is shaded black. A 10-bp polycytidine tract (C) is present in aspA. The primers used in this study are indicated. AspAFor and AspARev were used to amplify aspA from meningococcal strain SD and MC58 for cloning and sequencing. tr-AspAFor and AspARev were used to amplify a 5′-truncated aspA from meningococcal strain MC58 for cloning into the expression vector pCRT7/NT-TOPO. The primers AspAfl-1 and AspAfl-2 were used to amplify the region indicated (containing a single, naturally occurring KpnI restriction site at position 137 in the aspA gene), which was cloned in pGEM-T Easy. The Ω cassette, amplified by using the primers indicated and cloned in pGEM-T Easy, was inserted into the KpnI restriction site. The complementary primers, AspAS426A-1.2 and AspAS426A-2.0, were used for the site-directed mutagenesis of S426.
Plasmids pTOPOtr-AspA and pTOPOAspA were constructed by ligating gel-purified, undigested tr-aspA and aspA PCR products from meningococcal strain MC58 to the expression T-vector pCRT7/NT-TOPO (Invitrogen) according to the manufacturer's instructions. pTOPOtr-AspA and pTOPOAspA were transformed into E. coli strain TOP10F′ by heat shock and plasmid preparations from the resultant transformants were analyzed by restriction enzyme digestion to ascertain the orientation of tr-aspA or aspA within the constructs. Constructs in which tr-aspA or aspA were appropriately orientated for translation were chosen and transformed into E. coli strain BL21(DE3)pLysS by heat shock according to the manufacturer's instructions. The pCRT7/NT-TOPO-derived constructs drive expression of the recombinant protein with an N-terminal six-histidine tag under the control of the phage T7 promoter. In the absence of the inducer, IPTG (isopropyl-β-d-thiogalactopyranoside), basal levels of T7 RNA polymerase are reduced by pLysS-derived T7 lysozyme. Anti-His.G HRP antibodies (Invitrogen) were used in Western blot experiments, according to the manufacturer's instructions, to detect recombinant proteins.
Construction of the AspA mutants.
A 2,437-bp region of DNA, encompassing ca. 1.4 kb of upstream DNA and the first ca. 1 kb of aspA (Fig. 1) and containing two complete copies of the neisserial uptake sequence (5′-GCCGTCTGAA-3′), was amplified by PCR from meningococcal strain MC58 with the primers AspAfl-1 and AspAfl-2 (Table 4 and Fig. 1). This DNA fragment was cloned into pGEM-T Easy (Promega) according to the manufacturer's instructions. The resulting plasmid (pGEMAspAfl) contained a single KpnI restriction enzyme site at position 137 from the aspA initiation codon.
The 2-kb Ω element of plasmid pHP45Ω (27) was amplified by using primers based on the T3 and T7 primers and designed to contain terminal KpnI restriction sites (T3Kpn1 and T7Kpn1; Table 4 and Fig. 1). The resulting DNA fragment was cloned into pCRT7/NT-TOPO, and the resulting construct (pTOPOΩ) was propagated in TOP10F′ cells. The Ω element was excised from pTOPOΩ by restriction digest by using KpnI (Promega).
pGEMAspAfl was digested by using KpnI and was ligated to the KpnI-digested Ω element by using T4 DNA ligase (Fermentas). The resulting construct (pTOPOAspAflΩ) was propagated in TOP10F′ cells according to the manufacturer's instructions. pTOPOAspAflΩ was used to mutate N. meningitidis strains MC58 and Z4181 (Tables 1 and 2) by natural transformation and allelic exchange. PCR and partial sequencing (see below) were used to confirm the presence of the Ω element at the desired site in putative meningococcal mutants.
Site-directed mutagenesis of serine-426 (S426) to alanine.
To construct plasmid pTOPOAspAS426A, pTOPOAspA was used as a template to amplify 1.2-kb 5′ and 2.0-kb 3′ PCR fragments that overlapped at a 23-base region containing the mutated site, with the primers AspAFor and AspAS426A-1.2 and the primers AspAS426A-2.0 and AspARev, respectively (Table 5 and Fig. 1). The two fragments were then combined in a 1:1 molar ratio to serve as a template in generating a mutated aspA (aspAS426A) PCR product, with the external primers AspAFor and AspARev plus a 1:100 dilution of AspAS426A-1.2. The recombinant PCR product, aspAS426A, was gel purified and ligated into pCRT7/NT-TOPO, thus generating pTOPOAspAS426A. An appropriately orientated pTOPOAspAS426A construct was chosen, partially sequenced to confirm the desired mutation (see below) and transformed into E. coli strain BL21(DE3)pLysS.
TABLE 5.
Autotransporter proteins used to search the meningococcal genome of Z2491 (serogroup A)
| Autotransporter | Organism | Reference | % Homology to AspA
|
||
|---|---|---|---|---|---|
| Overall | N-terminal | C-terminal | |||
| AutA | N. meningitidis | 2 | 24 | 26 | 24 |
| NhhA | N. meningitidis | 25 | 22 | 24 | 21 |
| IgA1 protease | N. meningitidis | 26 | 20 | 20 | 22 |
| Hap | H. influenzae | 30 | 18 | 18 | 19 |
| Hia | H. influenzae | 5 | 20 | 21 | 20 |
| Hsf | H. influenzae | 29 | 22 | 21 | 21 |
| Aida-1 | E. coli | 6 | 21 | 22 | 24 |
| EspC | E. coli | 31 | 18 | 16 | 21 |
| Pet | E. coli | 10 | 20 | 17 | 21 |
| Pic | E. coli | 13 | 19 | 17 | 23 |
| PrtS | S. marcescens | 23 | 22 | 25 | 18 |
| Ssp-H1 | S. marcescens | 24 | 22 | 24 | 19 |
| Ssp-H2 | S. marcescens | 24 | 26 | 29 | 19 |
DNA sequencing.
The aspA gene, cloned from meningococcal strain SD, was fully sequenced in both directions, as previously described (2), by primer walking with internal primers based on the Z2491 genomic sequence data (Sanger Centre). The aspA gene from meningococcal strain Z4181 was fully sequenced in both directions at the School of Biomedical Sciences (University of Nottingham) on an ABI 377 automated DNA sequencer, with the T7 forward primer, and the same internal primers. Confirmation of the presence of the Ω element at the desired site in putative meningococcal mutants was achieved by partial sequencing of DNA fragments generated by PCR with the primers AspAFor and AspAfl-2 (Table 4). The PCR product was purified by using the High Pure PCR Product Purification Kit (Boehringer Mannheim) according to the manufacturer's instructions. The sequencing was performed at the School of Biomedical Sciences (University of Nottingham) with the AspAFor primer. Confirmation of the mutagenesis of S426 was confirmed by partial sequencing of pTOPOAspAS426A, with two appropriately positioned internal primers described above.
Purification of histidine-tagged recombinant tr-AspA, SDS-PAGE, and Western blotting.
E. coli BL21(DE3)pLysS containing pTOPOtr-AspA was grown overnight in 10 ml of Luria-Bertani (LB) broth containing ampicillin and chloramphenicol. One milliliter of the culture was added to 50 ml of fresh LB broth and allowed to grow to an optical density at 600 nm (OD600) of 0.5 to 0.6 before the addition of IPTG to 1 mM. After 3 h, cells were harvested and the recombinant protein was purified under denaturing or nondenaturing conditions as previously described (2). To provide appropriate negative controls for Western blot and dot immunoblot experiments, elutes from E. coli strain BL21(DE3)pLysS transformed with pCRT7/NT-TOPO (with no meningococcal insert) were used. Proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting, as previously described (4, 20).
Rabbit antiserum to purified protein.
Rabbit monospecific polyclonal antibodies to tr-AspA (RαAspA) were raised against the denatured purified protein in a New Zealand White female rabbit. Preimmune serum was obtained before immunization. The animals were immunized four times at 2-week intervals with ca. 20 μg of protein emulsified in Freund complete (first immunization only) or incomplete adjuvant. After three injections, the animals were test bled, boosted once more, and sacrificed 10 days later. The serum was stored at −20°C. Before use in Western blots and in immunogold electron microscopy, RαAspA was preadsorbed overnight at 4°C with a 10% (vol/vol) suspension of MC58AspA cells before being cleared by centrifugation (11,000 × g for 10 min) and filtered through a 0.2-μm-pore-size Minisart filter (Sartorius). RαAspA was used for Western blots at a dilution of 1:500.
Preparation of E. coli supernatant (secreted) proteins.
E. coli strain BL21(DE3)pLysS containing pTOPOAspA or pTOPOAspAS426A was grown overnight in 10 ml of LB broth containing ampicillin and chloramphenicol. Half a milliliter of the culture was added to 5 ml of fresh LB broth and allowed to grow to an OD600 of 0.5 to 0.6 before addition of IPTG to 1 mM. After 3 h, cells were removed by centrifugation (11,000 × g for 10 min), and the supernatant was filtered by using a 0.2-μm-pore-size Minisart filter (Sartorius). The supernatant (secreted) proteins were concentrated by ultrafiltration by using a Vivaspin-2 protein concentrator (30,000-molecular-weight cut off; Vivascience) according to the manufacturer's instructions.
Preparation of meningococcal supernatant and cellular protein fractions.
N. meningitidis strains were grown overnight in MHB/V at 37°C with shaking. The cells were harvested by centrifugation (11,000 × g for 10 min), and the supernatant was filtered through a Minisart filter. Supernatant (secreted) proteins were concentrated by ultrafiltration by using a Vivaspin-2 protein concentrator (30,000-molecular-weight cut off; Vivascience) according to the manufacturer's instructions. The pellet was resuspended in water, and the cells were lysed by sonication. Unlysed cells were removed by centrifugation at 250 × g for 2 min. The supernatant was collected and centrifuged at 11,000 × g for 30 min. The supernatant (cytosolic and periplasmic protein fraction) was collected. The pellet was resuspended in 25 μl of phosphate-buffered saline (PBS) by vigorous pipetting, 25 μl of 4% Triton X-100 in PBS was added, and the mixture was incubated at 37°C for 30 min. Triton-insoluble proteins (outer-membrane-protein [OMP]-enriched fraction) were pelleted by centrifugation at 11,000 × g for 30 min. The supernatant containing Triton-soluble proteins (cytoplasmic-membrane-protein-enriched fraction) was collected. All samples were stored at −20°C until further processing. Supernatant and cellular protein fractions were probed with RαAspA antiserum as previously described (4, 20).
Immunogold labeling and electron microscopy.
Meningococcal strains MC58 and MC58AspA were grown overnight in MHB/V (5 ml) and prepared for immunogold staining and electron microscopy by using a modification of a previously described method (3). Briefly, meningococcal cells from 1 ml of the overnight culture were resuspended in fresh MHB/V, grown for 2 h at 37°C, and then resuspended in 150 μl of distilled water. Formvar-carbon-coated copper grids (200 mesh) treated with poly-l-lysine were floated on drops of the meningococcal suspensions for 1 h. The grids were washed with three drops of distilled water, fixed in 3% glutaraldehyde (Agar Scientific) for 5 min, and placed in blocking solution (PBS containing 5% bovine serum albumin and 0.1% Tween) for 30 min. The grids were then floated on drops of RαAspA (preadsorbed with MC58AspA cells and diluted 1:20 in blocking solution) for 1 h, washed with three drops of blocking solution, and floated on goat anti-rabbit immunoglobulin G (whole molecule) conjugated to 5-nm gold particles (Sigma), diluted 1:20 in blocking solution, for 2 h. The grids were washed with three drops of blocking solution, fixed in 3% glutaraldehyde in distilled water for 15 s, washed in three drops of distilled water, and air dried. The grids were examined by using a JEOL 1010 transmission electron microscope. Photographs were taken by using a Kodak Megaplus camera. Antiserum against meningococcal transferrin-binding proteins was used as a positive control as previously described (3). Preimmune serum from the same rabbit as RαAspA, preadsorbed with MC58AspA cells, was used as a negative control.
Nucleotide sequences.
Accession numbers for the nucleotide sequences of the aspA gene from the meningococcal strains SD (serogroup B) and Z4181 (serogroup C) are AJ277537 and AJ311654, respectively.
RESULTS
Identification of a novel meningococcal autotransporter protein.
Known autotransporter proteins from N. meningitidis, E. coli, S. marcescens, and H. influenzae were used to search the predicted CDSs of N. meningitidis Z2491 (Table 5). Identified meningococcal putative proteins were analyzed for the characteristic features exhibited by autotransporter proteins including: size (>60 kDa); a low cysteine content (two to five); and the presence of a signal sequence and a C-terminal consensus sequence, (Y/V/I/F/W)-x-(F/W) (15, 17). The search and analysis identified a previously uncharacterized CDS (NMA0478) with a deduced molecular mass of ca. 112 kDa. This putative protein showed significant overall and N-terminal homology to the autotransported serine proteases from S. marcescens (Table 5). It also showed homology to SphB1 (30% over 956 amino acids) from Bordetella pertussis (9), Ssa1 (30% identity over 384 amino acids) from Mannheimia (Pasteurella) haemolytica A1 (11, 21), PspA/PspB (21 and 25% identity over 474 and 485 amino acids, respectively) from Pseudomonas fluorescens (19), and multiple other serine proteases from the subtilase family (data not shown). Therefore, we designated this CDS autotransported serine protease A (AspA). Subsequent release of the genome sequence of the serogroup B strain MC58 (32) confirmed that AspA is also present in this serogroup (NMB1969) and that it is highly conserved (96 and 99% identity at the N and C termini, respectively). A possible orthologue of AspA (97% identity at the nucleotide level) was found in the available gonococcal genomic sequence database. Although a predicted initiation codon, ribosome-binding site and promoter sequence were present, the sequence contained 14 termination codons, suggesting that it may be a pseudogene in N. gonorrhoeae.
In strain Z2491, the aspA gene is 3,204 bp long. A predicted ribosome-binding site (nucleotides −14 to −9) and a promoter sequence (nucleotides −159 to −112) were found upstream of the putative ATG initiation codon (Fig. 1). The predicted transcriptional start site is at nucleotide −121. A 10-bp polycytidine tract is present (nucleotides 287 to 296). A 13-bp inverted repeat sequence is present 15 to 27 and 34 to 46 nucleotides downstream of the termination codon, which might represent a transcriptional termination signal. Analysis of the DNA sequence flanking the aspA gene, revealed that aspA is downstream of a gene encoding a putative enzyme involved in folic acid synthesis (para-aminobenzoate synthase component I) and upstream of a gene involved in fermentation (aldehyde dehydrogenase A; aldA). In addition, the aspA gene is immediately flanked by “Correia elements” (Fig. 1).
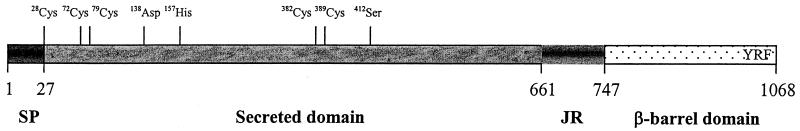
The aspA gene encodes a deduced peptide comprising 1,068 amino acids (Fig. 2 and 3). A 27-amino-acid long predicted signal sequence is found at the N terminus. AspA contains only five cysteine residues; a single cysteine at the N terminus of the predicted mature protein after cleavage of the signal sequence (28Cys) and two pairs of cysteines, each separated by six amino acids (72Cys-79Cys and 382Cys-389Cys). Conserved aspartic acid (138Asp), histidine (157His), and serine (412Ser) residues are present, which presumably compose the serine protease catalytic triad. An ATP/GTP binding site (P-loop) motif (1010Ala-1017Thr), which is present in many autotransporter proteins (15) was also predicted. Unlike the serine proteases (and Ssp-H1/H2) of S. marcescens, however, an RGD motif was absent (15). In comparison with PrtS and Ssp-H1/H2 of S. marcescens, an N-terminal domain (from 28Cys-661Asp; ∼67 kDa), a junctional domain (662Ser-747Thr), and a C-terminal domain (748Phe-1068Phe; ∼33 kDa) were identified.
FIG. 2.
Alignment of the deduced amino acid sequences of AspA from meningococcal strains Z2491 (Sanger Centre), MC58 (TIGR), and Z4181 (this study). CDS and accession numbers are as follows: Z2491, NMA0478; MC58, NMB1969; and Z4181, AJ311654.
FIG. 3.
Schematic diagram of the AspA protein showing the putative domains and conserved motifs (see the text), based on homology to PrtS from S. marcescens (accession number P09489). SP, signal peptide; JR, junctional region.
Sequencing of aspA.
The aspA gene of meningococcal strains SD (accession number AJ277537) and Z4181 (accession number AJ311654) were fully sequenced. A 15-bp polycytidine tract was present in strain SD (nucleotides 287 to 301), which placed a premature termination codon in-frame. Apart from this, the aspA gene from strain SD was almost identical to that of strain MC58 (4-nucleotide difference). The polycytidine tract in the aspA gene from strain Z4181, as in strains Z2491 and MC58, was 10 bp. The aspA sequence from the serogroup C strain (FAM18) genomic sequence assembly (Sanger Centre) contains a 9-bp polycytidine tract, placing a premature termination codon in-frame as in strain SD. Apart from this difference, the sequence was virtually identical to that of Z4181 (1-nucleotide difference).
The AspA amino acid sequences from strains Z2491 and Z4181 are 99% identical. The AspA sequence from strain MC58 was 97 and 98% identical to Z2491 and Z4181, respectively (Fig. 2).
Cloning and expression of truncated AspA.
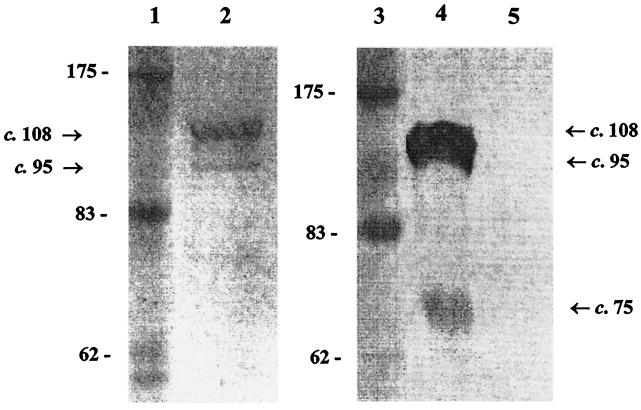
Prior to determination of the sequence of aspA from strain SD, several attempts were made to express the complete aspA gene of strain SD. Despite successful cloning into the pQE30 expression vector, no protein expression was found (not shown). Failed attempts at affinity purification excluded low-level expression. A 5′-truncated aspA sequence (excluding the polycytidine tract and its upstream DNA) was subsequently amplified from meningococcal strain MC58 and cloned into the expression vector pCRT7/NT-TOPO. After induction with IPTG, cells containing this new plasmid construct expressed a protein of the predicted size (ca. 108 kDa), which includes the truncated AspA (tr-AspA), and an additional 35 amino acids, including the six-histidine tag, added by the vector. Figure 4 shows affinity-purified tr-AspA electrophoresed on an SDS gel. Two additional lower-molecular-mass proteins (c. 95 and 75 kDa) copurified with tr-AspA, which were absent from the negative control (not shown). These two additional proteins reacted with anti-His.G antibodies in Western blot experiments (not shown) and probably represent C-terminally truncated forms of tr-AspA.
FIG. 4.
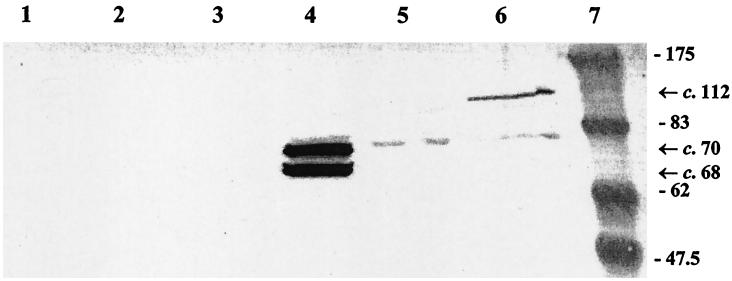
Coomassie brilliant blue-stained SDS-polyacrylamide gel (left) and Western blot probed with RαAspA (right). Lanes 1 and 3, molecular mass markers; lanes 2 and 4, affinity-purified tr-AspA (∼108 kDa) and the copurifying proteins of ca. 95 and 75 kDa, respectively (the ∼75 kDa is not visible on the SDS gel); lane 5, elute from the host E. coli strain not expressing tr-AspA. The numbers indicate the size in kilodaltons.
Mutation of aspA.
A 2,437-bp fragment of DNA flanking the initiation codon of aspA (including 1.4 kb of upstream DNA) was amplified by PCR from meningococcal strain MC58 and cloned into pGEM-T Easy (Fig. 1). The resulting plasmid was digested with the KpnI restriction enzyme and ligated to the KpnI-digested Ω element (a cassette encoding resistance to spectinomycin and streptomycin). This construct was used to mutate meningococcal strains MC58 and Z4181 by natural transformation and allelic exchange to yield MC58aspA and Z4181aspA, respectively. PCR was used to confirm the presence of a 2-kb insert in the aspA gene of both mutants (not shown). Partial sequencing of PCR-amplified fragments confirmed that the Ω element was inserted into aspA immediately after the KpnI restriction enzyme site at position 137 (Fig. 1).
Expression and secretion of AspA and AspAS426A in E. coli.
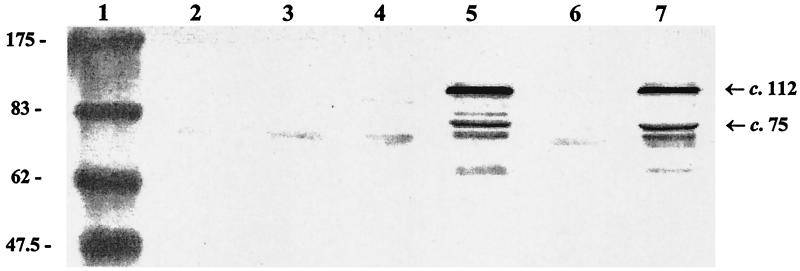
Affinity-purified tr-AspA was used to raise rabbit polyclonal monospecific anti-AspA antibodies (RαAspA). This antiserum reacted strongly with tr-AspA and the two additional copurified proteins (Fig. 4). The complete aspA gene was amplified from meningococcal strain MC58 and ligated into the expression vector pCRT7/NT-TOPO, generating pTOPOAspA. After induction with IPTG, cells containing pTOPOAspA expressed a protein of approximately 112 kDa as predicted, which reacted with RαAspA in Western blots (Fig. 5). Concentrated E. coli supernatant (secreted) protein preparations contained two IPTG-inducible, RαAspA-reactive proteins of ∼68 (rAspA68) and 70 (rAspA70) kDa, respectively, which were absent from whole-cell protein preparations (Fig. 6).
FIG. 5.
Western blot of RαAspA probed against whole-cell proteins from E. coli BL21(DE3)pLysS containing pCRT7/NT-TOPO (with no insert; lanes 2 and 3), pTOPOAspA (lanes 4 and 5), and pTOPOAspAS426A (lanes 6 and 7). Lanes 2, 4, and 6 without IPTG and lanes 3, 5, and 7 with IPTG induction. Lane 1, molecular mass markers. (The numbers indicate the size in kilodaltons.) Arrows to the right indicate the ∼112-kDa rAspA precursor protein (together with a ∼75-kDa breakdown product).
FIG. 6.
Western blot of RαAspA probed against concentrated supernatant (secreted) proteins from E. coli BL21(DE3)pLysS containing pCRT7/NT-TOPO (no insert; lanes 1 and 2), pTOPOAspA (lanes 3 and 4), and pTOPOAspAS426A (lanes 5 and 6). Lanes 1, 3, and 5 without IPTG and lanes 2, 4 and 6 with IPTG induction. Lane 7, molecular weight markers. The numbers indicate the size in kilodaltons. Arrows to the right indicate the secreted forms of rAspA (68 and 70 kDa) and the 112-kDa precursor protein.
Site-directed mutagenesis of S426 to alanine was accomplished by a three-step PCR protocol. The mutated aspA recombinant PCR product (aspAA426A) was ligated into pCRT7/NT-TOPO. The resultant construct, pTOPOAspAS426A, was partially sequenced to confirm the desired mutation. After induction with IPTG, cells containing pTOPOAspAS426A expressed a c. 112 kDa, RαAspA-reactive protein, which was indistinguishable from the protein expressed by E. coli cells containing pTOPOAspA (Fig. 5). Concentrated E. coli supernatant (secreted) protein preparations, however, lacked both rAspA68 and rAspA70 but contained a trace of the 112-kDa rAspA precursor protein (Fig. 6).
Expression of AspA in meningococci.
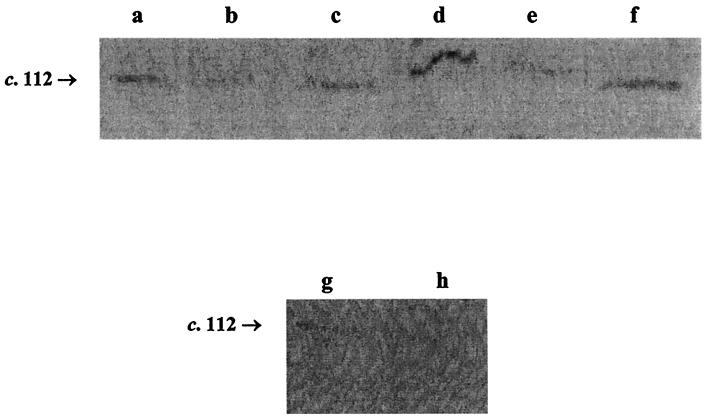
To determine whether AspA is expressed in meningococci, whole-cell extracts from a panel of meningococcal strains (Tables 1 and 2) were probed in Western blots with RαAspA. RαAspA reacted with a 112-kDa protein in 12 of 20 meningococcal strains examined, including Z2491, AS, H44/76, and MC58 and in 8 of 10 strains from hypervirulent lineages (Table 2), but not in the AspA mutant strains Z4181aspA (Fig. 7) and MC58aspA (not shown).
FIG. 7.
Western blots of RαAspA probed against whole-cell proteins from meningococcal strains Z1035 (a), Z1503 (b), Z5035 (c), Z4696 (d), Z6424 (e), 6414 (f), Z4181 (g), and Z4181AspA (h). The AspA precursor protein (∼112 kDa) is present in the wild-type stains but absent from Z4181AspA.
PCR confirmed the presence of the aspA gene in all those meningococcal strains in which AspA expression could not be detected (not shown). The aspA gene from three of these strains (BT, Z1269, and Z1001) was partially sequenced to determine the length of the polycytidine tract. The polycytidine tract was 9 bp long in all three stains, thus placing premature stop codons in frame.
Localization of AspA in meningococci.
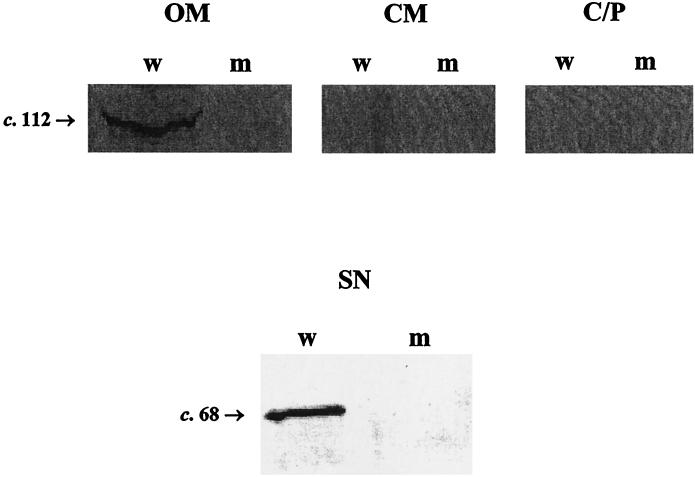
To further localize AspA in the meningococcus and to determine whether AspA is transported, cleaved, and secreted in meningococci, cellular fractions and supernatant protein preparations were probed with RαAspA. RαAspA reacted against the 112-kDa AspA precursor polypeptide in the OMP-enriched fraction of the wild-type meningococcal strain MC58 but not in MC58aspA (Fig. 8). The AspA precursor polypeptide was not present in cytoplasmic-membrane-enriched or cytosolic-periplasmic protein fractions (Fig. 8). Similar results were obtained for meningococcal strains Z4181 and Z4181aspA (not shown). When probed against supernatant proteins, RαAspA reacted with proteins of 68 kDa (AspA68) in strain MC58 (Fig. 8) and 70 kDa (AspA70) in strain Z4181 (not shown). As a control for the purity of the cellular fractions, the OMP-enriched, cytoplasmic-membrane-enriched, and cytosolic-periplasmic fractions were probed in immunoblots with a monoclonal antibody to the OMP PorA. PorA was readily detected (in equal quantities) in the OMP-enriched fractions from meningococcal strains MC58 and MC58AspA but was undetectable in cytoplasmic-membrane-enriched and cytoplasmic-periplasmic fractions (not shown). In addition, the cytoplasmic-membrane and periplasmic protein, ferric binding protein (FbpA), was readily detected in both the cytoplasmic-membrane-enriched and cytoplasmic-periplasmic fractions, but not the OM-enriched fraction, by immunoblot (not shown).
FIG. 8.
Western blot of RαAspA probed against outer-membrane-enriched (OM), cytoplasmic-membrane-enriched (CM), cytosolic-periplasmic (C/P), and concentrated supernatant (SN) fractions from meningococcal strains MC58 (w) and MC58AspA (m). The AspA precursor protein (∼112 kDa) is only present in the outer-membrane-enriched fraction of meningococcal strain MC58. The secreted form of AspA (∼68 kDa) is present in the concentrated supernatant fraction from MC58 but not MC58AspA.
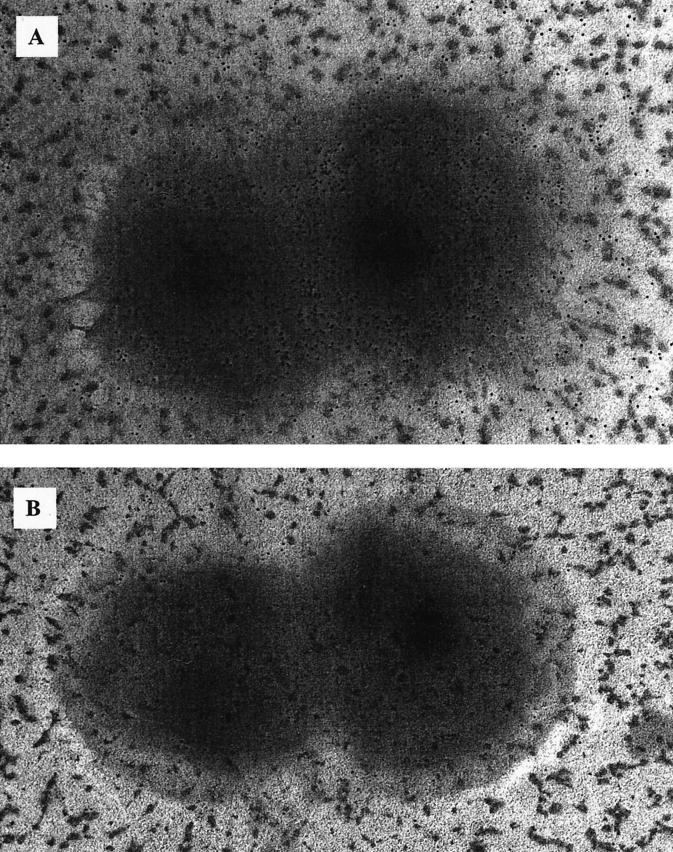
Surface exposure of AspA was investigated by immunogold labeling of meningococcal cells from strains MC58 and MC58aspA. Figure 9 shows an electron micrograph demonstrating the typical immunogold labeling of the wild-type and mutant meningococcal cells. Most MC58 cells were strongly immunogold-labeled (an average of 11 gold particles per cm2 [compared to <1 for MC58aspA]). The spaces between MC58 cells were also immunogold-labeled, albeit at a lower density (an average of 2 gold particles per cm2 [compared to < 0.5 for MC58aspA]), a finding consistent with the secretion of AspA. Positive control cells (incubated with antiserum against meningococcal transferrin-binding proteins) were strongly labeled; negative control cells (incubated with preimmune serum) were not labeled (not shown).
FIG. 9.
Electron micrographs of meningococcal cells (A, strain MC58; B, strain MC58AspA) after incubation in RαAspA and labeling with anti-rabbit immunoglobulin G conjugated to 5-nm gold particles (magnification, ×80,000).
In vivo expression and antigenicity of AspA.
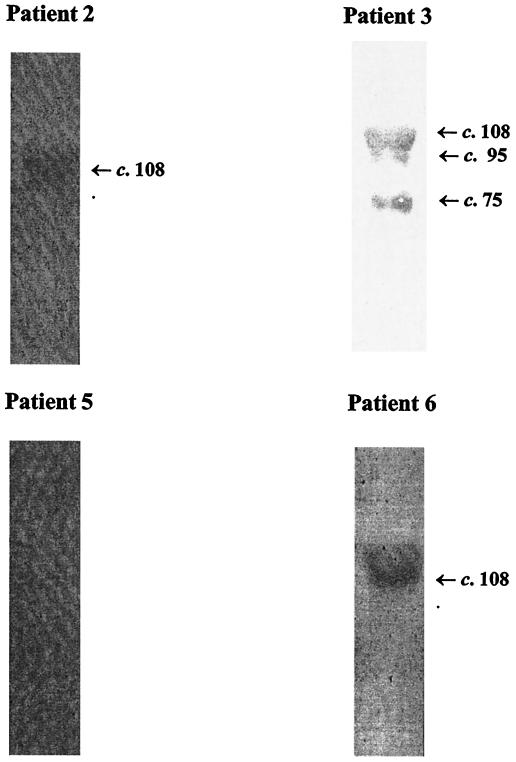
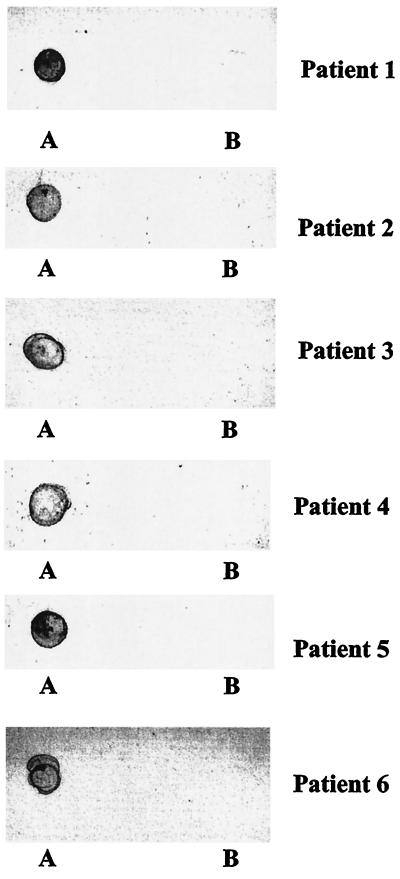
To characterize the in vivo B-cell antigenicity of AspA after invasive meningococcal disease, acute- and convalescent-phase sera from six patients (Table 3) infected with meningococci of different serogroups and types were examined by Western blotting with purified, denatured tr-AspA. Convalescent-phase sera from three of the patients (patients 2, 3, and 6) reacted to tr-AspA (∼108 kDa); one patient (patient 3) also reacted with the two copurified proteins (presumed to be truncated forms of tr-AspA) of 95 and 75 kDa, respectively (Fig. 10). Acute-phase sera were unreactive (not shown). Attempts were made to determine whether the lack of antibody reaction in patients 1, 4, and 5 was due to protein denaturing and hence the loss of conformational epitopes. tr-AspA purified under nondenaturing conditions was used to screen the six patients' sera in dot immunoblot experiments. All six patients' sera reacted to the native protein (Fig. 11). The reactions were specific to tr-AspA since none of the sera reacted with the negative control, which consisted of elutes from the E. coli host strain not expressing tr-AspA.
FIG. 10.
Western blots of denatured, affinity-purified tr-AspA probed with convalescent-phase serum from patients 2, 3, 5, and 6 (see Table 3). Arrows to the right indicate the position of tr-AspA (or major breakdown products). Sera from patients 1 and 3 (not shown) and patient 5 were unreactive. None of the sera reacted to the control, which consisted of an elute from the host E. coli strain not expressing tr-AspA (not shown). Numbers refer to sizes in kilodaltons.
FIG. 11.
Dot immunoblots of tr-AspA purified under nondenaturing conditions (A) and elutes from the host E. coli strain not expressing tr-AspA (B), probed with convalescent-phase sera from patients 1 to 6 (Table 3).
DISCUSSION
Since the first description of IgA1 protease of N. gonorrhoeae by Pohlner et al. (26), an increasing number of proteins have been discovered in various gram-negative organisms, which share the same mechanism of transportation, sometimes referred to as type V secretion (15). Together, they form a large family named autotransporters (15). Most, if not all, of the studied autotransporter proteins are known or putative virulence determinants that are important in bacterial pathogenesis and/or protection from the host's defenses. It is interesting that apart from the mechanism of secretion, which is largely determined by the C-terminal domain of the proteins, members of this family are highly diverse, with unrelated N-terminal “functional” domains.
Ait-Tahar et al., Abdel-Hadi et al., and others have recently identified at least four different autotransporter proteins in N. meningitidis. These include IgA1 protease, App, AutA, AutB (a pseudogene), and NhhA (1, 2, 25, 26). We investigated the possibility that N. meningitidis might utilize this common mode of protein transportation to secrete additional, functionally diverse autotransporter proteins. The availability of the meningococcal genomic sequence data allowed a screening strategy to be devised by using a selection of autotransporter proteins from N. meningitidis itself and other gram-negative organisms. Proteins that were identified by this screening process were then examined for the presence of the characteristic features of autotransporter proteins and for homology to known autotransporter proteins. We identified a novel putative autotransporter protein, which was designated AspA in view of its N-terminal homology to the autotransported serine proteases PrtT and PrtS, and their homologues, Ssp-H1 and Ssp-H2, in S. marcescens.
AspA also shows 30% N-terminal homology to the autotransported serotype-1 specific antigen (Ssa1) in M. haemolytica A1, which is recognized as the principal etiological agent causing fibrinonecrotizing pleuropneumonia in cattle. In contrast to M. haemolytica serotype 2, which is a common upper respiratory tract commensal in healthy cattle, serotype 1 is rarely cultured in the absence of disease (11, 21). Although both serotypes possess the gene encoding Ssa1, expression of Ssa1 cannot be detected serologically in M. haemolytica serotype 2. Intriguingly, cattle that exhibit immunity to pneumonic pasteurellosis, have been found to possess protective, Ssa1-specific antibodies (33). In this context, it is interesting to speculate whether anti-AspA antibodies offer protection against invasive meningococcal disease.
Recently, an autotransporter protein, designated SphB1, was described in B. pertussis, which has 30% overall homology to AspA (9). SphB1 serves as a maturation protease in B. pertussis; it is specifically required for the maturation of the filamentous hemagglutinin (FHA) precursor, FhaB, thereby allowing the extracellular release of mature FHA (9). Furthermore, preliminary evidence suggests that the sphB1 mutation affects the ability of B. pertussis to colonize the respiratory tract in a mouse model of infection (9). The meningococcal genomic databases for strains Z2491 and MC58, both contain putative homologues of FhaB (NMA0688 and NMB0497/1779, respectively); we are currently investigating whether AspA is involved in the maturation and secretion of these putative proteins.
Having identified AspA, the main aim of this study was to characterize the molecular features of the protein: demonstrating that it is indeed an autotransporter protein, determining whether it is finally surface exposed and/or secreted, and determining its in vivo expression and fully characterizing its immunochemical properties. The genes flanking aspA encode housekeeping enzymes, which are unlikely to be transcriptionally or functionally related to AspA. The aspA gene is immediately flanked by “Correia elements,” which may act to increase the rate of horizontal transfer of the aspA locus (8). A promoter sequence was predicted with a high probability upstream of aspA, further suggesting that aspA is independently transcribed. The derived amino acid sequence of aspA, in common with the majority of autotransporter proteins, was predicted to contain a 27-amino-acid signal sequence. This sequence is the same length as that predicted for PrtS and PrtT (although not homologous) but shorter than those found in the subgroup of autotransporter proteins that possess extended signal peptides (15). In common with all other autotransporter proteins thus far described, AspA from meningococcal strains Z2491, MC58, and Z4181 contains few (five) cysteine residues. It is postulated that a high cysteine content may interfere with translocation through the outer membrane due to the formation of disulfide bonds (18). Of the five cysteine residues present in AspA, one is located at the predicted N terminus of the mature peptide and the remaining four are present as two pairs. Two pairs of cysteine residues are also present in PrtS and PrtT from S. marcescens, suggesting that they may represent conserved motifs. Conserved aspartic acid, histidine, and serine residues were also present although, as exemplified by the PrtS homologues, Ssp-H1 and Ssp-H2, the presence of a potential serine protease catalytic triad does not necessarily imply that the protein will exhibit serine protease activity. Furthermore, the natural substrate(s) and biological significance of the S. marcescens secreted proteases are yet to be determined (14).
The aspA genes of meningococcal strains SD and Z4181 were fully sequenced. Aside from the difference in the length of polycytidine tract, the aspA genes from strains MC58 and SD, are virtually identical, which is not surprising since the two strains belong to the same clonal subgroup (ET-5). Similarly, the aspA genes from the serogroup C strains, FAM18 and Z4181, which are both from the clonal subgroup ET37, are virtually identical. The derived AspA amino acid sequences of the serogroup A strain Z2491 and the serogroup C strain Z4181 were >99% identical (10 amino acids different). Overall, the three meningococcal AspA sequences were >96% identical, suggesting that AspA is a well-conserved protein in meningococci. Sequencing of the aspA gene from additional strains from other clonal subgroups will be required to further assess the degree of AspA conservation. The gonococcal aspA gene, which is 97% identical to the meningococcal aspA gene, contains multiple termination codons, suggesting that it may be a pseudogene in N. gonorrhoeae.
The presence of the polycytidine tract in aspA, 287 bp from the putative ATG initiation codon, suggests that aspA may be phase variable due to the “slipped-strand mispairing” mechanism (16, 28). The length of the polycytidine tract in the aspA gene was identical (10 bp) in strains Z2491, MC58, and Z4181, but was 15 bp in strain SD, thus placing a premature termination codon in-frame. A 9-bp polycytidine tract (placing the same premature stop codon in-frame) was found in three strains in which AspA expression could not be detected by Western blot analysis, and in strain FAM18, which was otherwise virtually identical to the clonally related strain Z4181 (which has a 10-bp polycytidine tract), suggesting that AspA is indeed phase variable.
Initial attempts to overexpress recombinant AspA in E. coli from meningococcal strain SD proved unsuccessful, presumably due to the presence of the polycytidine tract placing the gene out-of-frame. This was not predictable before the aspA gene of strain SD was fully sequenced, and hence, a truncated aspA (referred to as tr-aspA) was cloned from meningococcal strain MC58, which lacked the 5′ sequence up to, and including, the polycytidine tract. The expressed tr-AspA migrated on SDS-PAGE gels with an apparent molecular mass of 108 kDa, which was in good agreement with the predicted molecular mass based on the nucleotide sequence.
Subsequently, the complete aspA gene was cloned and expressed from meningococcal strain MC58. The expressed recombinant AspA (rAspA), migrated on SDS-PAGE gels with an apparent molecular masses of 112 kDa as predicted. Furthermore, secreted forms of rAspA, rAspA68, and rAspA70, with molecular masses of ca. 68 and 70 kDa, respectively, were identified in E. coli supernatant (secreted protein) preparations after IPTG induction. This provides further evidence that AspA is an autotransporter protein and suggested the possibility that AspA might exhibit autoproteolysis. In order to clarify the latter point, the putative active serine residue (S426 in AspA from strain MC58) was mutated to an alanine residue. Although a 112-kDa recombinant precursor protein was still expressed, secretion of rAspA68 and rAspA70 was abolished, confirming that AspA is a self-processing autotransporter protein. The 112-kDa AspA precursor protein was never detected in the culture supernatant from E. coli expressing the wild-type aspA gene, but a trace amount was detected in the culture supernatant from E. coli expressing the S426A mutant aspA gene. This probably reflects a degree of E. coli cell lysis in the presence of the overexpressed but unprocessed AspA precursor protein.
Western blot experiments confirmed that AspA is indeed expressed in nature and is antigenically conserved among meningococci. Predicted precursor proteins migrated at the estimated 112-kDa level on SDS gels. We could not detect AspA in strain SD, the sequence of which contained a 15-bp polycytidine tract with a premature stop codon. Furthermore, the rabbit antiserum confirmed that AspA was expressed in 12 of 20 wild-type (serogroup A, B, and C) meningococcal strains examined (including 8 of 10 strains from hypervirulent lineages). However, the relevance of the observed phase variation during invasive disease is not clear. All six patient's sera contained specific anti-AspA antibodies. It is possible that the organism might switch AspA expression off when not needed, e.g., in vitro or in the nasopharynx, only to express it later in other body compartments during invasion. As highlighted in the case of capsular gene expression, phase variation does not necessarily suggest that the gene is dispensable or that these antigens are not good vaccine targets.
It was clear from cell fractionation that the 112-kDa protein was present in outer membrane-enriched meningococcal fractions but absent from the cytoplasmic membrane and cytosolic fractions, confirming that the AspA precursor polypeptide is translocated to the outer membrane. In the culture supernatants, a smaller 68-kDa (or 70-kDa) immunoreactive protein was detected, suggesting that the larger precursor protein is cleaved before the secreted (presumed N-terminal) domain is released. The precursor and secreted proteins were absent in the mutant strains derived from the MC58 and Z4181 parents. The molecular masses of the secreted forms of AspA are in good agreement with the calculated molecular mass (67 kDa) of the hypothetically deduced secreted domain and consistent with the secretion of rAspA68 and rAspA70 that was observed in E. coli. Curiously, however, whereas in E. coli two secreted forms of AspA (rAspA68 and rAspA70) were clearly observed, in a given meningococcal strain either AspA68 or AspA70 was detected in the culture supernatant, but not both. The variability in size of the secreted form of AspA from MC58 and Z4181 cannot be due to sequence variation as the two sequences are almost identical. Also, the AspA precursor protein from MC58 is clearly capable of self-processing at two distinct cleavage sites when expressed in E. coli. It is possible that overexpression of rAspA in E. coli promotes autoproteolysis at a secondary cleavage site. In wild-type meningococci, the secondary cleavage site may only be processed in the absence of the preferential cleavage site, which might occur as a result of a single amino acid change. This hypothesis is currently under investigation.
As a protein corresponding to the C-terminal β-barrel domain after cleavage of the precursor polypeptide could not be detected in the outer-membrane-enriched fraction, it is presumably either rapidly degraded or poorly immunogenic.
Immunogold labeling confirmed that the AspA precursor polypeptide is present in the meningococcal outer membrane and is surface exposed. Gold labeling was also apparent in the extracellular milieu of the wild-type but not AspA mutant meningococcal cells, a finding consistent with the proposed secretion of the N-terminal domain of AspA.
The presence of anti-AspA antibodies in patients' sera confirms not only that the protein is naturally expressed in vivo but also that it is antigenic and generates cross-reactive antibodies. It is interesting that, similar to our previous observation with AutA, native three-dimensional folding of the protein may be important in detecting cross-reactive antibodies.
In conclusion, we have identified AspA, a novel autotransporter protein in meningococci, which shares homology with serine proteases from B. pertussis and S. marcescens. Its precursor polypeptide is translocated to the meningococcal outer membrane, becomes surface exposed, and a smaller 68- to 70-kDa fragment of AspA is secreted. The aspA gene is phase variable, but its amino acid sequence appears to be well conserved in serogroup A, B, and C meningococci. AspA is expressed in vivo and generates cross-reactive antibodies in patients. We propose that AspA merits further investigation as a potential vaccine antigen and novel virulence determinant.
Acknowledgments
D.P.J.T. is a Medical Research Council Fellow supported by grant G84/5370.
Editor: V. J. DiRita
REFERENCES
- 1.Abdel-Hadi, H., K. G. Wooldridge, K. Robinson, and D. A. A. Ala'Aldeen. 2001. Identification and characterization of App: an immunogenic autotransporter protein of Neisseria meningitidis. Mol. Microbiol. 41:611-623. [DOI] [PubMed] [Google Scholar]
- 2.Ait-Tahar, K., K. G. Wooldridge, D. P. J. Turner, M. Atta, I. Todd, and D. A. A. Ala'Aldeen. 2000. Autotransporter A protein of Neisseria meningitidis: a potent CD4+ T-cell and B-cell stimulating antigen detected by expression cloning. Mol. Microbiol. 37:1094-1105. [DOI] [PubMed] [Google Scholar]
- 3.Ala'Aldeen, D. A. A., N. B. Powell, R. A. Wall, and S. P. Borriello. 1993. Localization of the meningococcal receptors for human transferrin. Infect. Immun. 61:751-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ala'Aldeen, D. A. A., P. Stevenson, E. Griffiths, A. R. Gorringe, L. I. Irons, A. Robinson, S. Hyde, and S. P. Borriello. 1994. Immune responses in humans and animals to meningococcal transferrin-binding proteins: implications for vaccine design. Infect. Immun. 62:2884-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barenkamp, S. J., and J. W. St. Geme III. 1996. Identification of a second family of high-molecular-weight adhesion proteins expressed by non-typable Haemophilus influenzae. Mol. Microbiol. 19:1215-1223. [DOI] [PubMed] [Google Scholar]
- 6.Benz, I., and M. A. Schmidt. 1992. AIDA-I, the adhesin involved in diffuse adherence of the diarrhoeagenic Escherichia coli strain 2787 (O126:H27), is synthesized via a precursor molecule. Mol. Microbiol. 6:1539-1546. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W. P., and T. T. Kuo. 1993. A simple and rapid method for the preparation of gram-negative bacterial genomic DNA. Nucleic Acids Res. 21:2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Correia, F. F., S. Inouye, and M. Inouye. 1988. A family of small repeated elements with some transposon-like properties in the genome of Neisseria gonorrhoeae. J. Biol. Chem. 263:12194-12198. [PubMed] [Google Scholar]
- 9.Coutte, L., R. Antoine, H. Drobecq, C. Locht, and F. Jacob-Dubuisson. 2001. Subtilisin-like autotransporter serves as maturation protease in a bacterial secretion pathway. EMBO J. 20:5040-5048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eslava, C., F. Navarro-Garcia, J. R. Czeczulin, I. R. Henderson, A. Cravioto, and J. P. Nataro. 1998. Pet, an autotransporter enterotoxin from enteroaggregative Escherichia coli. Infect. Immun. 66:3155-3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gonzalez, C. T., S. K. Maheswaran, and M. P. Murtaugh. 1995. Pasteurella haemolytica serotype 2 contains the gene for a noncapsular serotype 1-specific antigen. Infect. Immun. 63:1340-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guyer, D. M., I. R. Henderson, J. P. Nataro, and H. L. Mobley. 2000. Identification of sat, an autotransporter toxin produced by uropathogenic Escherichia coli. Mol. Microbiol. 38:53-66. [DOI] [PubMed] [Google Scholar]
- 13.Henderson, I. R., J. Czeczulin, C. Eslava, F. Noriega, and J. P. Nataro. 1999. Characterization of pic, a secreted protease of Shigella flexneri and enteroaggregative Escherichia coli. Infect. Immun. 67:5587-5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 16.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 17.Jose, J., F. Jahnig, and T. F. Meyer. 1995. Common structural features of IgA1 protease-like outer membrane protein autotransporters. Mol. Microbiol. 18:378-380. [DOI] [PubMed] [Google Scholar]
- 18.Jose, J., J. Kramer, T. Klauser, J. Pohlner, and T. F. Meyer. 1996. Absence of periplasmic DsbA oxidoreductase facilitates export of cysteine-containing passenger proteins to the Escherichia coli cell surface via the Iga beta autotransporter pathway. Gene 178:107-110. [DOI] [PubMed] [Google Scholar]
- 19.Kawai, E., A. Idei, H. Kumura, K. Shimazaki, H. Akatsuka, and K. Omori. 1999. The ABC-exporter genes involved in the lipase secretion are clustered with the genes for lipase, alkaline protease, and serine protease homologues in Pseudomonas fluorescens no. 33. Biochim. Biophys. Acta 1446:377-382. [DOI] [PubMed] [Google Scholar]
- 20.Kizil, G., I. Todd, M. Atta, S. P. Borriello, K. Ait-Tahar, and D. A. A. Ala'Aldeen. 1999. Identification and characterization of TspA, a major CD4+ T-cell- and B-cell-stimulating Neisseria-specific antigen. Infect. Immun. 67:3533-3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lo, R. Y., C. A. Strathdee, P. E. Shewen, and B. J. Cooney. 1991. Molecular studies of Ssa1, a serotype-specific antigen of Pasteurella haemolytica A1. Infect. Immun. 59:3398-3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiden, M. C., J. A. Bygraves, E. Feil, G. Morelli, J. E. Russell, R. Urwin, Q. Zhang, J. Zhou, K. Zurth, D. A. Caugant, I. M. Feavers, M. Achtman, and B. G. Spratt. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. USA 95:3140-3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miyazaki, H., N. Yanagida, S. Horinouchi, and T. Beppu. 1989. Characterization of the precursor of Serratia marcescens serine protease and COOH-terminal processing of the precursor during its excretion through the outer membrane of Escherichia coli. J. Bacteriol. 171:6566-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi, Y., T. Beppu, and S. Horinouchi. 1997. Two genes encoding serine protease homologues in Serratia marcescens and characterization of their products in Escherichia coli. J. Biochem. 121:902-913. [DOI] [PubMed] [Google Scholar]
- 25.Peak, I. R., Y. Srikhanta, M. Dieckelmann, E. R. Moxon, and M. P. Jennings. 2000. Identification and characterization of a novel conserved outer membrane protein from Neisseria meningitidis. FEMS Immunol. Med. Microbiol. 28:329-334. [DOI] [PubMed] [Google Scholar]
- 26.Pohlner, J., R. Halter, K. Beyreuther, and T. F. Meyer. 1987. Gene structure and extracellular secretion of Neisseria gonorrhoeae IgA protease. Nature 325:458-462. [DOI] [PubMed] [Google Scholar]
- 27.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 28.Saunders, N. J., A. C. Jeffries, J. F. Peden, D. W. Hood, H. Tettelin, R. Rappuoli, and E. R. Moxon. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37:207-215. [DOI] [PubMed] [Google Scholar]
- 29.St. Geme, J. W., III, D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178:6281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St Geme, J. W., III, M. L. de la Morena, and S. Falkow. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol. Microbiol. 14:217-233. [DOI] [PubMed] [Google Scholar]
- 31.Stein, M., B. Kenny, M. A. Stein, and B. B. Finlay. 1996. Characterization of EspC, a 110-kilodalton protein secreted by enteropathogenic Escherichia coli which is homologous to members of the immunoglobulin A protease-like family of secreted proteins. J. Bacteriol. 178:6546-6554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 33.Weldon, S. K., D. A. Mosier, K. R. Simons, R. C. Craven, and A. W. Confer. 1994. Identification of a potentially important antigen of Pasteurella haemolytica. Vet. Microbiol. 40:283-291. [DOI] [PubMed] [Google Scholar]