Abstract
Shiga toxin (Stx) types 1 and 2 are encoded within intact or defective temperate bacteriophages in Stx-producing Escherichia coli (STEC), and expression of these toxins is linked to bacteriophage induction. Among Stx2 variants, only stx2e from one human STEC isolate has been reported to be carried within a toxin-converting phage. In this study, we examined the O91:H21 STEC isolate B2F1, which carries two functional alleles for the potent activatable Stx2 variant toxin, Stx2d, for the presence of Stx2d-converting bacteriophages. We first constructed mutants of B2F1 that produced one or the other Stx2d toxin and found that the mutant that produced only Stx2d1 made less toxin than the Stx2d2-producing mutant. Consistent with that result, the Stx2d1-producing mutant was attenuated in a streptomycin-treated mouse model of STEC infection. When the mutants were treated with mitomycin C to promote bacteriophage induction, Vero cell cytotoxicity was elevated only in extracts of the Stx2d1-producing mutant. Additionally, when mice were treated with ciprofloxacin, an antibiotic that induces the O157:H7 Stx2-converting phage, the animals were more susceptible to the Stx2d1-producing mutant. Moreover, an stx2d1-containing lysogen was isolated from plaques on strain DH5α that had been exposed to lysates of the mutant that produced Stx2d1 only, and supernatants from that lysogen transformed with a plasmid encoding RecA were cytotoxic when the lysogen was induced with mitomycin C. Finally, electron-microscopic examination of extracts from the Stx2d1-producing mutant showed hexagonal particles that resemble the prototypic Stx2-converting phage 933W. Together these observations provide strong evidence that expression of Stx2d1 is bacteriophage associated. We conclude that despite the sequence similarity of the stx2d1- and stx2d2-flanking regions in B2F1, Stx2d1 expression is repressed within the context of its toxin-converting phage while Stx2d2 expression is independent of phage induction.
Stx-producing Escherichia coli (STEC) causes a spectrum of diseases in humans, from mild diarrhea to hemorrhagic colitis and the potentially fatal hemolytic uremic syndrome (10). The prototype STEC, Escherichia coli O157:H7, is typically acquired by ingestion of contaminated undercooked hamburger, water, or vegetables tainted with manure from asymptomatically infected cattle (2, 9). In the United States alone, the incidence of STEC infection is estimated to be 110,000 cases per year, and approximately 4% of these cases result in the hemolytic uremic syndrome (29). The more severe systemic consequences of STEC infection are attributed to the Shiga toxins (Stxs) produced by these microbes (18, 47).
The two major types of Stx's, Stx1 and Stx2, also referred to as Vero toxins 1 (VT1) and VT2, have the same structure and enzymatic activity but are antigenically distinct (35). While Stxl's made by diverse STEC isolates are essentially the same toxin, several related variants of Stx2 have been described that include Stx2c, Stx2d, Stx2e, and Stx2f (16, 25, 31, 43, 44). These toxins are highly homologous to Stx2 and are cross-neutralizable with anti-Stx2 antibody, but they differ in biological activity (e.g., preferred cellular receptor and/or relative cytotoxicity for Vero and HeLa cells) or the host range of strains that produce them. For example, Stx2c and Stx2d are made by STEC strains isolated from both humans and animals, whereas Stx2e is primarily made by STEC responsible for edema disease of swine. Nevertheless, E. coli bacteria that produce the Stx2e variant are infrequently isolated from humans with gastrointestinal illness (38). Stx2f is made by STEC strains isolated from feral pigeons but has also been associated with diarrhea in a child (7, 43).
Although the pathogenicities of various STEC strains that produce different types of Stx2s cannot be compared directly because the strains are not isogenic, we have found that an O91:H21 strain that produces Stx2 is not virulent in the streptomycin-treated mouse model for STEC infection, whereas O91:H21 strains and an O91 nonmotile isolate that produce Stx2d are highly virulent in those mice (30). Indeed, STEC bacteria that make Stx2d are lethal for orally challenged streptomycin-treated CD-1 mice at very low doses (21, 22), but STEC bacteria that synthesize Stx2 or the variants Stx2c or Stx2e (assessment of virulence of Stx2f-producing STEC not reported) have an oral 50% lethal dose (LD50) of 1010 CFU/CD-1 mouse or greater. The lower LD50 for mice of STEC that produce Stx2d correlates with the capacity of Stx2d to be activated by elastase derived from murine intestinal mucus (19, 31). Activation of Stx2d by elastase, which cleaves two amino acids from the C terminus of the Stx2d A2 peptide, results in increased cytotoxicity of Stx2d to Vero cells (30).
Until recently, Stx2 variants from human isolates of E. coli were thought to be chromosomally encoded; however, an Stx2e toxin-converting phage has been isolated from an O-nontypeable, H-negative human STEC isolate (32). In contrast, Stx1 and Stx2 were shown to exist on inducible bacteriophages as early as 1983 (36, 46; S. M. Scotland, H. R. Smith, G. A. Willshaw, and B. Rowe, Letter, Lancet ii:216, 1983), and subsequent reports demonstrated the homologies of these phages with lambda phage (15). More recent data demonstrate that Stx1 and Stx2 expression is strongly influenced by the phage lytic cycle (33, 50). Circumstances that trigger the host cell SOS response upregulate transcription of recA. The RecA protease cleaves CI, the repressor of the lytic cycle, which, in turn, leads to transcription of the gene for the antitermination factor Q. Q modifies RNA polymerase at the late gene promoter pR′, and transcription proceeds beyond the strong transcription termination site tR′. The toxin genes are then transcribed along with the late phage genes downstream of tR′ (6, 39, 52). In addition, the toxin genes are amplified through bacteriophage genome replication, and host cell lysis promotes toxin release (33, 50).
In this study we began to examine the regulation of activatable Stx2d produced by the O91:H21 Escherichia coli strain B2F1. B2F1 produces two Stx2ds, Stx2d1 and Stx2d2 (formerly designated VT2vha and VT2vhb, respectively, by Ito et al. in 1990 [16] and Shiga-like toxin II-vha [SLTII-vha] and SLTII-vhb by Lindgren et al. in 1993 [21]). These activatable toxins differ by only one amino acid in the noncatalytic portion of the A subunit (16, 30) and are equally toxic to Vero cells and in the mouse model when expressed from plasmids in a K-12 strain (22). Stx2d1 and Stx2d2 also appear to be activated to the same degree by mouse intestinal mucus (31). Examination of DNA sequences that flank stx2d1 and stx2d2 for putative regulatory elements of toxin expression revealed regions of homology with inducible as well as noninducible bacteriophages. Therefore, we hypothesized that regulation of toxin expression is influenced by bacteriophage elements or that the toxin genes are actually bacteriophage borne. To test that theory, we assessed whether factors known to induce bacteriophages influenced toxin production from B2F1 in vitro or virulence in vivo and isolated a toxin-converting bacteriophage that carries stx2d1.
MATERIALS AND METHODS
Bacterial strains, plasmids, phage induction, and growth conditions.
The strains and plasmids used in this investigation are summarized in Table 1. Antibiotics were added at the following concentrations as needed for selection: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; and streptomycin, 50 μg/ml. E. coli strain DH5α served as the host strain for recombinant plasmids. E. coli strain B2F1 was kindly supplied by M. A. Karmali, and a streptomycin-resistant mutant was spontaneously derived from this strain (21). Strains 395-1, C600, and DH5α were challenged with bacteriophage lysates from B2F1 as described below for the detection of bacteriophage plaques and to isolate potential lysogens of Stx2d-converting phages. Strain C600 lysogenized with 933W (36) was challenged with phage lysates from B2F1 to assess phage immunity as an indicator of phage relatedness. All E. coli strains were routinely grown overnight at 37°C in Luria Bertani (LB) broth with aeration or on LB agar. Reduced-salt (2.5 g/liter) LB media supplemented with 10 mM CaCl2 (hereafter called modified LB media) were used for bacteriophage induction and plaque detection. Bacteriophages were induced with 0.5 μg of mitomycin C (Sigma, St. Louis, Mo.)/ml, which was added after broth cultures had been incubated for 1 h. To test for antibiotic induction of bacteriophages, 25 ng of ciprofloxacin (Bayer, Westport, Conn.)/ml or 800 ng of fosfomycin (Sigma)/ml was incorporated into the modified LB broth. Induced cultures were grown with aeration for 4 h at 37°C. Bacteriophages were harvested by chloroform lysis of the host bacterial strain in suspension, centrifugation of the lysate, and filter sterilization (0.45 μm) of the resulting supernatant (25). This clarified cell supernatant was then serially diluted in 10-fold increments in LB broth. Samples (100 μl) of each dilution were incubated at 37°C for 20 min with 200 μl of log-phase indicator cells. These phage-bacterial cell cultures were then added to 2.8 ml of warm, liquid modified LB top agar, the mixtures were overlaid onto LB agar in petri dishes, and the top layer was permitted to solidify at room temperature (double-layer method). After overnight incubation of these double-layer plates at 37°C, the top agar was examined for plaques.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| B2F1 Strr | Spontaneous streptomycin mutant of human isolate of E. coli O91:H21 STEC, encodes stx2dl, stx2d2 | 21 |
| DH5α | K-12 strain recA-E. coli strain | 12 |
| B2F1 mutant 1-1 | stx2d1 toxin knockout, expresses Stx2d2, Strr | This study |
| B2F1 mutant 7-4 | stx2d2 toxin knockout, expresses Stx2d1, Strr | This study |
| 395-1 | K-12 E. coli strain | 41 |
| C600 | K-12 E. coli strain | 47 |
| C600(933W) | Lysogen of Stx2-converting phage 933W (O157:H7 strain EDL933) | 36 |
| PUC18 | Amr | 34 |
| PCM4 | Amr Chr Tcr | Pharmacia |
| pMAK705 | Kmrorits | 11 |
| pJES210 | stx2d2 cosmid clone in pHC79 | This study |
| pSQ12 | stx2d1 cosmid clone in pHC79 | 21 |
| pSQ343 | stx2d1 in Bluescript, Amr | 21 |
| pSQ544 | stx2d2 in Bluescript, Amr | 20 |
| pMB100 | stx2d2 in Bluescript with blunt-ended cat inserted into EcoRV site, Amr | This study |
| pMB101 | pUC18 with stx2d1 | This study |
| pMB102 | stx2d1 in pUC18 with blunt-ended cat inserted into blunted Ava1 and Acc1 sites, Amr | This study |
| pPSTAMP | Derived from pMAK705, Kmr was replaced with Amr, orits | This study |
| pAM450 | pSTAMP with sacB/R cloned in at PstI site | 28 |
| pMB103 | pAM450 with mutated stx2d2 inserted into SalI and BamHI sites | This study |
| pLT10 | pSTAMP with mutated stx2d1 inserted into KpnI and PstI sites | This study |
| pIM10 | Escherichia coli recA clone | 6 |
Recombinant DNA techniques.
Plasmid DNA was isolated by the Miniprep procedure (Qiagen, Valencia, Calif.) according to the manufacturer's instructions. Restriction enzymes were purchased from New England Biolabs (Beverly, Mass.). Restriction DNA fragments were purified by agarose gel electrophoresis and eluted from the gel with GENECLEAN SPIN columns and reagents (Bio 101, Carlsbad, Calif.). T4 ligase was purchased from U.S. Biochemicals (Cleveland, Ohio).
DH5α was made competent for transformation by calcium chloride treatment and heat shock (23). B2F1 or its derivatives were made competent for transformation as described previously by Sizemore (45) or by the procedure of Chuang et al. (5) with the following modifications. These bacteria were grown in LB at 30°C, subjected to heat shock at 37°C, harvested by centrifugation, and resuspended in 10% glycerol prior to being frozen at −80°C. Samples of these frozen treated organisms were thawed as needed and transformed with mutagenic suicide vectors by electroporation with a Bio-Rad Gene Pulser (25 μF, 1.25 kV, 1,000 Ohms).
Nucleic acid sequencing of the stx2d-flanking regions was done with the ABI Prism or Big Dye Sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) with primers synthesized on the ABI Nucleic Acid Synthesizer model 394 or 3948. The products were separated and analyzed with the Applied Biosystems model 377 or 3100, and the sequence results were aligned and compared with those in GenBank of the National Center for Biotechnology and Informatics by the BLAST program, version 2.2.1 (1) or the Wisconsin Package, version 10.1 (Genetics Computer Group, Madison, Wis.).
The PCR was used to amplify a portion of the Stx2d toxin genes with the following primers: LT2 (CAGATAATCAGTGCGAGC) or LT10 (GTCAGAACGGATGATATTGC) and JCS2 (ACTCCGGAAGCACATTGC). The full-length Stx2 or Stx2d gene was amplified with primers CKS1 (TGAGAGCGATCGACTCATAAT) and CKS2 (GACTGAATTGTGACACAGATTA).
AmpliTaq and GeneAmp reagents by Perkin-Elmer (Roche, Branchburg, N.J.) were used for the PCRs done in a Perkin-Elmer DNA Thermocycler (Perkin-Elmer, Norwalk, Conn.) or MJ Research Minicycler (Watertown, Mass.).
Construction of individual toxin-producing mutants of B2F1.
Individual mutants of B2F1 Strr that produced either Stx2d1 or Stx2d2 were derived by allelic exchange as follows. First, a suicide vector, pSTAMP, was constructed in which the kanamycin (kan) resistance marker from the temperature-sensitive vector pMAK705 (11) was replaced with the beta-lactamase gene from pUC18. This step was necessary because B2F1 gives rise to kanamycin-resistant derivatives at a high frequency (A. R. Melton-Celsa, unpublished observation). A sucrose sensitivity allele, sacB/R, was then inserted into the PstI site in pSTAMP to give rise to pAM450 (28). The original purpose of introducing the sacB/R allele into pSTAMP was to create a generalized suicide vector for use in STEC in which putative mutants that retained the sacB/R allele (such as an unresolved cointegrate) could be selected against by the addition of sucrose to the media, as described by Blomfield et al. (4). We subsequently found, however, that B2F1 was not sensitive to high concentrations of sucrose even in the presence of sacB/R. Nevertheless, both pSTAMP and pAM450 (without benefit of the sucrose selection) were used to facilitate allelic exchange in B2F1.
Next, the chloramphenicol acetyltransferase (cat) cassette was released from pCM4 (Amersham Pharmacia Biotech, Piscataway, N.J.) by digestion with BamHI. The DNA polymerase I Klenow fragment (Boehringer Mannheim, Indianapolis, Ind.) was used to blunt the staggered ends that resulted from cleavage with BamHI. The cat cassette was then ligated into pMB101 (stx2d1) that had been digested with AvaI and AccI and treated with Klenow to yield compatible blunt ends, or into pSQ544 (carries stx2d2) at the EcoRV site to make pMB102 and pMB100, respectively. The mutated toxin genes were then subcloned into pSTAMP or pAM450, respectively. B2F1 was transformed with the resulting clones (pLT10 or pMB103) by electroporation (as described above). Putative cointegrates were selected for vigorous growth during incubation at 44°C in the presence of 100 μg of ampicillin/ml. Cointegrates were then resolved by several rounds of growth at 30°C in LB broth with or without chloramphenicol (15 μg/ml). Putative mutants (sensitive to ampicillin with low-level resistance to chloramphenicol) were screened for insertions in the toxin genes by PCR and confirmed by Southern blot analysis as described in the next section.
DNA hybridization studies.
Southern analyses were used to verify that mutational insertions of the appropriate size had been made within the individual toxin genes of B2F1 following mutation by allelic exchange. Chromosomal DNA was isolated by phenol-chloroform extraction (3) from broth-grown wild-type B2F1 and its toxin gene mutants. The DNA was then digested with PstI. The resulting DNA fragments were separated by electrophoresis in 0.8% agarose gels and transferred by capillary action to nitrocellulose with the Turboblotter system (Schleicher & Schuell, Keene, N.H.) for subsequent detection with labeled gene probes.
Dot blot hybridization was used to assess differences in toxin gene copy number in the B2F1 toxin mutants with and without phage induction. The bacteria were grown in LB broth for 4 h in the presence or absence of mitomycin C to induce bacteriophage and then disrupted by sonication. The lysates were clarified by centrifugation, serially diluted, and applied to nitrocellulose membranes with a vacuum manifold. The membranes were probed with a PCR-derived stx2d1 DNA or the cat DNA restricted from pCM4 with BamHI and labeled as described below, and the intensity of the signal was visually compared to detect gene amplification with induction.
Colony hybridization was used to detect DH5α colonies transduced with the stx2d1-bearing phage. Colonies of putative lysogens were lifted from agar plates onto nitrocellulose filters. The filters were treated with 0.5 M NaOH to lyse the colonies and denature the DNA. The membranes were then washed in 5× SSC (75 mM sodium citrate, 0.75 M sodium chloride, [pH 7.0]) and probed with stx2d1 gene DNA. The toxin gene probes for each experiment were generated from PCR-derived DNA products (described above) that were labeled with the ECL Direct Nucleic Acid Labeling System reagents (Amersham Life Science, Buckinghamshire, England). DNA-DNA hybridization was detected with the ECL detection system according to the manufacturer's instructions (Amersham Life Science).
Cytotoxicity measured by Vero cell assay.
Bacterial cell lysates were prepared by sonically disrupting cells from broth cultures. The lysates were then centrifuged to remove cellular debris. To determine toxin levels in mouse feces, pellets were collected, weighed, and suspended in sterile saline to make a 1:10 dilution (wt/vol). The suspensions were homogenized with a vortex mixer, and the supernatants sterilized by filtration. The supernatants from cultures or fecal extracts were serially diluted in tissue culture medium and inoculated into wells of microtiter plates that had been seeded with 104 Vero cells per well 24 h prior to addition of the toxin-containing materials (8). After 48 h of incubation, the viable adherent cells in each well were fixed in 10% formalin and stained with crystal violet, and the absorbance at 600 nm was measured in each well with an automated ELx800 microtiter plate reader (Bio-Tek Instruments Inc., Winooski, Vt.). The reciprocal of the dilution that caused death of 50% of the cells in the monolayer compared with control wells was expressed as the 50% cytotoxic dose (CD50) per milliliter of culture lysate or CD50 per milliliter of fecal extract from a gram of feces. Assays were done at least three times, and the geometric means were calculated from the log values of CD50/ml of lysate or fecal extract. The 95% confidence intervals were determined from the standard errors of the geometric mean of each group.
Mouse model of STEC infection.
The streptomycin-treated mouse model of STEC infection (49) was used to assess virulence of B2F1 Strr and the individual toxin-producing mutants. Briefly, juvenile CD-1 male mice were fed streptomycin water (5 g/liter), and food was withheld overnight to reduce normal gut flora. The following day, bacterial strains that had been grown overnight in LB broth were diluted to the desired concentration in saline and then suspended in a 20% sucrose solution that was fed to the mice in a 25-μl volume. The mice were then permitted food ad libitum but maintained on streptomycin water for the duration of the experiment. To conduct a comparative LD50 study of orally administered B2F1, the Stx2d1-producing mutant, and the Stx2d2-producing mutant, inocula ranging from 102 to 108 CFU were fed to groups of five mice each.
To assess the influence of subinhibitory doses of ciprofloxacin on virulence of the Stx2d1-producing B2F1 mutant, we used a modified version of the protocol described by Zhang et al. (53). These investigators tested the influence of ciprofloxacin therapy on in vivo Stx2 expression by E. coli O157:H7. In our studies, the subinhibitory dose of ciprofloxacin for the Stx2d1-producing B2F1 mutant was defined as that concentration that decreased fecal bacterial counts by 1 to 3 logs. The timing of the dosing of ciprofloxacin to achieve this reduction in CFU/g of feces was determined in a pilot study and was different from that used by Zhang and colleagues (53). The requirement for such an adjustment in the dosing schedule probably reflects the fact that E. coli O157:H7, unlike strain B2F1 and the Stx2d1-producing B2F1 mutant, readily lyses after ciprofloxacin induction in vitro. For these mouse experiments, 20 animals were fed approximately 107 organisms (day zero). Ten of the mice were then treated intraperitoneally on days 2, 3, 4, and 5 with 40 μg of ciprofloxacin (in 100 μl of sterile water), while the other 10 received 100-μl intraperitoneal injections of sterile saline according to the same schedule. Five control mice received ciprofloxacin injections but no bacteria. The actual dose of bacteria given to each group of animals was calculated retrospectively on the basis of CFU/ml of the original overnight broth culture. The mice were assessed daily for signs of illness and death over a 3-week period, and fecal pellets were obtained on days 2 through 5 and again on day 9 to quantitate CFU/g of feces. Note that moribund animals typically stopped producing fecal pellets. The LD50 was calculated by the Reed and Meunch method for computation of 50% endpoints (40). Fecal cytotoxicity levels were assayed as detailed above.
Isolation and identification of an stx2d1 lysogen.
Cultures of wild-type B2F1 were induced with mitomycin C and grown for 4 h. The cells were then treated with 0.5 ml of chloroform per 3.0 ml of broth culture, and the resulting phage lysate was filter sterilized and used to infect indicator strains as described above. The indicator cell-phage mixture was suspended in soft top agar and plated. Samples of the surface agar that contained plaques were excised, suspended in broth, and emulsified, and the supernatant was diluted 106-fold. A 100-μl sample of the diluted broth was plated onto agar and incubated overnight. Isolated colonies that appeared were first subcultured onto LB agar and then transferred onto nitrocellulose membranes to be screened for toxin gene acquisition. Potential lysogens were identified from the colony blots by hybridization with an stx2d1 DNA probe. Probe-positive isolates were transformed with a clone of the recA gene (pIM10, generously provided by T. Oelschlaeger and J. Hacker) to complement the recA defect in DH5α (6). The putative lysogens were grown in broth with and without addition of mitomycin C, and the culture lysates were tested for Vero cell cytotoxic activity. To determine which Stx2d gene had been transduced into DH5α, the toxin gene was amplified by PCR with primers LT2 or LT10 and JCS2 from chromosomal DNA of the lysogen, and the resulting PCR products were digested with EcoRV and AccI. The stx2d1 fragment contains no EcoRV site and one AccI site, while the stx2d2 fragment contains one EcoRV site and two AccI sites (16). The restriction fragments were separated by agarose gel electrophoresis and compared to corresponding restriction digests of PCR products amplified from chromosomal DNA from B2F1 and purified plasmid DNA from Stx2d toxin clones.
Electron microscopy.
Five-hundred-milliliter cultures of the Stx2d1-producing B2F1 mutant or the RecA-complemented DH5α lysogen were induced with mitomycin C and incubated for 4 h, and the cellular material was removed by centrifugation. Bacteriophages were collected from the supernatant by precipitation with polyethylene glycol 8000 (Fisher Biotech, Fair Lawn, N.J.). Chloroform was used to extract the polyethylene glycol 8000 and cell debris, and bacteriophages were harvested from the aqueous phase by centrifugation as described previously (24). The bacteriophage pellet was resuspended in SM buffer (0.1 M NaCl, 10 mM MgSO4, 50 mM Tris·Cl, 0.01% gelatin [pH 7.5]) with gentle agitation at 4°C overnight. Approximately 15 μl of the suspension was applied to Formvar-coated copper grids (Ladd Industries, Burlington, Vt.). After 20 min, excess liquid was absorbed from the edges of the grids with a paper towel, and 15 μl of 2% uranyl acetate (Sigma) was applied to the grids for negative staining. Excess stain was removed by absorption as described above. The dried grids were viewed in a Philips electron microscope, model CM100, under ×94,000 magnification.
Nucleotide sequence accession numbers.
The DNA sequences upstream and downstream of stx2d1 and stx2d2 were determined and submitted to GenBank under the accession numbers AF479828 and AF479829, respectively.
RESULTS
Cytotoxicity and virulence of the B2F1 toxin mutants.
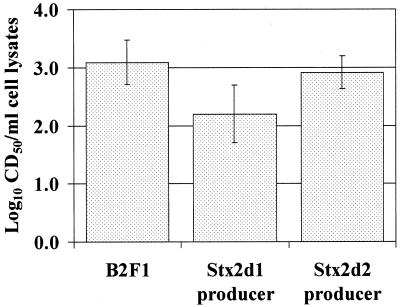
Mutants of B2F1 were generated in which one or the other Stx2d gene was disrupted. The individual mutants grew at the same rate as wild-type B2F1 (data not shown). The single-toxin-producing mutants of B2F1 did not produce equivalent levels of cytotoxin. Rather, the geometric mean CD50/ml of sonically disrupted broth culture of Stx2d1-producing mutant was approximately ninefold lower than that of wild-type B2F1 (Fig. 1). Conversely, the Stx2d2 producer yielded essentially the same levels of cytotoxin as the wild type. When the two mutants were compared, a sevenfold difference in geometric mean CD50/ml of sonically disrupted broth culture was noted (Fig. 1). Although the 95% confidence intervals of the geometric means of the groups overlapped, paired comparisons of these mutants in different experiments, always showed that the Stx2d1-producing mutant was less cytotoxic than the Stx2d2-producing mutant. In contrast, when the individual stx2d1 and stx2d2 genes were separately ligated into the same type vector, the clones expressed comparable levels of toxin as determined by the Vero cell cytotoxicity assay (data not shown). The latter result, combined with the lower toxicity of the mutant that produced Stx2d1, suggests that Stx2d1 expression is repressed in strain B2F1.
FIG. 1.
Cytotoxicities of sonicated lysates of overnight cultures of B2F1 and single-toxin-producing mutants. Columns depict log10 of geometric means of seven or eight experiments, and error bars represent the 95% confidence intervals for the log10 of geometric mean for each group.
The toxicity difference between the mutants was even more pronounced in vivo, as measured in comparative lethal dose studies in mice. The oral LD50 of wild-type B2F1 in streptomycin-treated mice was less than 20 CFU. The Stx2d2-producing mutant was still highly virulent (LD50 = 2 × 102 CFU), but the Stx2d1-producing mutant was almost completely attenuated (LD50 = 108 CFU). Both mutants colonized the mice equally well (data not shown). These results further support the hypothesis that the individual toxin genes are intact but differentially regulated in B2F1 and that Stx2d2 contributes more to cytotoxicity and pathogenicity in mice than does Stx2d1.
Comparison of DNA sequences flanking stx2d1 and stx2d2.
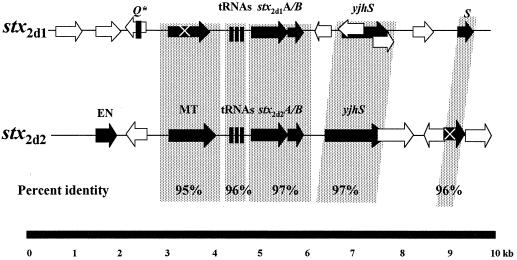
Previous attempts in this laboratory to induce bacteriophages from B2F1 were not successful (20). Therefore, we initially examined the DNA sequences directly upstream of both stx2d1 and stx2d2 in an effort to identify other possible regulatory mechanisms that might influence the differential expression of either toxin. No obvious transcriptional regulatory elements were detected. Instead, we found sequences homologous to lambdoid bacteriophage genes upstream of both toxin genes. We continued to sequence 4 kb upstream and downstream from each stx2d gene and compared the sequences flanking stx2d1 to those flanking stx2d2 (Fig. 2). The DNA sequences upstream of each were very similar to one another (95% identical) over a distance of 1.9 kb. This homologous region contained three putative tRNA genes, ileZ, argN, and argO, directly upstream of each toxin gene and a putative DNA methyl transferase gene just upstream of the tRNA genes. There was a complete open reading frame for the putative methyl transferase upstream of stx2d2, whereas the methyl transferase gene upstream of stx2d1 contained an internal stop codon that would result in a truncated protein product. Upstream beyond the methyl transferase genes the sequences diverged and shared no significant homology.
FIG. 2.
Comparison of stx2d1- and stx2d2-flanking regions to one another. Arrows show relative lengths and directions of open reading frames. Black arrows represent ORFs with homologues in GenBank. White arrows indicate that no homologues were identified. The regions of greatest DNA homology between the stx2d-associated sequences appear on a stippled background. Abbreviations for putative genes are as follows: EN, endonuclease; MT, methyl transferase; Q∗, 50-bp fragment homologous to the 5′ end of the Q gene sequence in 933W; tRNAs, ileZ, argN, argO, S, holin lysis gene, yjhS, E. coli K-12 homologue with unknown function. ORFs containing X encode proteins truncated by stop codons.
Downstream of both stx2d1 and stx2d2 we detected an open reading frame (ORF) homologous to yjhS and of a size (1 kb) comparable to that of the K-12 gene. The function of the protein that yjhS encodes has not yet been defined. The stx2d genes were followed further downstream by sequences homologous to the lambdoid bacteriophage holin gene,S. The intervening DNA sequences between the toxin genes, the yjhS homologues, and the holin genes were less than 75% identical. In sum, the genetic arrangement and DNA sequences of the stx2d1- and stx2d2-flanking regions were very similar from approximately 2 kb upstream to 4 kb downstream of the toxin genes, a finding that suggests they share a common origin. Because of their similarity we could not predict a mechanism for their differential expression from the sequences we studied.
Similarity of the stx2d-flanking DNA to that of other toxin-converting phages.
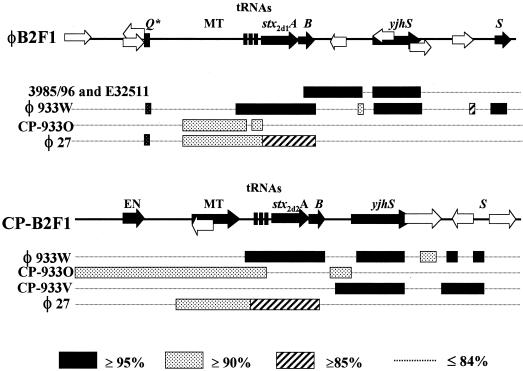
Next we compared the genetic organization and DNA sequences of the stx2d-flanking regions to those of other toxin-converting phages and DNA sequences in GenBank (Fig. 3.). Stx2 toxin-converting phages encode tRNA genes immediately upstream of their toxin genes, and the transcriptional antiterminator-encoding Q gene is directly upstream of the tRNA genes (39, 42). In contrast, both stx2d genes had sequences homologous to those of a bacteriophage-associated DNA methyl transferase gene instead of Q upstream of the tRNA genes. Furthermore, we did not identify a Q gene homologue within the 4 kb that we sequenced upstream of either toxin allele. However, we did find a 50-bp sequence 3 kb upstream of stx2d1 that was 90% identical to the 5′ portion of the Q gene from the stx2-bearing bacteriophage 933W (39), an observation that suggests that a Q gene homologue may have once existed in that region (Fig. 3). Although the stx2-converting phages encode a similar DNA methyl transferase gene, it is generally found further upstream, about 3 kb from the start of the toxin genes.
FIG. 3.
Diagram of DNA sequences flanking stx2d1 and stx2d2 compared with the most closely related sequences from GenBank. Abbreviations are the same as those used in Fig. 2. CP, cryptic phage. Sequences near stx2 in E. coli strain 3985/96 and stx2c in E32511 were available only downstream of the B-subunit toxin genes. Strains 3985/96 and E32511 both exhibited the same extent of homology to φB2F1 over the downstream region shown. A key to percent identities is shown at bottom.
DNA sequences upstream of both stx2d alleles most closely resembled the non-toxin-bearing cryptic phage (CP) CP-933O of strain EDL 933 (37) and a homologue in the Sakai strain of E. coli O157:H7 (13). Specifically, neither the sequences upstream of stx2d1 and stx2d2 nor the region upstream of CP-933O encode a homologue of the 933W Q gene. Furthermore, the DNA sequence 5′ of stx2d2 was homologous to CP-933O over the entire 4 kb sequenced and contains an ORF homologous to the CP933O. The comparable region of stx2d1 was homologous to CP-933O for 1.9 kb and diverged from CP-933O at the same site where the upstream stx2d2 and stx2d1 sequences also began to differ. The region up to 1.9 kb upstream of each toxin gene was also 89% identical to the corresponding region in the stx2e-bearing phage φ27 (32). Beyond the point at which the stx2d1-flanking sequence diverged from stx2d2, we identified one ORF that was homologous in part to a putative cytoplasmic protein, STM2240, in Salmonella enterica serovar Typhimurium (27).
The regions downstream of stx2d1 and stx2d2 have an organizational structure like that of stx1- and stx2-bearing phages (48), both of which also encode an ORF homologous to the yjhS gene of E. coli K-12 (albeit larger, 2 kb). The sequences downstream of the two Stx2d genes showed some highly conserved regions between them, but overall they resembled different toxin-associated phages. The region downstream of stx2d1 most closely resembled the corresponding regions downstream of stx2 in E. coli strain 3985/96 and stx2c in strain E32511 (48) and was somewhat less similar to the region downstream of stx2 in 933W. The sequence 3′ of stx2d2 most closely resembled the stx1-bearing cryptic phage of strain EDL933, CP-933V, but also shared regions of strong homology with 933W. Although the DNA 5′ to both of the toxin genes was similar to φ27, neither of the toxin genes stx2d1 showed homology to φ27 in the 3′ direction. In sum, the stx2d-flanking regions resembled other toxin-converting phages in organization, but these DNA sequences did not show strong identity to any one previously described phage. Rather, the sequences surrounding the stx2d alleles appeared as a patchwork with strong similarities over short distances to a variety of specific genes associated with both inducible and cryptic phages.
Results of bacteriophage induction.
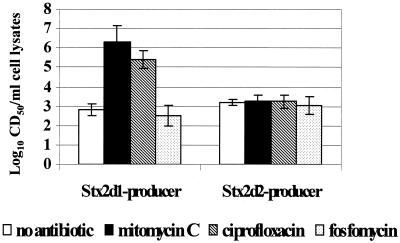
As a first step to determine whether an inducible, toxin-converting phage was indeed present in strain B2F1 (as implied by the sequence data), we measured the cytotoxicity of clarified cell lysates from each toxin mutant after growth in the presence or absence of mitomycin C. Although the broth cultures treated with mitomycin C remained relatively turbid, the levels of cytotoxicity of wild-type B2F1 (data not shown) and the Stx2d1-producing mutant were greatly enhanced by exposure to mitomycin C. The cytotoxicity of the Stx2d2-producing mutant was unchanged by mitomycin C treatment (Fig. 4). These results suggested that Stx2d1, but not Stx2d2, is associated with an inducible bacteriophage. Tiny turbid plaques were observed on E. coli strains 395-1, C600, and DH5α that were treated with cell lysates from either toxin mutant. Because the plaques were barely discernible it was not possible to determine the actual numbers of PFU; however, the most concentrated samples applied to the host bacterial lawn resulted in a very mottled appearance on the host culture surface, and this effect was eliminated by dilution of the phage inoculum.
FIG. 4.
Cytotoxicity of clarified sonic lysates from overnight cultures of single-toxin-producing mutants of B2F1 treated with subinhibitory concentrations of agents that induce bacteriophages. Each columns represents the log10 of the geometric mean for three experiments, and error bars depict 95% confidence intervals for the log10 geometric means.
Dot blots of clarified cell lysates of both mutants probed with stx2d1 DNA revealed an increase in gene dosage upon mitomycin C treatment (data not shown). However, the gene probe used to detect stx2d in these dot blots could not differentiate between mutant and wild-type toxin alleles. When the dot blots were probed with the chloramphenicol acetyltransferase gene to distinguish the mutated from functional toxin allele, only the mutated stx2d1::cat copy number was increased with induction. Mitomycin C induction increased the stx2d1 and stx2d1::cat gene dose and cytotoxicity in the B2F1 mutant with an intact Stx2d1 gene, while Stx2d2 gene copy number and expression were unchanged by this treatment. These findings provide further evidence that stx2d1 expression is bacteriophage associated but stx2d2 expression is not.
Phage immunity.
The DNA sequence immediately upstream of each stx2d was 95% identical to the Stx2 toxin-converting phage 933W. Therefore, we asked whether the putative stx2d1-bearing phage shared lysogenic immunity with phage 933W. For this purpose, we tested the capacity of lysates from B2F1, the individual toxin-producing mutants, and a C600 lysogen of 933W to form plaques on one another. Plaques were observed on C600 and C600(933W) cocultured with extracts from B2F1 or either of the individual toxin-producing mutants of B2F1, a finding that indicates that a phage released from B2F1 was not of the same immunity group as 933W. As expected, the B2F1 phage lysates failed to produce plaques on B2F1, and lysates of C600(933W) did not produce plaques on C600(933W). We anticipated that lysates from C600(933W) would produce plaques on strain B2F1 because lysogenic immunity is generally reciprocal, but no plaques were observed. Other factors, such as capsule type or lack of appropriate receptor molecules, may have prevented infection of B2F1 with 933W. It is also possible that B2F1 is lysogenized with another phage of the same immunity group as phage 933W.
Influence of ciprofloxacin on toxin expression.
Quinolone antibiotics induce bacteriophages and increase toxin production in strains that harbor Stx2-bearing bacteriophages (26, 53). The investigators who reported this finding speculated that the inhibitory effect of these antibiotics on DNA gyrase probably results in the accumulation of single-stranded DNA fragments that trigger the SOS response and a subsequent RecA-mediated conversion of the phage from the lysogenic cycle to the lytic cycle. Based on the observation of quinolone-mediated induction of Stx2-expressing phage, we decided to test whether subinhibitory concentrations of ciprofloxacin exert a similar inductive effect on Stx2d1 expression in vitro and in infected animals. First, we compared cytotoxicity of the single-toxin-producing mutants grown in broth alone to those grown in broth supplemented with mitomycin C, ciprofloxacin, or fosfomycin, an antibiotic that does not induce stx2-converting phages from STEC O157:H7 (53). The results of those antibiotic studies are summarized in Fig. 4. Ciprofloxacin induced Stx2d1 production nearly as well as mitomycin C, while fosfomycin did not cause an increase in cytotoxicity from the mutants. As was the case with mitomycin C induction, increased cytotoxicity with ciprofloxacin induction was seen only with B2F1 (not shown) or the mutant that carried a functional stx2d1 gene.
Next, we examined whether the enhanced toxicity of the Stx2d1 producer that we observed in vitro upon treatment with ciprofloxacin would render this strain virulent for mice treated with ciprofloxacin (Fig. 5). Therefore we fed the Stx2d1-producing mutant to 20 mice and, after 48 h, treated half of the mice with subclinical doses of ciprofloxacin according to a modified version of the protocol used to increase Stx2 production in vivo in antibiotic-treated mice (53). The remainder of the infected mice received intraperitoneal injections of phosphate-buffered saline (PBS) to serve as controls. None of the 10 infected mice that received PBS and none of the 5 uninfected mice given ciprofloxacin died. However, 6 out of 10 of the ciprofloxacin-treated mice infected with the Stx2d1-producing mutant died, compared to 9 of the 10 control mice fed wild-type B2F1.
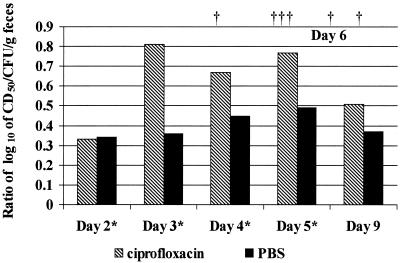
FIG. 5.
Comparison of the ratios of the geometric mean log10 values of cytotoxicity (CD50) per gram of feces to the bacterial counts (CFU) per gram of feces pooled for each group of mice fed the Stx2d1-producing mutant of B2F1. Stippled columns represent samples from mice treated with ciprofloxacin, and black columns depict those treated with PBS. Asterisks indicate the days on which the animals were treated, and daggers (†) represent time points when the six ciprofloxacin-treated animals died.
Fecal pellets from the infected mice were pooled by group and cultured to verify that the mice had become colonized. After antibiotics were administered, fecal pellets were tested for cytotoxicity and cultured for CFU/gram of feces. In the ciprofloxacin-treated group, the number of colony counts per gram of feces decreased with treatment, as expected, while the CD50/gram of feces increased sharply. With the PBS-treated controls the number of CFU/gram remained relatively constant, while the number of CD50/gram increased less dramatically during infection than for the ciprofloxacin-treated mice. The log values of CD50 per CFU for each group at each sample day are depicted in Fig. 5. The ciprofloxacin-treated mice showed up to twofold-higher fecal toxin levels per CFU per gram of feces compared to the PBS-treated controls. The results of this mouse experiment strongly suggest that ciprofloxacin upregulated toxin expression in the B2F1 mutant that only expresses Stx2d1. The implication of this conclusion, in the context of the in vitro induction studies, is that ciprofloxacin therapy induced an Stx2d1-converting phage in vivo.
Isolation of a DH5α lysogen-bearing stx2d1.
The increased cytotoxicity and mouse virulence of the B2F1 mutant that produces Stx2d1 when exposed to ciprofloxacin, together with the formation of bacteriophage plaques when these mutant bacteria were induced with mitomycin C, indicated coordinate Stx2d1 expression and production of phage(s). To assess whether the simultaneous increase in toxin expression and phage plaques occurred because stx2d1 was actually borne on a phage, we attempted to transduce the toxin gene into other E. coli strains. Cultures of wild-type B2F1 were induced with mitomycin C, and sterile cell lysates were prepared and used to heavily infect C600 or 395-1. Samples of the plaqued soft agar were harvested and subcultured for potential lysogens. We reasoned that organisms which did not lyse when challenged with the B2F1 cell extract either were uninfected by phages or were lysogenized and therefore protected from lysis. To identify potential lysogens, we screened the colonies that we recovered from the cultured plaques by PCR for toxin genes or colony blot hybridization with a toxin gene probe. Despite the appearance of incomplete plaques on 395-1 and C600, repeated efforts failed to yield any lysogens of these RecA+ strains. Therefore, we challenged DH5α with B2F1 phage lysates because that RecA-negative strain had been used successfully to isolate a lysogen of the stx2e-bearing phage φ27 (32). Of the 538 DH5α colonies we screened, 38 reacted with the toxin gene probe. Chromosomal DNA of two representative probe-positive colonies was subjected to PCR with two sets of primer pairs that would yield internal toxin gene sequences of two different lengths. Fragments of the appropriate sizes for stx2d were obtained. To verify that the stx2d1 allele and not the stx2d2 allele had been transduced, the PCR products were treated with EcoRV that cleaves stx2d2 but not stx2d1 and AccI that cuts once in stx2d1 and twice in stx2d2. The PCR products were resistant to digestion with EcoRV but cut with AccI, yielding fragments consistent with digests of control PCR products obtained from an stx2d1 clone and distinguishable in size from those obtained from a clone of stx2d2.
To test whether a functional toxin gene had been transduced, we sought to demonstrate that the putative lysogens were cytotoxic. Preliminary Vero cell assays showed that the transductants were not cytotoxic. However, DH5α lacks the RecA protease necessary to cleave the phage repressor of delayed early gene expression, a step that is required for the lytic cycle to be induced in lysogens that contain the lambda-like 933W phage and for concomitant Stx2 expression (6, 33). Therefore, in an effort to promote expression of Stx2d1 from the putative lysogen, we complemented that lysogen of DH5α with a cloned recA gene contained in pIM10. These transformants produced 2.9 × 102 CD50/ml of broth without mitomycin C induction and 104 CD50/ml with induction, a finding that supports the hypothesis that Stx2d1 expression is linked to phage induction. Additionally, phage preparations derived from the induced RecA-complemented lysogen produced plaques on DH5α and 395-1 host cells.
Electron-microscopic examination of phage from the Stx2d1-producing mutant of B2F1 and from the RecA-complemented lysogen of DH5α
Finally, we were able to visualize bacteriophage-like particles by transmission electron microscopy from lysates of induced broth cultures of the Stx2d1-producing mutant of B2F1 (Fig. 6). The stx2d1-converting bacteriophage, which we designated φB2F1, appeared morphologically similar to the stx2-converting phage, 933W. The head appeared to be a regular hexagonal shape. Filaments were also seen that were quite long and did not appear to be attached to the hexagonal particles. We speculated that the strands were flagella, because this preparation was made from the motile B2F1 toxin mutant that produces Stx2d1 and were absent in a preparation made from the nonmotile RecA-complemented lysogen of DH5α (data not shown).
FIG. 6.
Transmission electron micrograph of bacteriophage φB2F1. Bacteriophage preparation was made from a broth culture of the Stx2d2-producing mutant of B2F1. Bar, 100 nm.
DISCUSSION
Four lines of evidence derived from this investigation indicate that the stx2d1 gene for the potent activatable Stx2d1 toxin is borne on an inducible toxin-converting bacteriophage in E. coli strain B2F1. First, the Stx2d1 toxin gene was transferred to DH5α via a protocol used for transduction, and the resulting lysogen was cytotoxic when recA was supplied in trans. We believe that stx2d1 moved by specific transduction rather than generalized transduction because of the high frequency of stx2d1 transductants isolated from bacteriophage plaques on DH5α, the presence of intact phage gene sequences flanking stx2d1, and the fact that Stx genes are often found on competent phages. Second, Stx2d1 expression was increased in vitro and in vivo under conditions that are known to induce bacteriophages. Although the in vivo induction of elevated toxin expression in the presence of ciprofloxacin therapy was not as pronounced as that which we observed in vitro, the biological variations in colonization levels and antibiotic uptake in the mouse model make it difficult to optimize the inductive effect of ciprofloxacin. Nonetheless, the normally attenuated mutant of B2F1 that produces only Stx2d1 became more virulent in the presence of ciprofloxacin. Third, the RecA-dependent nature of Stx2d1 expression in DH5α suggested that Stx2d1 is coregulated with the bacteriophage late genes involved in the lytic cycle. The low level of expression of Stx2d1 without induction implies that φB2F1 is not readily induced spontaneously in vitro or in mice. Fourth, electron-microscopic examination of lysates from the Stx2d1-producing mutant of B2F1 (Fig. 6) or the RecA-complemented lysogen of DH5α revealed particles with a morphology similar to that of the stx2-converting bacteriophage 933W.
Comparison of the stx2d1-flanking regions with the GenBank database revealed genetic as well as structural similarities between φB2F1 and 933W. However, unlike the Stx2-converting phages, no Q gene homologue, Q binding site, or tR′ termination sequence was identified in the corresponding upstream region of the stx2d1-bearing phage. The antiterminator Q enhances the transcription of stx1 and stx2 (33), and it is puzzling that we did not identify a Q homologue in the stx2d1 phage. Perhaps a Q gene homologue is encoded further upstream beyond the 4 kb that we sequenced, or a protein analogous to Q, not readily apparent by DNA sequence, serves as an antiterminator factor. Alternatively, late gene expression in φB2F1 may not be regulated by the same terminator-antiterminator control found in other toxin-converting phages. Roughly 3 kb upstream of the start of the stx2d1 gene there is a 50-bp region that is 90% identical to a portion of the 933W Q gene. This finding suggests a third possibility, that the Q gene was present at some time in the evolution of φB2F1 and was truncated or replaced during a recombination event. The immunity regions of φB2F1 and 933W differ as well, as evidenced by the formation of plaques on a 933W lysogen challenged with lysates from B2F1.
The expression of the other toxin allele in B2F1, stx2d2, was not influenced by bacteriophage induction. Paradoxically, studies with the single-toxin-expressing mutants of B2F1 revealed that the cytotoxicity and virulence in mice of wild-type B2F1 was predominantly attributable to Stx2d2. In the wild-type background, expression from the stx2d1 gene remained relatively low while Stx2d2 expression was apparently constitutive or regulated by some other yet-to-be-defined host or phage factor. The DNA sequence upstream of the stx2d2 gene was similar to that region upstream of stx2d1 and shared homology with the cryptic phage CP-933O. However, the abrupt divergence of these 5′-flanking regions 1.9 kb upstream of stx2d1 and stx2d2 suggests that an insertion, deletion, and/or recombination event occurred during the evolution of B2F1 that either provided a mechanism for Stx2d1 phage repression or disengaged Stx2d2 expression from phage-mediated repression (or both). The sequence downstream of stx2d2 was most similar in structure and sequence to CP-933V, the cryptic stx1-bearing phage of O157:H7. We observed from the DNA sequence that the holin lysis gene homologue downstream of stx2d2 was truncated. If transcription of the stx2d2 gene along with the late phage genes is constitutive, then defects in the late genes that prevent the lytic phase, coupled with tight regulation of Stx2d1 phage induction, would likely be necessary to maintain a lysogenized population of E. coli. Therefore, we conclude that stx2d2 is encoded within a defective or cryptic phage. Additional sequencing and regulation studies have been undertaken in our laboratory to define mechanisms of regulation of Stx2d2 expression.
The sequences flanking stx2d1 and stx2d2 showed homology to more than one bacteriophage. The recombinational promiscuity of lambdoid phages both within and among species has been well documented (14). Moreover, Johansen et al. analyzed stx2-encoding phages from various O157:H7 strains and described them as mosaics in which the toxin genes and basic lambdoid phage organization are conserved but the heterogeneity of individual phage genes reflects exchanges among a broader gene pool (17). Unkmeier and Schmitt have shown that the chromosomally encoded variant toxin genes stx2c and stx2f as well as stx in Shigella are flanked with DNA of phage origin and hypothesized that all Stxs are bacteriophage associated, whether or not they are actually inducible (48). Genomic sequencing has demonstrated that up to 20% of the E. coli chromosome is comprised of bacteriophage DNA that could provide many opportunities for intragenomic homologous recombination as well as recombinational exchange with newly acquired and cryptic bacteriophages (37). We speculate that a recombination event occurred that resulted in the duplication of the 2d toxin gene in B2F1 but that only one of the toxin alleles was situated within a phage that could be induced. Although many STEC strains encode more than one toxin type, B2F1 is the first STEC organism described to our knowledge where two Stx2 variants are differentially regulated.
We previously hypothesized that Stx2d may be more toxic than Stx2 based on our observations that it is activatable and because Stx2d-producing organisms kill mice, but Stx2 producers do not. These studies with the individual toxin-producing mutants of B2F1 show that Stx2d2 is expressed at lethal levels independently of induction, whereas a Stx2d1-only-producing B2F1 derivative is lethal for mice under bacteriophage-inducing conditions. The isolation of an Stx2d toxin-converting phage demonstrates that this activatable toxin has the potential to be transferred horizontally and reinforces the notion that quinolone therapy is contraindicated for treatment of E. coli O157:H7 infections (51, 53).
Acknowledgments
We gratefully acknowledge Tom Baginski for his assistance with electron microscopy and Steve Darnell and Edda Twiddy for their technical assistance with the cytotoxicity assays. We also acknowledge Marian Batts and Belinda Davis for their contributions to the production of the B2F1 toxin mutants.
This work was funded by a Public Health Service grant from the National Institutes of Health (AI20148-18).
The opinions or assertions contained herein are the private views of the authors and are not to be construed as the views of the Department of Defense.
Editor: J. T. Barbieri
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong, G. L., J. Hollingsworth, and J. G. Morris, Jr. 1996. Emerging foodborne pathogens: Escherichia coli O157:H7 as a model of entry of a new pathogen into the food supply of the developed world. Epidemiol. Rev. 18:29-51. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1989. Current protocols in molecular biology. Greene Publishing Associates and Wiley Interscience, New York, N.Y.
- 4.Blomfield, I. C., V. Vaughn, R. F. Rest, and B. I. Eisentein. 1991. Allelic exchange in Escherichia coli using the Bacillus subtilis sacB gene and a temperature-sensitive pSC101 replicon. Mol. Microbiol. 5:1447-1457. [DOI] [PubMed] [Google Scholar]
- 5.Chuang, S.-E., A.-L. Chen, and C.-C. Chao. 1995. Growth of E. coli at low temperature dramatically increases the transformation frequency by electroporation. Nucleic Acids Res. 23:1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchs, S., I. Muhldorfer, A. Donohue-Rolfe, M. Kerenyi, L. Emody, R. Alexiev, P. Nenkov, and J. Hacker. 1999. Influence of RecA on in vivo virulence and Shiga toxin 2 production in Escherichia coli pathogens. Microb. Pathog. 27:13-23. [DOI] [PubMed] [Google Scholar]
- 7.Gannon, V., C. Teerling, S. A. Masri, and C. Gyles. 1990. Molecular cloning and nucleotide sequence of another variant of the Escherichia coli Shiga-like toxin II family. J. Gen. Microbiol. 136:1125-1135. [DOI] [PubMed] [Google Scholar]
- 8.Gentry, M. K., and J. M. Dalrymple. 1980. Quantitative microtiter cytotoxicity assay for Shigella toxin. J. Clin. Microbiol. 12:361-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffin, P. M. 1995. Escherichia coli O157:H7 and other enterohemorrhagic Escherichia coli, p. 739-761. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, Ltd., New York, N.Y.
- 10.Griffin, P. M., S. M. Ostroff, R. V. Tauxe, K. D. Greene, J. G. Wells, J. H. Lewis, and P. A. Blake. 1988. Illnesses associated with Escherichia coli O157:H7 infections. Ann. Intern. Med. 109:705-712. [DOI] [PubMed] [Google Scholar]
- 11.Hamilton, C. M., M. Aldea, B. K. Washburn, P. Bapitzke, and S. R. Kushner. 1989. New method for generating deletions and gene replacements in Escherichia coli. J. Bacteriol. 171:4617-4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanahan, D. 1983. Studies on transformation of E. coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi, T., K. Makino, K. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C.-G. Han, E. Ohtsubo, T. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, Y. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, A. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 14.Hendrix, R., M. C. M. Smith, R. N. Burns, M. E. Ford, and G. Hatfull. 1999. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96:2192-2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang, A., S. De Grandis, J. Friesen, M. Karmali, M. Petric, R. Congi, and J. Brunton. 1986. Cloning and expression of the genes specifying Shiga-like toxin production in Escherichia coli H19. J. Bacteriol. 166:375-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ito, H., A. Terai, H. Kurazono, Y. Takeda, and M. Nishibuchi. 1990. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb. Pathog. 8:47-60. [DOI] [PubMed] [Google Scholar]
- 17.Johansen, B. K., Y. Wasteson, P. E. Granum, and I. Brzuszczak. 2001. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology 147:1929-1936. [DOI] [PubMed] [Google Scholar]
- 18.Karmali, M. A., M. Petric, C. Lim, P. C. Fleming, G. S. Arbus, and H. Lior. 1985. The association between idiopathic hemolytic uremic syndrome and infection by verotoxin-producing Escherichia coli. J. Infect. Dis. 151:775-782. [DOI] [PubMed] [Google Scholar]
- 19.Kokai-Kun, J. F., A. R. Melton-Celsa, and A. D. O'Brien. 2000. Elastase in intestinal mucus enhances the cytotoxicity of Shiga toxin type 2d. J. Biol. Chem. 275:3713-3721. [DOI] [PubMed] [Google Scholar]
- 20.Lindgren, S. W. 1993. Characterization and virulence assessment of two O91:H21 enterohemorrhagic Escherichia coli isolates. Ph.D. dissertation. Uniformed Services University of the Health Sciences, Bethesda, Md.
- 21.Lindgren, S. W., A. R. Melton, and A. D. O'Brien. 1993. Virulence of enterohemorrhagic Escherichia coli O91:H21 clinical isolates in an orally infected mouse model. Infect. Immun. 61:3832-3842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindgren, S. W., J. E. Samuel, C. K. Schmitt, and A. D. O'Brien. 1994. The specific activities of Shiga-like toxin type II (SLT-II) and SLT-II-related toxins of enterohemorrhagic Escherichia coli differ when measured by Vero cell cytotoxicity but not by mouse lethality. Infect. Immun. 62:623-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandel, M., and A. Higa. 1970. Calcium dependent bacteriophage DNA infection. J. Mol. Biol. 53:154-162. [DOI] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Marques, L. R. M., J. S. M. Peiris, S. J. Cryz, and A. D. O'Brien. 1987. Escherichia coli strains isolated from pigs with edema disease produce a variant of Shiga-like toxin II. FEMS Microbiol. Lett. 44:33-38. [Google Scholar]
- 26.Matsushiro, A., K. Sato, H. Miyamoto, T. Yamamura, and T. Honda. 1999. Induction of prophages of enterohemorrhagic Escherichia coli O157:H7 with norfloxacin. J. Bacteriol. 181:2257-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McClelland, M., K. E. Sanderson, J. Spieth, S. W. Clifton, P. Latreille, L. Courtney, S. Porwollik, J. Ali, M. Dante, F. Du, S. Hou, D. Layman, S. Leonard, C. Nguyen, K. Scott, A. Holmes, N. Grewal, E. Mulvaney, E. Ryan, H. Sun, L. Florea, W. Miller, T. Stoneking, M. Nhan, R. Waterston, and R. K. Wilson. 2001. Complete genome sequence of Salmonella enterica serovar typhimurium LT2. Nature 413:852-856. [DOI] [PubMed] [Google Scholar]
- 28.McKee, M. L., A. R. Melton-Celsa, R. A. Moxley, D. H. Francis, and A. D. O'Brien. 1995. Enterohemorrhagic Escherichia coli O157:H7 requires intimin to colonize the gnotobiotic pig intestine and to adhere to HEp-2 cells. Infect. Immun. 63:3739-3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melton-Celsa, A. R., J. F. Kokai-Kun, and A. D. O'Brien. 2002. Activation of Shiga toxin type 2d (Stx2d) by elastase involves cleavage of the C-terminal two amino acids of the A2 peptide in the context of the appropriate B pentamer. Mol. Microbiol. 43:207-215. [DOI] [PubMed] [Google Scholar]
- 31.Melton-Celsa, A. R., and A. D. O'Brien. 1996. Activation of Shiga-like toxins by mouse and human intestinal mucus correlates with virulence of enterohemorrhagic Escherichia coli O91:H21 isolates in orally infected, streptomycin-treated mice. Infect. Immun. 64:1569-1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muniesa, M., J. Recktenwald, M. Bielaszewska, H. Karch, and H. Schmidt. 2000. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect. Immun. 68:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neely, M. N., and D. I. Friedman. 1998. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol. Microbiol. 28:1255-1267. [DOI] [PubMed] [Google Scholar]
- 34.Norander, J., T. Kempe, and J. Messing. 1983. Construction of improved M13 vectors using oligonucleotide-directed mutagenesis. Gene 26:101-106. [DOI] [PubMed] [Google Scholar]
- 35.O'Brien, A. D., and R. K. Holmes. 1987. Shiga and Shiga-like toxins. Microbiol. Rev. 51:206-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Brien, A. D., J. W. Newland, S. F. Miller, R. K. Holmes, H. W. Smith, and S. B. Formal. 1984. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science 226:694-696. [DOI] [PubMed] [Google Scholar]
- 37.Perna, N. T., G. Plunkett III, V. Burland, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Postfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. Grotbeck, N. W. Davis, A. Lim, E. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. Welch, and F. Blattner. 2001. Genomic sequence of enterohemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 38.Pierard, D., G. Muyldermans, L. Moriau, D. Stevens, and S. Lauwers. 1998. Identification of new verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J. Clin. Microbiol. 36:3317-3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plunkett, G., III, D. J. Rose, T. J. Durfee, and F. R. Blattner. 1999. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7 Shiga toxin as a phage late-gene product. J. Bacteriol. 181:1767-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. 27:493-497. [Google Scholar]
- 41.Sansonetti, P., T. L. Hale, D. J. Dammin, C. Kapfer, H. Collins, Jr., and S. B. Formal. 1983. Alterations in the pathogenicity of E. coli K-12 after transfer of plasmid and chromosomal genes from S. flexneri. Infect. Immun. 39:1392-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmidt, H., J. Scheef, C. Janetzki-Mittmann, M. Datz, and H. Karch. 1997. An ileX tRNA gene is located close to the Shiga toxin II operon in enterohemorrhagic Escherichia coli O157 and non-O157 strains. FEMS Microbiol. Lett. 147:39-44. [DOI] [PubMed] [Google Scholar]
- 43.Schmidt, H., J. Scheef, S. Morabito, A. Caprioli, L. H. Wieler, and H. Karch. 2000. A new Shiga toxin 2 variant (Stx2f) from Escherichia coli isolated from pigeons. Appl. Environ. Microbiol. 66:1205-1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schmitt, C. K., M. L. McKee, and A. D. O'Brien. 1991. Two copies of Shiga-like toxin II-related genes common in enterohemorrhagic Escherichia coli strains are responsible for the antigenic heterogeneity of the O157:H− strain E32511. Infect. Immun. 59:1065-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sizemore, D. R., P. S. Fink, J. T. Ou, L. Baron, D. J. Kopecko, and R. L. Warren. 1991. Tn5 mutagenesis of the Salmonella typhimurium 100 kb plasmid: definition of new virulence regions. Microb. Pathog. 10:493-499. [DOI] [PubMed] [Google Scholar]
- 46.Smith, H. W., P. Green, and Z. Parsell. 1983. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens, and pigs. J. Gen. Microbiol. 129:3121-3137. [DOI] [PubMed] [Google Scholar]
- 47.Tesh, V. L., and A. D. O'Brien. 1991. The pathogenic mechanisms of Shiga toxin and the Shiga-like toxins. Mol. Microbiol. 5:1817-1822. [DOI] [PubMed] [Google Scholar]
- 48.Unkmeir, A., and H. Schmidt. 2000. Structural analysis of phage-borne Stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect. Immun. 68:4856-4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wadolkowski, E. A., J. A. Burris, and A. D. O'Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 58:2438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wagner, P., M. N. Neely, C.-O. Zhang, D. Acheson, M. K. Waldor, and D. I. Friedman. 2001. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J. Bacteriol. 183:2081-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wong, C. S., S. Jelacic, R. L. Habeeb, S. L. Watkins, and P. I. Tarr. 2000. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N. Engl. J. Med. 342:1930-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yarnell, W. S., and J. W. Roberts. 1992. The phage λ gene Q transcription antiterminator binds DNA in the late gene promoter region as it modifies RNA polymerase. Cell 69:1181-1189. [DOI] [PubMed] [Google Scholar]
- 53.Zhang, X., A. D. McDaniel, L. E. Wolf, G. T. Keusch, M. K. Waldor, and D. W. K. Acheson. 2000. Quinolone antibiotics induce Shiga toxin encoding bacteriophages, toxin production, and death in mice. J. Infect. Dis. 181:664-670. [DOI] [PubMed] [Google Scholar]