Abstract
RfaH is a regulatory protein in Escherichia coli and Salmonella enterica serovar Typhimurium. Although it enhances expression of different factors that are proposed to play a role in bacterial virulence, a direct effect of RfaH on virulence has not been investigated so far. We report that inactivation of rfaH dramatically decreases the virulence of uropathogenic E. coli strain 536 in an ascending mouse model of urinary tract infection. The mortality rate caused by the wild-type strain in this assay is 100%, whereas that of its isogenic rfaH mutant does not exceed 18%. In the case of coinfection, the wild-type strain 536 shows higher potential to colonize the urinary tract even when it is outnumbered 100-fold by its rfaH mutant in the inoculum. In contrast to the wild-type strain, serum resistance of strain 536rfaH::cat is fully abolished. Furthermore, we give evidence that, besides a major decrease in the amount of hemin receptor ChuA (G. Nagy, U. Dobrindt, M. Kupfer, L. Emody, H. Karch, and J. Hacker, Infect. Immun. 69:1924-1928, 2001), loss of the RfaH protein results in an altered lipopolysaccharide phenotype as well as decreased expression of K15 capsule and alpha-hemolysin, whereas levels of other pathogenicity factors such as siderophores, flagella, Prf, and S fimbriae appear to be unaltered in strain 536rfaH::cat in comparison to the wild-type strain. trans complementation of the mutant strain with the rfaH gene restores wild-type levels of the affected virulence factors and consequently restitutes virulence in the mouse model of ascending urinary tract infection.
Pathogenic strains of Escherichia coli produce virulence factors that differentiate them from commensal variants of the same species and enable them to cause disease. Constitutive expression of virulence determinants, however, would be needless and energetically exhausting for bacteria. Moreover, the presence of some virulence factors could even be disadvantageous at certain points of the infectious process. For effective pathogenesis, bacteria sense their environment and regulate the expression of genes encoding virulence factors. This response to environmental signals is usually mediated by specific or global regulators, which often form a correlative, complex network (for a review, see reference 15). A regulatory protein has been described in gram-negative bacteria which was first demonstrated to have an influence on lipopolysaccharide (LPS) core synthesis and therefore was named RfaH (23).
The RfaH protein acts as a transcriptional regulator in E. coli, Salmonella enterica serovar Typhimurium, and possibly in other gram-negative bacteria (3). During the last two decades, several operons that are dependent on RfaH for full expression were identified in various strains. These include rfa, rfb, hly, tra, chu, cps, and kps, whose altered expressions in the absence of RfaH result in a decreased amount of LPS core (23), O-antigen (42), alpha-hemolysin (5, 22), F-factor (35), hemin receptor (29), and group I (32), group II (39), and group III (9) capsules, respectively. The exact mechanism by which RfaH enhances expression of these components is not yet fully understood. According to the present view, RfaH regulation takes place at the level of transcriptional antitermination, hence suppressing operon polarity (3). The effect of RfaH is highly dependent on a cis-acting region termed JUMPStart sequence. Recently, the association of RfaH with an 8-bp motif located within the JUMPStart sequence (the ops element) was described (4).
Interestingly, all determinants affected by RfaH encode components that are exported from the bacterial cell or anchored in the outer membrane. Furthermore, they all are somehow related to bacterial virulence: they provide shelter against host defense mechanisms (capsules and intact LPS), supply bacteria with essential substances (hemin receptor), or serve as a cytotoxin (alpha-hemolysin) during infection.
However, observations on RfaH-dependent regulation of individual operons are derived from various strains belonging to different pathogroups of E. coli and S. enterica serovar Typhimurium. Furthermore, there have been no data available on the impact on virulence of the altered expression of these determinants. In this paper we show that disruption of the rfaH gene in uropathogenic E. coli strain 536 results in a significant decrease in virulence. Moreover, evidence is provided that RfaH is a global regulator which modifies expression of several virulence factors within a single strain.
MATERIALS AND METHODS
Animal experiments.
Animal experiments were conducted according to the principles set forth in the Guide for the Care and Use of Laboratory Animals (30) in a laboratory authorized by the Hungarian rule (decree no. XXVII, 1998) and by the subsequent regulation (order no. 243/1998).
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used are described in Table 1. Uropathogenic E. coli strains RZ430, RZ451, and RZ532 were provided by Gabriele Blum-Oehler (Institut für Molekulare Infektionsbiologie, Würzburg, Germany). E. coli strain 536 was isolated from a patient suffering from acute pyelonephritis (7). Construction of its rfaH mutant is described below. Bacteria were grown routinely in Luria-Bertani (LB) broth or LB solidified with 1.5% agar (Biolab, Budapest, Hungary). For hemolytic activity tests, supernatants were obtained from cultures grown in 2xYT medium. For the determination of siderophores, bacteria were grown in M9 medium. When appropriate, media were supplemented with the following concentrations of antibiotics: ampicillin (Ap), 100 μg/ml; chloramphenicol (Cm), 30 μg/ml; and streptomycin (Sm), 50 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| 536 | O6:K15:H31, Smr | 7 |
| 536rfaH::cat | rfaH inactivated by the insertion of a cat cassette, Smr Cmr | This study |
| HB101 | K-12 laboratory strain, F−ara-14 galK2 hsdS20 (hsr hsm mutant) recA13 supE44 lacZ4 leuB6 proA2 thi-1 rspL20 (Smr) xyl-5 mtl-1 λ− | 8 |
| RZ430 | O6:K−:H31 | 44 |
| RZ451 | O6:K18/22:H31 | 44 |
| RZ532 | O6:K+:H31 | 44 |
| Plasmids | ||
| pRF1 | prf determinant of strain 536 in pUC18 | 20 |
| pANN801-13 | sfa1 determinant of strain 536 in pBR322 | 13 |
| pGEM T-Easy | orif1 lacZ Apr | Promega |
| pSMK1 | rfaH gene together with flanking up- and downstream regions of E. coli 536 cloned in pGEM T-Easy, Apr | This study |
| pCVD442 | sacB oriColE1 Apr | 11 |
| pSMK5 | cat-inactivated rfaH gene cloned into pCVD442, Apr Cmr | This study |
| pMSC1 | cat cassette cloned into pUC18, Apr Cmr | 27 |
Inactivation of the rfaH gene in E. coli strain 536.
In order to inactivate the rfaH gene in E. coli strain 536 by gene disruption with a chloramphenicol acetyltransferase (cat) cassette, the entire rfaH gene together with approximately 0.5 kb of each the flanking up- and downstream regions was amplified by PCR. The resulting 1.6-kb E. coli strain 536-specific PCR product obtained with primer pair rfaH1 (5′-GTC GGC ATG TTC AAT ACT TGC-3′) and rfaH2 (5′-TAC ATC CTC ACG ACA GCA GC-3′) was cloned into plasmid pGEM T-Easy (Promega, Heidelberg, Germany), giving rise to pSMK1 (Table 1). The sequence analysis of rfaH536 revealed the presence of a PstI restriction site at position +131 to +136 with respect to the rfaH translational start. The subcloned rfaH gene of E. coli strain 536 was subsequently disrupted by the integration of a cat cassette—obtained by PstI restriction of the plasmid pMSC1 (27)—into the PstI restriction site located within rfaH536. The resulting rfaH536::cat::rfaH536 construct flanked by rfaH536 up- and downstream sequences was cloned into suicide vector pCVD442 for allelic exchange with the chromosomal rfaH up- and downstream regions of E. coli strain 536. The resulting plasmid was designated pSMK5. Allelic exchange was performed as previously outlined (26). Western blot analysis confirmed that in the resulting strain, 536rfaH::cat, no functional RfaH protein was expressed due to the rfaH gene disruption (data not shown). The mutant strain was supplemented either with pGEM T-Easy or the cloned rfaH gene (pSMK1). Introduction of pSMK1 into the mutant restored expression of RfaH, which was verified by Western blotting (data not shown).
Antisera.
Specific antiserum against RfaH was kindly provided by Vassilis Koronakis (Department of Pathology, Cambridge University, United Kingdom). P-related fimbriae (Prf) and SfaI fimbriae were harvested from recombinant strains HB101(pRF1) and HB101(pANN801-13) by using a commercial blender (Omnimixer; Waring) and were purified essentially as described by Khan and Schifferli (18). In order to obtain purified flagella, wild-type E. coli strain 536 was cultured in petri dishes for 36 h. From these bacteria, flagella were harvested and purified by the same procedure described above for fimbriae. Polyclonal antibodies against purified Prf, SfaI fimbriae, and flagella were raised in rabbits (EUROGENTEC, Herstal, Belgium). Antisera were exhaustively absorbed with HB101 nonfimbriated host strain before use.
Capsule-specific serum was produced according to the methods described by Kiesewalter and Seltmann (19). Briefly, bacteria (E. coli strain 536) were incubated in 50% ethanol overnight at 4°C to denature protein antigens. After washing, bacteria were diluted to 2 × 108 CFU/ml in phosphate-buffered saline (PBS). Rabbits were immunized with 0.2, 0.5, 1.0, and twice 2.0 ml of bacterial suspensions at 4-day intervals. Four days following the last inoculation, serum samples were taken and tested by tube agglutination. Afterwards, rabbits were sacrificed and sera were collected and diluted to 1:10 in sterile saline containing 0.3% phenol as preservative. Removal of nonspecific antibodies was achieved by absorption with live and boiled cells of uropathogenic E. coli strain RZ532 (O6:K+:H31) (19). A 0.5-g aliquot of wet bacteria was suspended in 1 ml of the diluted serum and the suspension was incubated for 1 h at 37°C and overnight at 4°C. The next day, bacterial cells were harvested, resuspended in saline, and boiled for 1 h. Following centrifugation, the pellet was resuspended in the same serum and suspensions were incubated again for 1 h at 37°C and subsequently overnight at 4°C. After centrifugation, aliquots of sera were stored at −20°C. Through this procedure sera became free of any immunoreactive antibodies except those against K15 antigen, as proven by enzyme-linked immunosorbent assay (ELISA).
Ascending urinary tract infection model.
Intravesical infection of 3-to-4-day-old CFLP mice (Gödöllõ, Hungary) was performed as previously described (1). Bacteria were grown overnight at 37°C in LB, harvested by centrifugation, washed once, and normalized to the required inoculum density (107 CFU/ml) in PBS by adjusting the suspension to the appropriate optical density at 600 nm (OD600) value justified by viable counts. A 25-μl aliquot of this bacterial suspension containing 0.05% Pontamin Sky Blue dye (Searle Pharmaceuticals, High Wycombe, United Kingdom) was introduced into the bladder directly through the abdominal wall. The stain, which had no toxic or antibacterial effects, served as an indicator for successful inoculation (i.e., the stain became localized only to the bladder, which was visible through the hairless skin). In order to exclude the possibility of vesicoureteral reflux caused by the inoculum, in an additional experiment mice were sacrificed immediately after inoculation. Neither dye nor bacteria were detectable in the kidneys, verifying this experiment to be a suitable model of ascending urinary tract infection. Six to 14 infant mice were injected simultaneously with each strain, and assays were repeated four times. Mice that survived infection were sacrificed 21 days postinfection. The bladder and both kidneys were removed under sterile conditions and homogenized in PBS, and aliquots were plated onto agar plates containing a selective antibiotic. Additionally, bacterial counts were determined from the blood obtained by puncturing the heart.
Coinfection experiments.
Mice were infected with a bacterial suspension containing 2 × 104 CFU/ml (150% lethal dose [LD50]) of the wild-type strain 536 mixed with either 2 × 105 or 2 × 106 CFU of its isogenic rfaH mutant/ml (providing 1:10 or 1:100 concentration ratios, respectively). The procedure of injecting the mixture into the bladder was as described above. Urine samples were taken daily for 20 days. Urine was diluted in saline and plated onto LB agar plates containing either streptomycin alone or in combination with chloramphenicol. Since both wild-type strain 536 and its rfaH mutant were resistant to streptomycin but only the mutant possessed chloramphenicol resistance, CFU counts for both strains could be established this way. Feces of dams were checked and found to be negative for streptomycin-resistant strains, indicating that no fecal contamination of the samples could occur. Furthermore, randomly taken colonies were identified by slide agglutination using specific antisera. On the 21st day postinfection, mice were sacrificed and colony counts were determined from the blood, the bladder, and the kidneys as described above.
Serum bactericidal test.
Bacteria grown in LB medium were washed in saline and diluted to 106 CFU/ml. One hundred-microliter aliquots of bacterial suspensions were mixed with an equal volume of human serum and incubated at 37°C for 4 h in microtiter plates. Samples were taken at 0, 0.5, 1, 2, 3, and 4 h. Viable cell counts were determined by plating aliquots onto LB plates and incubating overnight at 37°C. The assays were performed both with normal and heat-inactivated (56°C for 30 min) serum. Triplicates were used for each strain, and assays were repeated three times.
Assay of hemolytic activity.
Hemolytic activities of culture supernatants were determined using a formerly described method (14) with minor modifications. Briefly, erythrocytes were obtained from a healthy individual and washed three times with 150 mM NaCl, 20 mM CaCl2. Two-percent suspensions of erythrocytes were incubated for 40 min at 42°C with 1:10 diluted cell-free supernatants of bacterial cultures grown at 37°C in 2xYT medium. After a short centrifugation, the amount of released hemoglobin was measured photometrically (A543). The degree of hemolysis was quantified as the percentage of total hemolysis induced by Triton X-100 (Sigma).
Lung toxicity assay.
The assay was performed as previously described (12). Briefly, 3-week-old CFLP mice (Gödöllõ) weighing 10 to 12 g were infected intranasally under superficial ether anesthesia with 50 μl of bacterial suspension (3 × 109 CFU/ml) grown overnight in LB medium. Twenty mice were treated for each strain. The animals were observed for 24 h and death rates were recorded.
LPS analysis.
Bacteria grown on solid LB were suspended in distilled water and boiled for 30 min. Suspensions were sonicated in a Realsonic Cleaner apparatus for 10 min. LPS was purified by the procedure of Hitchcock and Brown (16). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed on a 12.5% gel according to the method of Laemmli (21). Gels were fixed overnight in a solution of 7% acetic acid and 25% 2-propanol and were silver stained as described by Nelson et al. (31).
ELISA.
The 96-well plates (CML-CEB, Nemours, France) were coated overnight with 0.2-ml aliquots of the bacterial suspensions (109 CFU/ml) in carbonate buffer (pH 9.5) at 4°C. The following day, plates were washed with PBS containing 0.05% Tween 20 and then blocked with PBS containing 2% bovine serum albumin (BSA; Sigma) for 1 h at 37°C. K15-specific serum was diluted in PBS containing 0.5% BSA and incubated with the antigen-coated plates for 90 min. Serial dilutions were conducted across the plates. After three washes, plates were probed with anti-rabbit immunoglobulin conjugated with horseradish peroxidase (DAKO A/S, Copenhagen, Denmark). The ELISA substrate was o-phenylenediamine (Sigma) dissolved in citric acid buffer containing H2O2. The OD was measured at 490 nm on a conventional ELISA plate reader. Duplicates were used for each strain and dilution, and the assays were repeated twice.
Western blotting.
Samples of LB cultures grown at 37°C were taken at different time points. Cells were pelleted, washed, and resuspended to the same OD (0.5 at 600 nm). Equal quantities of bacterial suspensions were centrifuged, resuspended in lysing buffer, and separated by SDS-PAGE as described by Laemmli (21). Samples were blotted onto a nitrocellulose membrane using a Mini Trans-Blot cell (Bio-Rad). Membranes were blocked for 90 min with 10% skimmed milk in Tris-buffered saline (pH 7.5) containing 0.05% Tween 20 (TBST). Separate membranes were incubated with Prf-, SfaI-, or H31-specific antisera diluted 1,000-, 1,500-, and 2,500-fold, respectively, in TBST containing 2% skimmed milk. Membranes were incubated for 1 h at room temperature. After washing, membranes were treated for 1 h at room temperature with anti-rabbit immunoglobulin-horseradish peroxidase conjugate (DAKO A/S) diluted 2,500-fold in TBST with 2% skimmed milk. Membranes were washed thoroughly and then developed by luminography using a Western blotting chemiluminescence kit (NEN Life Science, Boston, Mass.).
Detection of siderophores.
The amount of secreted siderophores was determined using a CAS (chrome azurol S) assay (37) with minor modifications. Bacteria were grown overnight in M9 medium. To induce siderophore production, the iron chelator 2,2′-dipyridyl was added at a final concentration of 0.1 to 0.4 mM. The cultures were incubated for an additional 3 h at 37°C. After centrifugation, bacteria-free supernatants were mixed with an equal volume of CAS assay solution (37). Samples were incubated for 2 h to reach equilibrium, and the absorbance was measured at 640 nm.
RESULTS
The rfaH mutant of E. coli 536 is reduced in urovirulence.
E. coli 536 is a virulent uropathogenic strain that was isolated from a patient suffering from acute pyelonephritis. The strain's urovirulence has already been proven in mouse uropathogenicity models (12). To assess whether RfaH plays any role in the uropathogenicity of the strain, we infected infant mice intravesically with the wild-type strain and its isogenic rfaH mutant, as well as with the mutant carrying either pGEM T-Easy or pSMK1 (rfaH gene cloned into pGEM T-Easy). The results are summarized in Table 2. While all mice injected with strain 536 died within 3 to 4 days, 82% of the animals receiving 536rfaH::cat survived for as long as 3 weeks without any sign of severe infection. Introduction of pGEM T-Easy (vector control) into the rfaH mutant did not influence the mortality rate. On the contrary, supplementation of the mutant with pSMK1 partially restored virulence. All mice surviving for 3 weeks were sacrificed and the bacterial counts were determined from the blood, the bladder, and the kidneys. The number of bacteria—if any—was found to be very low (data not shown), indicating no or moderate bacterial colonization in these mice.
TABLE 2.
Mortality rate of infant mice infected intravesically
| Bacterial strain injecteda | No. of mice infectedb | Mortality rate (%) |
|---|---|---|
| 536 | 38 | 100 |
| 536rfaH::cat | 39 | 18c |
| 536rfaH::cat (pGEM T-Easy) | 41 | 15c |
| 536rfaH::cat (pSMK1) | 35 | 63d |
Mice were infected intravesically with 25 μl of bacterial suspension (107 CFU/ml) directly through the abdominal wall.
Six to 14 mice were infected simultaneously, and four independent experiments were performed.
Significantly different in comparison to mortality rate elicited by the wild-type strain (χ2 probe, P < 0.001).
Significantly different in comparison to mortality rate elicited by strain 536rfaH::cat (χ2 probe, P < 0.001).
Coinfection experiments.
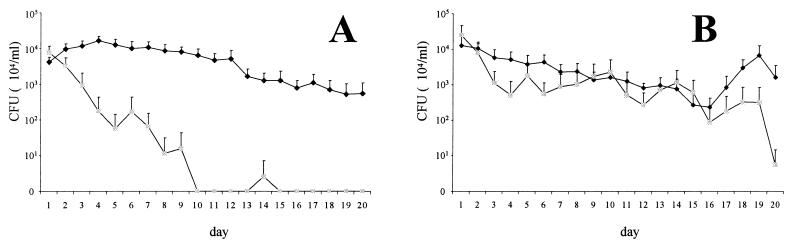
Wild-type strain 536 and its isogenic rfaH mutant were used to coinfect infant mice intravesically. The inoculum contained a standard concentration of the wild-type strain (2 × 104 CFU/ml), which was complemented with either 2 × 105 or 2 × 106 CFU of its rfaH mutant/ml, leading to bacterial ratios of 1:10 (experiment 1) or 1:100 (experiment 2), respectively. Several of the infected mice died within a few days. Mortality rates were 25% (3 of 12) and 77% (17 of 22), in experiments 1 and 2, respectively. From the surviving animals, urine samples were taken daily for 20 days and viable counts of strains 536 and 536rfaH::cat were determined. The obtained results are shown in Fig. 1. Wild-type strain 536 was permanently present in the urine of infected mice throughout the study period. Although its concentration was constantly decreasing, it still exceeded 107 CFU/ml on day 20 postinoculation in both experiments. On the contrary, mutant strain 536rfaH::cat disappeared from the urinary tract within 10 days, even when inoculated at a 10-times-higher concentration (experiment 1) (Fig. 1A). Higher doses of the mutant strain in the inoculum (experiment 2) resulted in tedious elimination; however, after 3 weeks the urine was virtually free of the mutant strain (Fig. 1B).
FIG. 1.
Number of E. coli 536 (♦) and 536rfaH::cat (░⃞) cells eliminated with the urine following intravesical coinjection of bacteria into mice. The inoculum contained 5 × 102 CFU of wild-type strain 536 complemented with either 5 × 103 CFU (A) or 5 × 104 CFU (B) of its isogenic rfaH mutant. Graphs represent means ± standard errors of the means of values originated from nine (A) and five (B) animals.
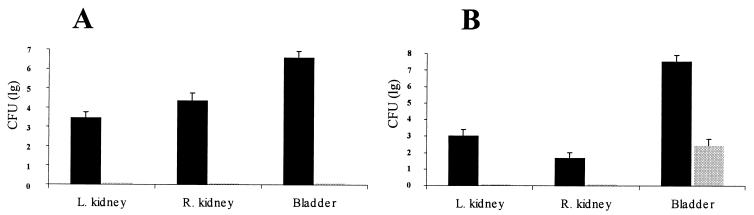
On the 21st day postinfection, mice were sacrificed and colony counts were determined from the blood, the bladder, and the kidneys. The results are shown in Fig. 2. The blood never contained any bacteria (data not shown). However, the bladder and the kidneys in all mice contained high numbers of the wild-type strain. In contrast, strain 536rfaH::cat was never present in the kidneys and was detectable in the bladder only in experiment 2. Even in this case, the bacterial counts for strain 536 exceeded that of its rfaH mutant by a factor of more than 105.
FIG. 2.
Viable counts in different organs on 21st day postinfection. Mice were inoculated intravesically with a mixture of 5 × 102 CFU of wild-type strain 536 complemented with either 5 × 103 CFU (A) or 5 × 104 CFU (B) of its isogenic rfaH mutant. Means ± standard errors of the means of values originated from nine (A) and five (B) mice are shown. Dark bars represent CFU of wild-type strain 536, and the light bar represents that of mutant strain 536rfaH::cat. L., left; R., right.
An intact RfaH protein is required for serum resistance.
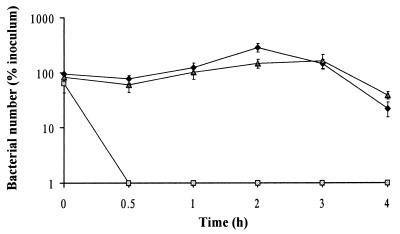
Serum resistance of uropathogenic E. coli strain 536 and its derivatives is shown in Fig. 3. The wild-type strain was able to survive in 50% normal human serum for at least 4 h, whereas its isogenic rfaH mutant survived no longer than 30 min. trans complementation of the mutant strain with the rfaH gene restored resistance to the bactericidal effect of human serum. All three strains could, however, survive for 4 h in a similar experiment when heat-inactivated serum was used (data not shown).
FIG. 3.
Serum resistance of E. coli strain 536 (♦), its isogenic mutant 536rfaH::cat (░⃞), and trans-complemented strain 536rfaH::cat (pSMK1) (▴). Bacteria were incubated in 50% serum obtained from a healthy individual. The graph represents means ± standard errors of the means of values originated from three similar assays.
Expression of several virulence factors is coregulated by RfaH.
RfaH has been reported to influence the expression of different operons in various E. coli strains. In order to investigate whether several virulence factors are coregulated by RfaH, we determined hemolytic activity, LPS structure, the amount of specific K-antigen, expression of flagella and various fimbriae, and siderophore-mediated iron binding in strain 536 and its derivatives. We have previously reported the regulatory influence of RfaH on the expression of hemin receptor ChuA in this strain (29).
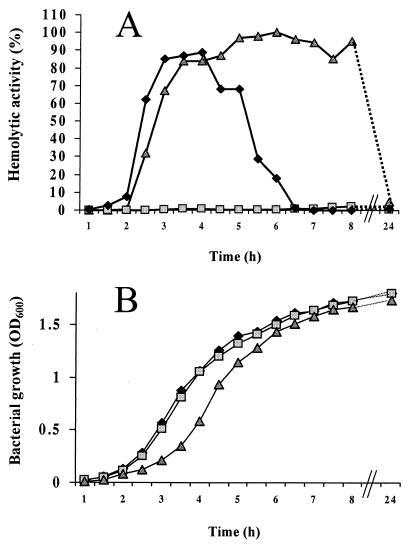
Figure 4A shows hemolytic activity derived from culture supernatants of wild-type strain 536, mutant strain 536rfaH::cat, and its trans-complemented variant. The wild-type strain exhibited strong hemolytic activity during the exponential phase of growth, reaching a plateau at mid-exponential phase (Fig. 4B). On the other hand, hemolysis elicited by the rfaH mutant was hardly detectable. Introduction of the cloned rfaH gene (pSMK1) into the mutant restored high hemolytic activity. In vivo hemolytic activity was tested in the lung toxicity assay, in which the acute toxicity of bacteria can be assessed (12). In this experimental model, mice die within a few hours from hemorrhagic lung edema elicited by alpha-hemolysin. The 100% mortality rate caused by wild-type strain 536 was reduced to 10% in the case of its isogenic rfaH mutant. Supplementation of the mutant with an intact rfaH gene on pSMK1 increased the death rate to 65% (data not shown).
FIG. 4.
(A) Hemolytic activities derived from E. coli strain 536 (♦), its isogenic mutant 536rfaH::cat (░⃞), and trans-complemented strain 536rfaH::cat (pSMK1) (▴). Values represent the percentage of total hemolysis relative to that induced by Triton X-100. (B) Growth curves of strain 536 (♦), 536rfaH::cat (░⃞), and 536rfaH::cat (pSMK1) (▴).
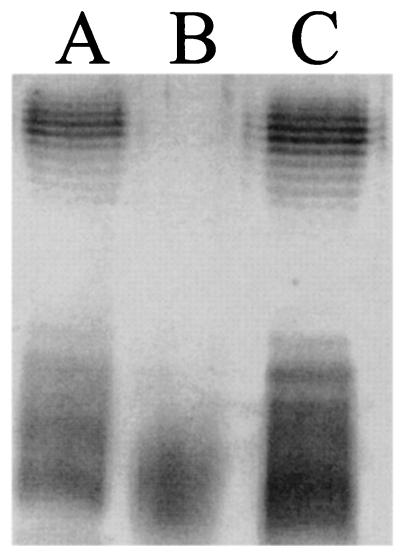
The LPS structure of strains 536 (Fig. 5, lane A), 536rfaH::cat (Fig. 5, lane B), and 536rfaH::cat (pSMK1) (Fig. 5, lane C) were compared with SDS-PAGE. The presence of a functionally active rfaH gene (lanes A and C) resulted in intact LPS structures, whereas the mutant strain (lane B) showed a rough phenotype.
FIG. 5.
LPS structures obtained from wild-type strain 536 (A), mutant 536rfaH::cat (B), and its trans-complemented variant 536rfaH::cat (pSMK1) (C). Equal quantities of purified LPS were silver stained following separation by SDS-PAGE.
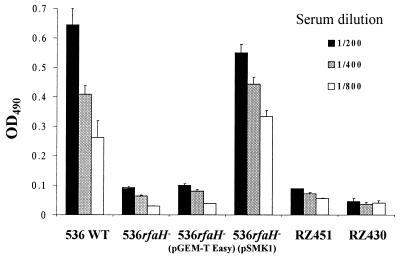
To evaluate the amount of K15 capsule expressed by strain 536 and its derivatives, we performed an ELISA using a K15-specific serum. The results are shown in Fig. 6. In comparison to the wild-type strain, the rfaH mutant showed decreased immunoreactivity to this serum. Introduction of pSMK1 into the mutant strain, but not that of the control vector pGEM T-Easy, restituted higher immunoreactivity to the K15 antiserum. In order to verify that these variations were due to differential expression of the capsule (i.e., the serum was specific for K15 antigen), we used additional O6:H31 uropathogenic E. coli strains possessing either K18/22 or no capsular antigens as negative controls. Indeed, these strains showed low immunoreactivity to the serum used.
FIG. 6.
Reactivity of E. coli strain 536 (O6:K15:H31) and its variants to different concentrations of a K15-specific serum. E. coli strains RZ430 (O6:K−:H31) and RZ451 (O6:K18/22:H31) were used as negative controls. The graph represents means ± standard errors of the means of values originated from two similar assays.
Expression of SfaI- and P-related (Prf) fimbriae as well as H31 flagella was determined by Western blotting at various phases of growth in each case. Expression of SfaI and Prf does not seem to be influenced by RfaH in strain 536. Similarly, no major differences could be shown in H31 flagellar expression between strain 536 and its rfaH mutant (data not shown).
Siderophore-mediated iron utilization was determined by a highly sensitive chemical assay. In this test, we assessed the total siderophore-mediated iron binding, which was activated by 2,2′-dipyridyl in a dose-dependent manner. However, we could show no significant difference between the iron binding capacities of strain 536 and its rfaH mutant (data not shown).
DISCUSSION
Since the first report describing RfaH as a regulator of the rfa locus (23), several other operons have been identified which depend for full expression on the presence of this regulatory protein (for a review, see reference 3). Nevertheless, a global regulatory function has not been attributed to RfaH, as distinct observations have been reported from different pathogenic isolates of S. enterica serovar Typhimurium and E. coli. In the present study, together with formerly described results (29), it is shown for the first time that expression of several components (LPS, K15 capsule, alpha-hemolysin, and hemin receptor ChuA) are coregulated by RfaH in a single E. coli strain. What could be the basis of common regulation of these structures? Interestingly, although these factors are different in their composition and function, they all are transported through both membranes of a bacterium; they are anchored in the outer membrane or transported out of the cell. Common regulatory systems for LPS and K15 (group II) capsule synthesis could be explained with similarities in synthesis, assembly, and transport mechanisms, in spite of the distinct genes involved. Furthermore, activity of secreted alpha-hemolysin was reported to be dependent on intact LPS (6, 38, 41). Similarly, the function of the hemin receptor ChuA is speculated to depend on the activity of alpha-hemolysin, i.e., the toxin that liberates heme by destroying erythrocytes (29). All these observations suggest that these components have evolved to coutilize the regulatory protein RfaH, as proposed by Bailey et al. (3).
Since all structures whose expression has been shown to be influenced by RfaH are potential virulence factors, we proposed that mutation of the rfaH gene results in a decrease in virulence. Indeed, in an ascending mouse model of urinary tract infection, the virulence of wild-type strain 536 was almost completely abolished through the loss of the regulatory protein RfaH. Decreased virulence of the rfaH mutant, however, could not be explained by inappropriate growth potential, since the in vitro growth curve of 536rfaH::cat did not differ from that of the wild-type strain 536. Moreover, both strains were shown to be able to grow equally ex vivo in the urine of healthy individuals (data not shown), indicating that there are no differences in growth abilities between the two strains at the beginning of the infectious process.
Colonization, however, is a more complex process requiring effective adhesion mechanisms and rapid multiplication through utilization of limited nutrients. By destroying epithelial cells, the possibility of tissue invasion and dissemination arises. On the other hand, bacteria have to face different host defense mechanisms. The inability of bacteria to overcome different attacks by the immune system results in localized infections only or, in most cases, total elimination of the infectious agent. The first line of defense against dissemination is the action of the complement system. In contrast to commensal gram-negative bacteria, extraintestinal clinical isolates of enterobacteria often show resistance to the killing effect of serum, suggesting that the abilities of bacteria to cause disease correlate with their resistance to the bactericidal effect of serum. Thus, serum resistance has been a virulence parameter for many pathogens, including extraintestinal isolates of E. coli. Resistance to serum killing is multifactorial and has been associated with several surface components of E. coli, among which capsule and LPS are thought to be the most important ones (17). We have shown that loss of the regulatory protein RfaH results in high susceptibility to human serum in strain 536rfaH::cat, which might be explained by altered expression of both LPS and K15 capsule due to inactivation of the rfaH gene.
The coinfection assay provides an objective model to compare the colonization capacity of the rfaH mutant with that of the wild-type strain without involving individual differences of the host. The usefulness of similar models has recently been shown in investigations aimed to clarify the pathogenic role of cytotoxic necrotizing factor type 1 (33) and different iron transport systems (40) in the virulence of uropathogenic E. coli strains. Even though we had to face a negative selection of our experimental animals (i.e., those mice having the most fulminant infections died within a few days), the difference in colonization potentials between the two strains has become clear. Our results provide evidence that, in contrast to the wild-type strain, the rfaH mutant is not able to cause ascending uroinfection, since it was never detectable in the upper urinary tract.
An important factor in effective pathogenesis within the urinary tract is the production of cytotoxins. Alpha-hemolysin has been proven to contribute to the virulence of E. coli strains causing extraintestinal infections (17, 24, 43). By destroying eukaryotic cells, the toxin not only provides an opportunity for deeper invasion, but in addition supplies bacteria with nutrients liberated from host cells (e.g., iron-containing substances, such as heme). Expression of alpha-hemolysin has been reported to be under the control of RfaH in E. coli strains (2, 5, 22). We have reported here that no considerable hemolysis derives from strain 536rfaH::cat at any phase of growth in vitro.
The lung toxicity assay serves as an ideal model for the assessment of in vivo hemolysin production (12). In this assay, mice die from the immediate toxic effect of bacteria before extensive bacterial multiplication. Since loss of the pathogenicity islands carrying the hly determinants of E. coli 536 results in an avirulent phenotype in this model (28), we consider hemolysin as the major factor to determine the outcome of intranasal instillation of bacteria. Mutation within rfaH significantly decreases death rates in this assay, suggesting decreased in vivo hemolytic activity elicited by the mutant strain.
In addition to a major loss in the level of effective HlyA toxin produced in the absence of RfaH, in vivo iron acquisition of bacteria could be further hindered by decreased expression of hemin receptor ChuA, as has been reported elsewhere by Nagy and colleagues (29). Although an essential role for this receptor in iron supply is questionable, its contribution to bacterial virulence was recently proven in uropathogenic E. coli (40). Utilization of hemin, however, is not an exclusive means of iron acquisition. E. coli strain 536 produces at least two different siderophore systems, namely, enterobactin (34) and yersiniabactin (36). The presence of an additional siderophore receptor (IroN), whose ligand is not yet identified, was recently described in strain 536 (10). The CAP assay used in the present study is based on the affinity of siderophores for iron and is therefore independent of the structure. Total siderophore-mediated iron acquisition does not seem to be under the regulatory control of RfaH; therefore, it may compensate the effect of decreased hemin receptor ChuA quantities on the in vivo iron supply of bacteria.
Bacterial pathogenesis is a complex phenomenon which is attained through a concerted action of virulence factors. An exclusive role of a single virulence factor is rare; they usually function synergistically, amplifying or complementing the effect of each other. Therefore, total abolishment of virulence is rarely caused by the loss of individual virulence factors (25). Nevertheless, mutation in one single gene can result in a significant loss of virulence if the related gene product has an impact on several virulence determinants simultaneously. We have presented evidence that the absence of functional RfaH protein results in parallel underexpression of several virulence factors, which probably all contribute to the complete virulence of E. coli strain 536. Further experiments are needed, however, to clarify the potential global regulatory role of RfaH in other pathogroups of E. coli and additional gram-negative bacteria.
Acknowledgments
We are grateful to Vassilis Koronakis (Department of Pathology, Cambridge University, Cambridge, United Kingdom) and Gabriele Blum-Oehler (Institut für Molekulare Infektionsbiologie, Würzburg, Germany) for providing the RfaH antiserum and the uropathogenic E. coli strains RZ430, RZ451, and RZ532, respectively. We thank Rózsa Lajkó for excellent technical assistance and Tibor Pál for critical reading of the manuscript.
This study was supported by grants OTKA T026019, T037833, and ETT 086/2001. The work of the Würzburg group was supported by the Deutsche Forschungsgemeinschaft (Ha1434/8-2 and Sonderforschungsbereich 479) and by the “Fonds der Chemischen Industrie.” G.N. was supported by a grant from the Bayerische Forschungsstiftung.
Editor: A. D. O'Brien
REFERENCES
- 1.Allison, C., L. Emody, N. Coleman, and C. Hughes. 1994. The role of swarm cell differentiation and multicellular migration in the uropathogenicity of Proteus mirabilis. J. Infect. Dis. 169:1155-1158. [DOI] [PubMed] [Google Scholar]
- 2.Bailey, M. J., C. Hughes, and V. Koronakis. 1996. Increased distal gene transcription by the elongation factor RfaH, a specialized homologue of NusG. Mol. Microbiol. 22:729-737. [DOI] [PubMed] [Google Scholar]
- 3.Bailey, M. J., C. Hughes, and V. Koronakis. 1997. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 26:845-851. [DOI] [PubMed] [Google Scholar]
- 4.Bailey, M. J., C. Hughes, and V. Koronakis. 2000. In vitro recruitment of the RfaH regulatory protein into a specialised transcription complex, directed by the nucleic acid ops element. Mol. Gen. Genet. 262:1052-1059. [DOI] [PubMed] [Google Scholar]
- 5.Bailey, M. J., V. Koronakis, T. Schmoll, and C. Hughes. 1992. Escherichia coli HlyT protein, a transcriptional activator of haemolysin synthesis and secretion, is encoded by the rfaH (sfrB) locus required for expression of sex factor and lipopolysaccharide genes. Mol. Microbiol. 6:1003-1012. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, M. E., and R. A. Welch. 1997. Pleiotropic effects of a mutation in rfaC on Escherichia coli hemolysin. Infect. Immun. 65:2218-2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, H., J. Hacker, A. Juarez, C. Hughes, and W. Goebel. 1982. Cloning of the chromosomal determinants encoding hemolysin production and mannose-resistant hemagglutination in Escherichia coli. J. Bacteriol. 152:1241-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 9.Clarke, B. R., R. Pearce, and I. S. Roberts. 1999. Genetic organization of the Escherichia coli K10 capsule gene cluster: identification and characterization of two conserved regions in group III capsule gene clusters encoding polysaccharide transport functions. J. Bacteriol. 181:2279-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dobrindt, U., G. Blum-Oehler, T. Hartsch, G. Gottschalk, E. Z. Ron, R. Funfstuck, and J. Hacker. 2001. S-fimbria-encoding determinant sfaI is located on pathogenicity island III536 of uropathogenic Escherichia coli strain 536. Infect. Immun. 69:4248-4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacker, J., H. Hof, L. Emody, and W. Goebel. 1986. Influence of cloned Escherichia coli hemolysin genes, S-fimbriae and serum resistance on pathogenicity in different animal models. Microb. Pathog. 1:533-547. [DOI] [PubMed] [Google Scholar]
- 13.Hacker, J., G. Schmidt, C. Hughes, S. Knapp, M. Marget, and W. Goebel. 1985. Cloning and characterization of genes involved in production of mannose-resistant, neuraminidase-susceptible (X) fimbriae from a uropathogenic O6:K15:H31 Escherichia coli strain. Infect. Immun. 47:434-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hardie, K. R., J. P. Issartel, E. Koronakis, C. Hughes, and V. Koronakis. 1991. In vitro activation of Escherichia coli prohaemolysin to the mature membrane-targeted toxin requires HlyC and a low molecular-weight cytosolic polypeptide. Mol. Microbiol. 5:1669-1679. [DOI] [PubMed] [Google Scholar]
- 15.Harel, J., and C. Martin. 1999. Virulence gene regulation in pathogenic Escherichia coli. Vet. Res. 30:131-155. [PubMed] [Google Scholar]
- 16.Hitchcock, P. J., and T. M. Brown. 1983. Morphological heterogeneity among salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154:269-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson, J. R. 1991. Virulence factors in Escherichia coli urinary tract infection. Clin. Microbiol. Rev. 4:80-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khan, A. S., and D. M. Schifferli. 1994. A minor 987P protein different from the structural fimbrial subunit is the adhesin. Infect. Immun. 62:4233-4243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiesewalter, J., and G. Seltmann. 1968. Herstellung der Diagnostischen Hyperimmun-Sera., p. 158-160. In J. Sedlak and H. Rische (ed.), Enterobacteriaceae-Infektionen. VEB Georg Thieme, Leipzig, Germany.
- 20.Knepper, B. 1990. Regulation of fimbrial adhesin determinants in the uropathogenic Escherichia coli strains 536 and J96. Ph.D. thesis. University of Würzburg, Würzburg, Germany.
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Leeds, J. A., and R. A. Welch. 1996. RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J. Bacteriol. 178:1850-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg, A. A., and C. G. Hellerqvist. 1980. Rough mutants of Salmonella typhimurium: immunochemical and structural analysis of lipopolysaccharides from rfaH mutants. J. Gen. Microbiol. 116:25-32. [DOI] [PubMed] [Google Scholar]
- 24.May, A. K., T. G. Gleason, R. G. Sawyer, and T. L. Pruett. 2000. Contribution of Escherichia coli alpha-hemolysin to bacterial virulence and to intraperitoneal alterations in peritonitis. Infect. Immun. 68:176-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mobley, H. L., M. D. Island, and G. Massad. 1994. Virulence determinants of uropathogenic Escherichia coli and Proteus mirabilis. Kidney Int. Suppl. 47:S129-S136. [PubMed] [Google Scholar]
- 26.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 27.Morschhauser, J., V. Vetter, L. Emody, and J. Hacker. 1994. Adhesin regulatory genes within large, unstable DNA regions of pathogenic Escherichia coli: cross-talk between different adhesin gene clusters. Mol. Microbiol. 11:555-566. [DOI] [PubMed] [Google Scholar]
- 28.Nagy, G., U. Dobrindt, G. Blum-Oehler, L. Emody, W. Goebel, and J. Hacker. 2000. Analysis of the hemolysin determinants of the uropathogenic Escherichia coli strain 536. Adv. Exp. Med. Biol. 485:57-61. [DOI] [PubMed] [Google Scholar]
- 29.Nagy, G., U. Dobrindt, M. Kupfer, L. Emody, H. Karch, and J. Hacker. 2001. Expression of hemin receptor molecule ChuA is influenced by RfaH in uropathogenic Escherichia coli strain 536. Infect. Immun. 69:1924-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.National Institutes of Health. 1985. Guide for the care and use of laboratory animals. Publication no. 85-23. National Institutes of Health, Bethesda, Md.
- 31.Nelson, D., W. Neill, and I. R. Poxton. 1990. A comparison of immunoblotting, flow cytometry and ELISA to monitor the binding of anti-lipopolysaccharide monoclonal antibodies. J. Immunol. Methods 133:227-233. [DOI] [PubMed] [Google Scholar]
- 32.Rahn, A., J. Drummelsmith, and C. Whitfield. 1999. Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J. Bacteriol. 181:2307-2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rippere-Lampe, K. E., A. D. O'Brien, R. Conran, and H. A. Lockman. 2001. Mutation of the gene encoding cytotoxic necrotizing factor type 1 (cnf1) attenuates the virulence of uropathogenic Escherichia coli. Infect. Immun. 69:3954-3964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritter, A., G. Blum, L. Emody, M. Kerenyi, A. Bock, B. Neuhierl, W. Rabsch, F. Scheutz, and J. Hacker. 1995. tRNA genes and pathogenicity islands: influence on virulence and metabolic properties of uropathogenic Escherichia coli. Mol. Microbiol. 17:109-121. [DOI] [PubMed] [Google Scholar]
- 35.Sanderson, K. E., and B. A. Stocker. 1981. Gene rfaH, which affects lipopolysaccharide core structure in Salmonella typhimurium, is required also for expression of F-factor functions. J. Bacteriol. 146:535-541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert, S., A. Rakin, D. Fischer, J. Sorsa, and J. Heesemann. 1999. Characterization of the integration site of Yersinia high-pathogenicity island in Escherichia coli. FEMS Microbiol. Lett. 179:409-414. [DOI] [PubMed] [Google Scholar]
- 37.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 38.Stanley, P. L., P. Diaz, M. J. Bailey, D. Gygi, A. Juarez, and C. Hughes. 1993. Loss of activity in the secreted form of Escherichia coli haemolysin caused by an rfaP lesion in core lipopolysaccharide assembly. Mol. Microbiol. 10:781-787. [DOI] [PubMed] [Google Scholar]
- 39.Stevens, M. P., P. Hanfling, B. Jann, K. Jann, and I. S. Roberts. 1994. Regulation of Escherichia coli K5 capsular polysaccharide expression: evidence for involvement of RfaH in the expression of group II capsules. FEMS Microbiol. Lett. 124:93-98. [DOI] [PubMed] [Google Scholar]
- 40.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wandersman, C., and S. Letoffe. 1993. Involvement of lipopolysaccharide in the secretion of Escherichia coli alpha-haemolysin and Erwinia chrysanthemi proteases. Mol. Microbiol. 7:141-150. [DOI] [PubMed] [Google Scholar]
- 42.Wang, L., S. Jensen, R. Hallman, and P. R. Reeves. 1998. Expression of the O antigen gene cluster is regulated by RfaH through the JUMPstart sequence. FEMS Microbiol. Lett. 165:201-206. [DOI] [PubMed] [Google Scholar]
- 43.Welch, R. A., E. P. Dellinger, B. Minshew, and S. Falkow. 1981. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature 294:665-667. [DOI] [PubMed] [Google Scholar]
- 44.Zingler, G., M. Ott, G. Blum, U. Falkenhagen, G. Naumann, W. Sokolowska-Kohler, and J. Hacker. 1992. Clonal analysis of Escherichia coli serotype O6 strains from urinary tract infections. Microb. Pathog. 12:299-310. [DOI] [PubMed] [Google Scholar]