Abstract
Two common pathogens known to cause bone infection, Salmonella and Staphylococcus aureus, were investigated to determine their abilities to induce chemokine expression in cultured mouse and human osteoblasts. While these cells are responsible for bone formation, we were surprised to find that they could respond to bacterial infection by upregulating expression of the chemokine CXCL10 (IP-10). However, there were significant differences in the abilities of the gram-negative bacterium Salmonella and the gram-positive bacterium S. aureus to induce expression of CXCL10. Reverse transcription-PCR and enzyme-linked immunosorbent assay analyses showed high levels of Salmonella-induced CXCL10 mRNA and protein expression, respectively, whereas the osteoblast response to S. aureus was significantly less. Consistent with these findings, Salmonella-derived lipopolysaccharide (LPS), but not S. aureus-derived peptidoglycan, could induce expression of CXCL10. An antibody against toll-like receptor 4 (TLR4) could block the LPS-induced CXCL10 production, demonstrating the functional expression of TLR4 by osteoblasts. Despite the inducible nature of TLR2 mRNA expression by bacterium-infected osteoblasts, peptidoglycan failed to stimulate CXCL10 secretion. Immunofluorescent staining of bacterium-infected calvaria (i.e., skull bone) demonstrated the presence of CXCL10 in osteoblasts. The fact that osteoblasts did not express CXCR3 mRNA, whereas T lymphocytes can express high levels of this receptor, suggests that osteoblast-derived CXCL10 may recruit T lymphocytes to the sites of bone infections.
T lymphocytes readily migrate to the sites of bone and joint diseases (12, 13, 54, 55, 58, 61). While these cells can contribute to the protective host response, they may also contribute to the destructive inflammatory response in these tissues (5, 7, 28, 32, 35, 56, 60, 63). The chemokines elaborated during bone and joint disease are not well defined, nor is it clear which cell populations contribute to the secretion of particular chemokines. This statement is especially true for bone and joint diseases caused by bacterial pathogens.
Surprisingly, bone-forming osteoblasts (6, 15) have the potential to secrete certain chemokines following their activation (10, 20, 25, 43). Recent studies suggest that stimulated osteoblasts may be responsible for recruitment of mononuclear leukocytes into damaged or diseased bone. Unfortunately, a comprehensive evaluation of osteoblast-derived chemokine production following interaction with bacterial pathogens has not been performed. In particular, it is not clear if infected osteoblasts can secrete select chemokines that preferentially recruit and activate T lymphocytes.
In the present study, we focused on the ability of osteoblasts to secrete CXCL10 (IP-10) following exposure of these cells to bacteria, bacterial products, or T-lymphocyte-derived cytokines. Traditionally, this chemokine can be induced in multiple cell types (36), especially macrophages (40), following exposure to gamma interferon (IFN-γ). However, bacterial products, such as lipopolysaccharide (LPS), can also induce macrophage-derived CXCL10 (40). The receptor for CXCL10, CXCR3, is expressed at high levels on T lymphocytes and has been reported to preferentially support TH1-type responses (8, 19, 44, 51). If osteoblasts elaborate CXCL10 following interaction with bacterial pathogens, this may help explain the presence of inflammatory T lymphocytes at the sites of bone infections.
Staphylococcus aureus and Salmonella are two common causative agents of bone and joint diseases (14, 34, 39, 42, 65) and were chosen for use in this investigation based on this fact. While these pathogens are both causes of bone diseases, they have very dissimilar characteristics and modes of invasion. S. aureus is an extracellular, gram-positive pathogen, and adherence to bone is an important component of the infectivity of this bacterium (18). Peptidoglycan, a component of the cell wall of S. aureus, can interact with toll-like receptor 2 (TLR2) expressed on cells such as macrophages to initiate a host response (1, 2). Conversely, Salmonella is an intracellular, gram-negative bacterium (3, 17), and Salmonella-derived LPS-CD14 complexes can bind to TLR4 to initiate host responses (1, 2). One recent report suggests that osteoblasts can express the mRNA encoding TLR2 and TLR4 (29).
Here we demonstrate for the first time the potential for differential regulation of CXCL10 secretion in response to S. aureus, Salmonella, bacterium-derived products, or certain cytokines. Surprisingly, Salmonella and LPS were potent inducers of CXCL10 in mouse and human osteoblast cultures, whereas S. aureus and peptidoglycan induced much less of this chemokine. Salmonella- or LPS-induced CXCL10 production could be reduced in the presence of an antibody against TLR4. Taken together, these results suggest that osteoblast-mediated chemokine production may augment TH1 lymphocyte recruitment and activation, especially during infection by intracellular bacterial pathogens.
MATERIALS AND METHODS
Isolation and culture of mouse calvaria and osteoblasts.
Two-day-old BALB/c or C57BL/6 mouse neonates (Charles River Laboratories, Wilmington, Mass.) were sacrificed, and calvaria (i.e., skull bones) were removed. Whole calvaria were used to isolate primary osteoblasts by sequential collagenase-protease digestion as previously described (9-11). Briefly, cell isolates were pooled in osteoblast growth medium consisting of Dulbecco modified Eagle medium containing 10% fetal bovine serum (Atlanta Biologicals, Atlanta, Ga.), 25 mM HEPES, 2 g of sodium bicarbonate per liter, 75 μg of glycine per ml, 100 μg of ascorbic acid per ml, 40 ng of vitamin B12 per ml, and 2 μg of p-aminobenzoic acid per ml (pH 7.4). Fourth-passage cells were seeded at a density of 106 cells per well in six-well plates or at a density of 2.5 × 105 cells per well in 24-well plates and incubated at 37°C in a 6% CO2 atmosphere until the cells reached confluence. More than 99% of these cells expressed markers of osteoblasts, including type I collagen, osteocalcin, and alkaline phosphatase, as determined by immunofluorimetric analyses performed as previously described (9-11).
Normal human osteoblast cultures.
Normal human osteoblasts (Clonetics, San Diego, Calif.) were purchased and propagated according to the guidelines provided by the vendor and as previously described by workers in our laboratory (9-11). Osteoblast growth medium (Clonetics) containing 10% fetal bovine serum, ascorbic acid, gentamicin, and amphotericin B was used to seed cells in 25-cm2 flasks, and this was followed by incubation at 37°C in the presence of 6% CO2. When the cultures reached confluence, cells were removed and seeded into the appropriate culture plates. These commercially available cells have been extensively characterized as authentic osteoblasts (21).
Exposure of cultured mouse and human osteoblasts to bacteria, LPS, or peptidoglycan.
S. aureus strain UAMS-1 (ATCC 49230) is a clinical osteomyelitis isolate and was grown overnight in 5 ml of Luria broth in a shaking water bath at 37°C. Salmonella enterica serovar Dublin strain SL 1363 is a wild-type, pathogenic strain and was also grown overnight in 5 ml of Luria broth at 37°C. Bacteria were then harvested by centrifugation for 10 min at 4,300 × g and washed with 5 ml of Hanks balanced salt solution. The pellet was resuspended in medium without antibiotics. Confluent layers of osteoblasts were infected with S. aureus or Salmonella suspensions at different ratios of bacteria to osteoblasts by incubation in medium without antibiotics for 45 min at 37°C. The ratios of Staphylococcus cells to osteoblasts used for exposure in these studies were 250:1 and 75:1, and the ratios of Salmonella cells to osteoblasts used were 30:1 and 10:1. These ratios were empirically determined to result in limited, but similar, intracellular infections (less than 2%) for each pathogen. Furthermore, these levels of exposure resulted in limited osteoblast cell death (less than 5%) during the times in culture used.
In some experiments, S. aureus and Salmonella cells were exposed to short-wavelength (250-nm) UV light for 5 and 3 min, respectively, prior to addition to cultured osteoblasts. The times used for UV inactivation were empirically determined to reduce the percentage of viable bacteria to less than 0.005%, as determined by colony counting (16). It should be noted that this low level of viable S. aureus or Salmonella cells in UV-inactivated preparations was not sufficient to induce osteoblasts to secrete any detectable CXCL10 (data not shown).
Following the brief exposure to bacteria, osteoblasts were washed three times with Hanks balanced salt solution to remove any extracellular bacteria, and cultures were incubated with medium supplemented with gentamicin (25 μg/ml) to kill any remaining extracellular bacteria. Supernatant collection, RNA isolation, and enumeration of intracellular bacteria occurred at the times indicated below as previously described (9-11).
In some experiments, osteoblasts were exposed to purified peptidoglycan or LPS (Sigma, St. Louis, Mo.) and incubated for different times before supernatant collection or RNA isolation.
Salmonella- and S. aureus-induced chemokine mRNA expression analyses performed by using macroarrays and reverse transcription (RT)-PCR.
To define chemokine expression following exposure of mouse osteoblasts to S. aureus or Salmonella, we began by using gene macroarrays. Poly(A)+ mRNA was isolated by using magnetic oligo(dT) beads (DYNAL, Olso, Norway) from uninfected, S. aureus-infected, or Salmonella-infected mouse osteoblast cultures 6 h postinfection. cDNA labeled with 32P (New England Nuclear, Boston, Mass.) was made from each mRNA preparation by using 200 U of RNase H−, Moloney leukemia virus reverse transcriptase (Superscript II; Gibco-BRL, Gaithersburg, Md.), and the primer mixture supplied by the manufacturer (CLONTECH, Palo Alto, Calif.). The labeled cDNAs were then hybridized to identical gene macroarrays (Atlas 1.2 mouse cancer array; CLONTECH) overnight at 65°C by using the hybridization buffer supplied by the manufacturer. The macroarrays each contained 1,176 mouse genes, including the CXCL10 gene and many of the known chemokine genes (CLONTECH). After unbound radiolabel was washed off, the blots were exposed to phosphorimaging screens, and hybridization was detected by using a TYPHOON 8600 image analyzer (Molecular Dynamics, Sunnyvale, Calif.). Differences in hybridization between blots were quantified by using ImageQuant software (Molecular Dynamics). These experiments were performed twice to demonstrate reproducibility and to allow comparison of results.
Results from the Atlas 1.2 mouse cancer arrays were used to direct more focused gene expression analyses performed with mouse chemokine GEArrays (SuperArray, Bethesda, Md.). Poly(A)+ RNA was isolated from uninfected, S. aureus-infected, or Salmonella-infected mouse osteoblast cultures, and 32P-labeled cDNA was made as described above by using the primer mixture supplied by the manufacturer (SuperArray). The labeled cDNAs were then hybridized to identical chemokine GEArrays (SuperArray) overnight at 65°C by using the hybridization buffer supplied by the manufacturer. After unbound radiolabel was washed off, the blots were exposed to phosphorimaging screens, and hybridization was detected and quantified as described above.
To confirm differences in CXCL10 mRNA expression that were detected in the macroarray analyses, semiquantitative RT-PCR was performed as previously described (9-11). Briefly, total RNA was extracted from uninfected, infected, or treated mouse or human osteoblasts (TRIZOL reagent; Gibco-BRL) and reverse transcribed. The PCR was performed with cDNA by using a denaturation temperature of 95°C, an annealing temperature of 59°C, and an extension temperature of 72°C (Robocycler 40; Stratagene, La Jolla, Calif.) with the first 3 of 27 cycles having extended denaturation and annealing times. The positive- and negative-strand PCR primers were derived from the previously published sequences of mouse (64) and human (37) CXCL10 and included GAAATCATCCCTGCGAGCCTATCC and GCAATTAGGACTAGCCATCCACTGGG (derived from the mouse CXCL10 sequence) and GCGATTCTGATTTGCTGCCTTATC and AGACATTTCCTTGCTAACTGCTTTCAG (derived from the human CXCL10 sequence).
RT-PCR were also performed to detect the mRNAs encoding mouse CD14 (38), CXCR3 (57), TLR2 (22), and TLR4 (45) by using the following positive- and negative-strand PCR primers pairs: CCTAGTCGGATTCTATTCGGAGCC and AACTTGGAGGGTCGGGAACTTG to amplify CD14; AACCACAAGCACCAAAGCAG and TGATGTTGAAGAGGGCACCT to amplify CXCR3; GCAGAATCAATACAATAGAGGGAGACGC and AAGTGAAGAGTCAGGTGATGGATGTCG to amplify TLR2; and TTATCCAGGTGTGAAATCCAGACAATT and TGAAAGGCTTGGTCTTGAATGAAGTCA to amplify TLR4. By using a portion of cDNA from each sample, a housekeeping gene, the glyceraldehyde-3-phosphate dehydrogenase (G3PDH) gene, (62), was also amplified to ensure equal RNA inputs and similar efficiencies of RT. The PCR primers used to amplify the G3PDH gene were CCATCACCATCTTCCAGGAGCGAG and CACAGTCTTCTGGGTGGCAGTGAT. Following PCR, 15% of each amplified sample was electrophoresed on ethidium bromide-stained agarose gels and visualized with UV illumination. In addition, DNA standards (PGC Scientific, Fredrick, Mass.) electrophoresed on each gel were used to verify the sizes of the amplified PCR fragments. Densitometric analysis of the amplified fragments was performed by using NIH Image (obtained from the National Institutes of Health website [http://rsb.info.nih.gov/nih-image]). A gel-plotting macro was used to outline the bands, and the intensity was calculated by using the Uncalibrated OD setting.
Quantification of CXCL10 secretion in culture supernatants by ELISA.
Capture enzyme-linked immunosorbent assays (ELISAs) were performed to quantify mouse and human CXCL10 secretion by using methods described previously (9-11). The capture and detection antibodies used to quantify mouse CXCL10 were monoclonal rat anti-mouse CXCL10 (clone A102-6) and biotinylated polyclonal rabbit anti-mouse CXCL10 (PharMingen, San Diego, Calif.), respectively. The capture and detection antibodies used to quantify human CXCL10 were monoclonal mouse anti-human CXCL10 (clone 4D5) and biotinylated monoclonal mouse anti-human CXCL10 (clone 6D4/D6/G2; PharMingen), respectively. Purified rat immunoglobulin G (10 μg/ml) was included in wash and incubation buffers to limit nonspecific binding. To detect the presence of bound antibody, strepavidin-horseradish peroxidase (PharMingen) was added. After a 45-min incubation at room temperature, wells were washed, and tetramethylbenzidine (BioFx Laboratories, Owings Mills, Md.) was added. Colorimetric reactions were subsequently stopped by addition of 0.5 M H2SO4, and A450 was measured (model 550 microplate reader; Bio-Rad, Hercules, Calif.). Standard curves were generated by determining absorbance values by using limiting dilutions of recombinant murine CXCL10 (PharMingen). Results are presented as means ± standard deviations based on triplicate determinations. ELISA sensitivity was 0.05 ng/ml as indicated in the respective figures (<0.05). Statistically significant differences in CXCL10 secretion were determined by using the Student t test.
Blocking of LPS-mediated or UV-killed Salmonella-mediated CXCL10 secretion by using an antibody against TLR4.
The ability of osteoblasts to respond to LPS through a TLR4-mediated mechanism was determined by using mouse osteoblasts cultured in the presence of an anti-mouse TLR4 antibody (clone MTS510; eBioscience, San Diego, Calif.), an isotype-matched control rat immunoglobulin G2a antibody (PharMingen). Briefly, mouse osteoblasts were cultured as described above, and antibodies (30 μg/ml) were added 30 min prior to the addition of LPS or UV-killed Salmonella. Supernatants were used 24 h later for quantification of CXCL10 secretion by ELISA as described above.
RESULTS
Salmonella and S. aureus induce CXCL10 mRNA expression by mouse osteoblasts.
Despite a limited number of studies which have shown that stimulated osteoblasts can secrete selected chemokines (10, 20, 25, 43), no comprehensive analysis of chemokine expression by osteoblasts following bacterial infection has been performed. Initially, we used macroarray analyses to define chemokine mRNAs which were differentially modulated following in vitro exposure of osteoblasts to S. aureus and following in vitro exposure of osteoblasts to Salmonella. Since these two bacteria can cause bone and joint diseases (14, 34, 39, 42, 65) but have very different properties and modes of invasion, we anticipated that the osteoblast responses to these bacteria might be different. As determined with two different macroarrays (see Materials and Methods), it was clear that Salmonella could induce rapid and substantial CXCL10 mRNA expression in cultured mouse osteoblasts, whereas the osteoblast response to S. aureus was modest (data not shown).
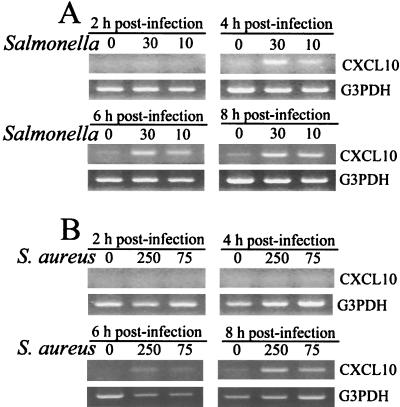
To confirm that differential mRNA induction occurred, semiquantitative RT-PCR was performed with RNAs isolated from S. aureus- and Salmonella-infected osteoblasts to detect CXCL10 mRNA expression. As shown in Fig. 1, constitutive expression of CXCL10 mRNA was absent in uninfected mouse osteoblasts. However, within 4 h after infection with Salmonella, CXCL10 mRNA expression was upregulated, and CXCL10 mRNA continued to be expressed at 6 and 8 h postinfection (Fig. 1A). In four separate RT-PCR analyses, the increases in CXCL10 mRNA expression were 11.3 (±2.2)-fold and 4.9 (±1.1)-fold (mean ± standard deviation) for Salmonella-to-osteoblast ratios of 30:1 and 10:1, respectively, at 6 h postinfection compared to the CXCL10 mRNA expression in uninfected cultures. A dose response was also observed; the higher ratio of Salmonella to osteoblasts (30:1) induced more CXCL10 mRNA expression than the lower ratio (10:1). This result was not observed when mouse osteoblasts were exposed to S. aureus (Fig. 1B). CXCL10 mRNA could be induced by exposure to S. aureus, but clear increases could not be detected until 6 to 8 h postinfection. These results confirmed what was observed when macroarray analyses were used and indicated that the CXCL10 response by mouse osteoblasts differed depending on the bacterial pathogen.
FIG. 1.
CXCL10 mRNA expression following infection of mouse osteoblasts with Salmonella or S. aureus. Cultured murine osteoblasts were not infected (lanes 0) or were exposed to viable Salmonella (A) or S. aureus (B) at bacterium-to-osteoblast ratios of 30:1 (lanes 30), 10:1 (lanes 10), 250:1 (lanes 250), or 75:1 (lanes 75) for 45 min; this was followed by removal of extracellular bacteria. RNA was isolated at different times following exposure to bacteria, and semiquantitative RT-PCR was performed to detect CXCL10 mRNA. The results are presented as amplified products electrophoresed on ethidium bromide-stained agarose gels. RT-PCR amplification of the G3PDH housekeeping gene was performed to ensure that similar amounts of input RNA and similar efficiencies of RT were being compared. The experiments were performed four times with similar results.
CXCL10 secretion following infection of mouse osteoblasts with S. aureus or Salmonella.
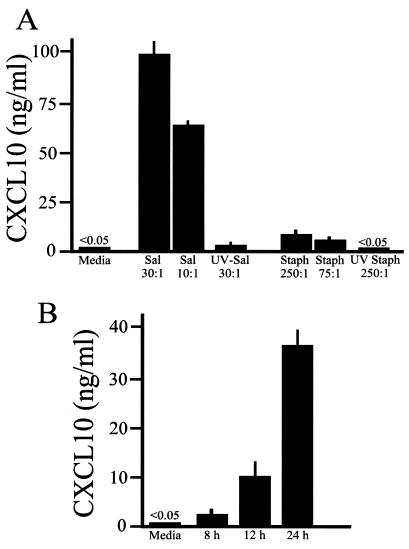
To quantify CXCL10 secretion following infection of mouse and human osteoblasts with Salmonella and S. aureus, capture ELISAs were performed. Figure 2A shows one representative dose response of mouse osteoblasts exposed to these two bacterial pathogens. Consistent with the different abilities of the organisms to induce mRNA expression, Salmonella was a potent stimulator of CXCL10 secretion when the stimulation was compared to that induced by S. aureus (Fig. 2A). Kinetic analysis showed that Salmonella could induce CXCL10 secretion as early as 8 h postinfection (Fig. 2B), indicating rapid induction of this chemokine by this pathogen.
FIG. 2.
CXCL10 secretion following infection of cultured mouse osteoblasts with Salmonella or S. aureus. (A) Cultured mouse osteoblasts were not infected (Media) or were exposed to viable Salmonella (Sal) or UV-killed Salmonella (UV-Sal) at a bacterium-to-osteoblast ratio of 30:1 or 10:1 or to viable S. aureus (Staph) or UV-killed S. aureus (UV Staph) at a bacterium-to-osteoblast ratio of 250:1 or 75:1 for 45 min; this was followed by removal of extracellular bacteria. Culture supernatants were taken 24 h postinfection, and specific capture ELISAs were performed to quantify CXCL10 secretion. (B) To investigate the kinetics of the response, mouse osteoblasts were infected with Salmonella (ratio of bacteria to osteoblasts, 10:1), and culture supernatants were removed 8, 12, and 24 h postinfection. ELISAs were performed to quantify CXCL10 secretion. The results are presented as means ± standard deviations based on triplicate determinations. The experiments were performed four times with similar results.
Of particular interest was the ability of UV-killed Salmonella to induce CXCL10 secretion (Fig. 2A). Nonviable Salmonella cannot invade osteoblasts, and these bone-forming cells have a limited ability to take up bacteria (16, 24, 47). Therefore, the finding that killed Salmonella could stimulate CXCL10 secretion, albeit at a lower level than the level stimulated by viable bacteria, suggested that osteoblasts might express cell surface molecules which have the ability to sense the presence of gram-negative bacteria or their products.
Salmonella and S. aureus induce CXCL10 mRNA expression and secretion by human osteoblasts.
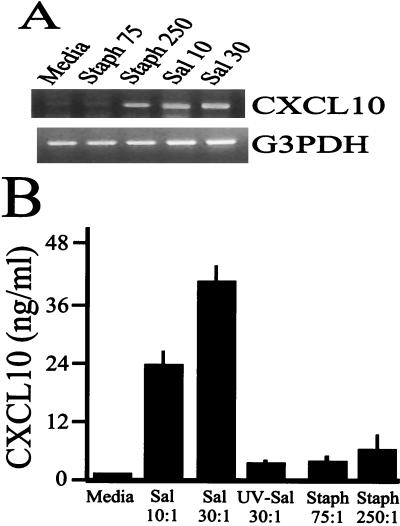
Similar results were observed when human osteoblasts were infected with Salmonella or S. aureus. At 6 h postinfection, Salmonella was a potent inducer of CXCL10 mRNA expression compared to the message expression in S. aureus-infected human osteoblasts (Fig. 3A). Consistent with the mRNA results, Salmonella induced high levels of CXCL10 secretion following infection of human osteoblasts compared with the levels of S. aureus-induced chemokine secretion (Fig. 3B). Together, these results demonstrate that the human and mouse osteoblast responses to a gram-negative intracellular pathogen and a gram-positive extracellular pathogen are quantitatively different. Furthermore, these results support the conserved nature of the response in human and mouse osteoblasts.
FIG. 3.
CXCL10 mRNA expression and secretion following infection of human osteoblasts with Salmonella or S. aureus. (A) Cultured human osteoblasts were not infected (Media) or were exposed to viable S. aureus at a bacterium-to-osteoblast ratio of 75:1 (Staph 75) or 250:1 (Staph 250) or to Salmonella at a bacterium-to-osteoblast ratio of 10:1 (Sal 10) or 30:1 (Sal 30) for 45 min; this was followed by removal of extracellular bacteria. RNA was isolated at 8 h after exposure to bacteria, and semiquantitative RT-PCR was performed to detect human CXCL10 mRNA expression. The results are presented as amplified products electrophoresed on ethidium bromide-stained agarose gels. RT-PCR amplification of the G3PDH housekeeping gene was performed to ensure that similar amounts of input RNA and similar efficiencies of RT were being compared. (B) To quantify human CXCL10 secretion, osteoblasts were not infected (Media) or were exposed to viable Salmonella (Sal) or UV-killed Salmonella (UV-Sal) at a bacterium-to-osteoblast ratio of 30:1 or 10:1 or to viable S. aureus (Staph) at a bacterium-to-osteoblast ratio of 250:1 or 75:1. Supernatants were taken 24 h postinfection, and specific ELISAs were performed. The experiments were performed twice with similar results.
Blocking of LPS-mediated CXCL10 secretion with an antibody against TLR4.
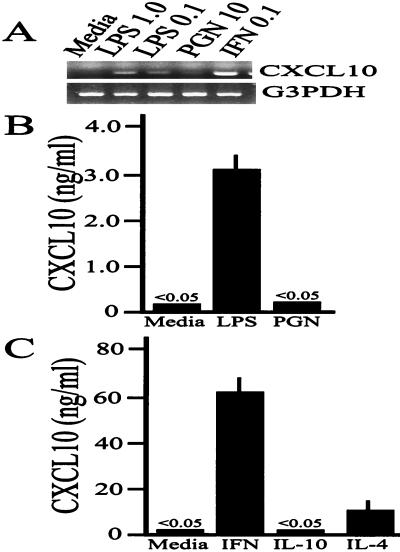
The finding that killed, noninvasive Salmonella could stimulate some CXCL10 secretion suggested that osteoblasts might express cell surface molecules which have the ability to sense the presence of gram-negative bacteria or their products. As shown in Fig. 4A, Salmonella-derived LPS, but not peptidoglycan, could stimulate cultured mouse osteoblasts to express the mRNA encoding CXCL10 and could induce secretion of this chemokine in a dose-dependent manner (Fig. 4B). Thus, it was not necessary to have intact, viable Salmonella cells to stimulate osteoblasts to express CXCL10. Furthermore, this result was consistent with the results of a limited number of previous publications (23, 25, 29, 31, 48, 49, 53) demonstrating that bacterial LPS can stimulate osteoblast secretion of select chemokines, cytokines, or nitric oxide. Interestingly, the TH1- and TH2-derived cytokines IFN-γ and IL-4 could also induce CXCL10 secretion (Fig. 4C).
FIG. 4.
Salmonella-derived LPS, IFN-γ, and IL-4, but not S. aureus-derived peptidoglycan or IL-10, can induce expression of CXCL10 by cultured mouse osteoblasts. (A) Mouse osteoblasts were not infected (Media) or were exposed to 1.0 μg of LPS per ml (LPS 1.0), 0.1 μg of LPS per ml (LPS 0.1), 10 μg of peptidoglycan per ml (PGN 10), or 0.1 μg of IFN-γ per ml (IFN 0.1) for 6 h, and RNA was isolated and used for RT-PCR to detect CXCL10 mRNA expression. The results are presented as amplified products electrophoresed on ethidium bromide-stained agarose gels. RT-PCR amplification of the G3PDH housekeeping gene was performed to ensure that similar amounts of input RNA and similar efficiencies of RT were being compared. The experiments were performed twice with similar results. (B) Similar studies were performed to quantify LPS and peptidoglycan (PGN)-induced CXCL10 secretion with an ELISA. The results are presented as the means ± standard deviations based on triplicate determinations. The experiments were performed twice with similar results. (C) Mouse osteoblasts were exposed to IFN-γ (IFN), IL-4, or IL-10 for 24 h, and culture supernatants were used for quantification of CXCL10 secretion by an ELISA. The results are presented as the means ± standard deviations based on triplicate determinations. The experiments were performed twice with similar results.
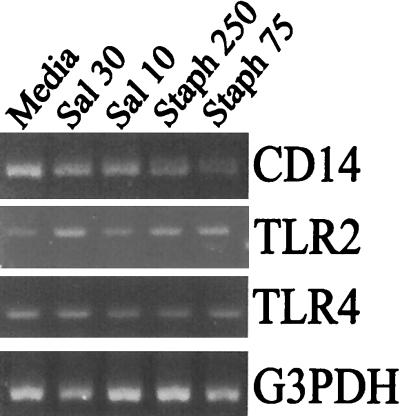
Bacterially derived LPS can complex with CD14 and bind to TLR4 to initiate a cellular response (1, 2). However, it is not altogether clear at present what the signaling mechanism for LPS-induced chemokine responses in osteoblasts is. There are conflicting reports concerning the ability of osteoblasts to express CD14 (31, 48), and there is only one recent publication which suggests that these cells express functional TLR4 (29). Therefore, we addressed the potential for CD14 and toll-like receptor mRNA expression during Salmonella infection of osteoblasts using RT-PCR. Mouse osteoblasts were not infected or were exposed to Salmonella or S. aureus, and then RNA was extracted and used for RT-PCR to detect expression of the mRNAs encoding TLR2, TLR4, CD14, and G3PDH. In mouse osteoblasts (Fig. 5), there was some constitutive mRNA for each of these mRNA species. The levels of mRNA encoding CD14 and TLR4 did not vary significantly following infection; however, the mRNA encoding TLR2 was induced by interaction with either bacterium (Fig. 5).
FIG. 5.
Infected osteoblasts express the mRNAs encoding CD14, TLR2, and TLR4. Mouse osteoblasts were not infected (Media) or were exposed to viable Salmonella at a bacterium-to-osteoblast ratio of 30:1 (Sal 30) or 10:1 (Sal 10) or to S. aureus at a bacterium-to-osteoblast ratio of 250:1 (Staph 250) or 75:1 (Staph 75) for 45 min; this was followed by removal of extracellular bacteria. RNA was isolated following exposure to the bacteria, and a semiquantitative RT-PCR was performed to detect CD14, TLR2, and TLR4 mRNAs. The results are presented as amplified products electrophoresed on ethidium bromide-stained agarose gels. RT-PCR amplification of the G3PDH housekeeping gene was performed to ensure that similar amounts of input RNA and similar efficiencies of RT were being compared. The experiments were performed twice with similar results.
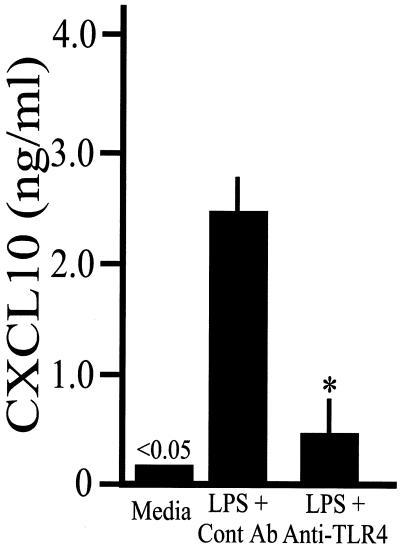
To investigate the functional expression of TLR4, mouse osteoblasts were treated with LPS in the presence of an antibody against TLR4 or an isotype-matched control antibody. Figure 6 demonstrates the ability of the TLR4 antibody to effectively block LPS-induced CXCL10 secretion. Taken together, the results suggest that gram-negative pathogens, like Salmonella, can stimulate CXCL10 production during infection by interacting with toll-like receptors on osteoblasts.
FIG. 6.
Blocking of LPS-mediated CXCL10 secretion with an antibody against TLR4. Mouse osteoblasts were not treated (Media) or were treated with an isotype-matched control antibody (Cont Ab) or an antibody against TLR4 (Anti-TLR4) for 30 min prior to addition of LPS. Twenty-four hours later, culture supernatants were taken, and CXCL10 secretion was quantified by an ELISA. The results are presented as means ± standard deviations based on triplicate determinations. The asterisk indicates that the value is statistically significantly different (P < 0.05) from the control value. The experiments were performed twice with similar results.
Lack of CXCR3 mRNA expression by infected osteoblasts.
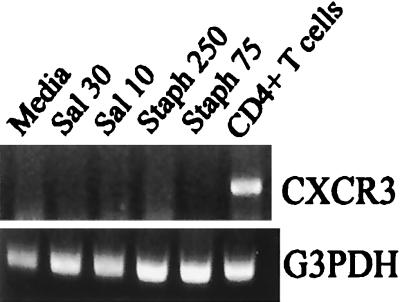
The receptor for CXCL10 is CXCR3; therefore, we questioned whether infected osteoblasts have the potential to express the mRNA encoding this protein. Mouse osteoblasts were infected, and RT-PCR was performed to detect CXCR3 mRNA. As shown in Fig. 7, no CXCR3 mRNA was observed in infected osteoblasts, despite the presence of CXCR3 mRNA in activated CD4+ T lymphocytes and G3PDH mRNA expression by all cells. Therefore, it is unlikely that CXCL10 produced by osteoblasts interacted with these bone cells in an autocrine or paracrine manner.
FIG. 7.
Mouse osteoblasts do not express CXCR3 mRNA. Cultured murine osteoblasts were not infected (Media) or were exposed to viable Salmonella at a bacterium-to-osteoblast ratio of 30:1 (Sal 30) or 10:1 (Sal 10) or to S. aureus at a bacterium-to-osteoblast ratio of 250:1 (Staph 250) or 75:1 (Staph 75) for 45 min; this was followed by removal of extracellular bacteria. RNA was isolated following exposure to the bacteria, and a semiquantitative RT-PCR was performed to detect CXCR3 mRNA. The results are presented as amplified products electrophoresed on ethidium bromide-stained agarose gels. RT-PCR amplification of the G3PDH housekeeping gene was performed to ensure that similar amounts of input RNA and similar efficiencies of RT were being compared. The experiments were performed twice with similar results.
DISCUSSION
Bone and joint diseases often involve excessive inflammatory responses with T-lymphocytic infiltrates (12, 13, 54, 55, 58, 61); however, it is not altogether clear which chemokines call these cells to the site of inflammation. The receptor for CXCL10 is CXCR3, which is found predominately on TH1 lymphocytes (8, 19, 44, 51). Surprisingly, we found that osteoblasts have the potential to be a significant source of CXCL10 following interaction with bacteria. Therefore, this result may help explain the recruitment of CXCR3-positive TH1 lymphocytes to the sites of bone infections via osteoblast-derived CXCL10.
Quantitative differences in the abilities of Salmonella and S. aureus to induce CXCL10 production may indicate fundamental differences in the host responses to these two very different bacterial pathogens. While S. aureus is the most frequent causative agent of bone infections (14, 34, 39), Salmonella spp. are also significant pathogens (4, 30, 34, 42, 65). In particular, Salmonella infection is one of the most frequent serious infections of the bone in sickle cell patients and can also be a significant cause of inflammatory joint diseases. Immunosuppressed patients are especially susceptible to Salmonella-induced bone diseases, and it has been known for some time that antibiotic therapy is becoming less effective in eliminating this bacterial pathogen (65). Salmonella spp. are intracellular pathogens (17, 26, 41), and TH1-driven cell-mediated immune responses are required to clear the pathogen (26, 27, 33). Therefore, successful resolution of Salmonella-mediated bone or joint infections would require recruitment of T lymphocytes capable of secreting IFN-γ. Salmonella-induced CXCL10 secretion by osteoblasts would be consistent with recruitment of TH1 lymphocytes to the site of bone infection, presumably to initiate cell-mediated immune responses to clear this intracellular pathogen. The fact that infected osteoblasts do not express CXCR3 mRNA (Fig. 7) further suggests that cells other than osteoblasts are the intended targets for this secreted chemokine. Therefore, the results presented here suggest that osteoblasts are capable of contributing to the initiation of cell-mediated immune responses that, if successful, would result in removal of the intracellular pathogen.
While S. aureus could induce some CXCL10 secretion (Fig. 2 and 3), an optimal host response may not require the same level of recruitment of TH1 lymphocytes for this extracellular pathogen. Interestingly, bacterial peptidoglycan from S. aureus was unable to induce detectable CXCL10 secretion (Fig. 4), despite expression of the mRNA encoding a potential receptor for this bacterial product. Peptidoglycan is a ligand for TLR2 (2, 52, 66), and we detected inducible TLR2 mRNA expression in infected osteoblasts (Fig. 5). However, if functional TLR2 is expressed by these cells, CXCL10 secretion is not one of the chemokines which are induced via this toll-like receptor. This result is consistent with the results of a recent report in which CXCL10 production could not be detected following stimulation through TLR2 in dendritic cells (46).
The fact that viable (i.e., invasive) Salmonella was a more potent stimulator of CXCL10 secretion than UV-killed Salmonella (Fig. 2A) or LPS (Fig. 4B) suggests that signaling through TLR4 is not the only mechanism responsible for chemokine induction during infection by this pathogen. This result was similar to the results observed for Salmonella-induced IL-6 (11), IL-12 (11), colony-stimulating factor (9), and CCL2 (10) secretion. In each case, UV-killed bacteria were not as potent an inducer of cytokine secretion as viable Salmonella, which is capable of invading and surviving intracellularly. The ability of Salmonella to invade osteoblasts is not surprising, since osteoblasts have been described as sophisticated fibroblasts (15) and since invasion of epithelial cells by Salmonella is well documented (26, 41, 50). Thus, the ability of Salmonella to interact with osteoblasts at the cell surface and intracellular interactions following invasion make investigating the signals mediating the osteoblast response somewhat complex. Future studies with Salmonella mutants that lack key genes involved in invasion and intracellular survival should provide insight into the mechanisms underlying the signals induced upon interaction of this pathogen with these sophisticated fibroblasts.
Perhaps the most important implication of the present study is the possibility that osteoblasts may represent an important contributor to T-lymphocyte recruitment and activation during bone infections. Formation and remodeling of bone are driven by bone-forming osteoblasts (15) and bone-resorbing osteoclasts (59). Osteoclasts are derived from a myeloid precursor and release various lysosomal enzymes to break down the organic matrix of bone. Conversely, osteoblasts are responsible for laying down new bone matrices of type 1 collagen and other proteins and also for directing the activity of osteoclasts. However, it is becoming apparent that leukocytes, especially T lymphocytes, can significantly modulate the activity of both osteoblasts and osteoclasts and their ability to remodel bone (5, 32, 35, 56, 58, 60). If osteoblasts can recruit T cells to the site of bone infection and activate these lymphocytes, such mechanisms would have important implications for the host response against the pathogen and for bone formation. The results presented here demonstrate the potential for osteoblasts to secrete high levels of CXCL10 in response to bacterial infection. This surprising result suggests that osteoblasts may have a new function beyond that of bone formation and may, in fact, be responsible for recruitment and activation of T lymphocytes at sites of bone disease.
Acknowledgments
This study was supported by National Institutes of Health grant AR47585.
Editor: R. N. Moore
REFERENCES
- 1.Aderem, A., and R. J. Ulevitch. 2000. Toll-like receptors in the induction of the innate immune response. Nature 406:782-787. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., K. Takeda, and T. Kaisho. 2001. Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2:675-680. [DOI] [PubMed] [Google Scholar]
- 3.Alpuche-Aranda, C. M., E. L. Racoosin, J. A. Swanson, and S. I. Miller. 1994. Salmonella stimulate macrophage macropinocytosis and persist within spacious phagosomes. J. Exp. Med. 179:601-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anand, A. J., and A. E. Glatt. 1994. Salmonella osteomyelitis and arthritis in sickle cell disease. Semin. Arthritis Rheum. 24:211-221. [DOI] [PubMed] [Google Scholar]
- 5.Arron, J. R., and Y. Choi. 2000. Bone versus immune system. Nature 408:535-536. [DOI] [PubMed] [Google Scholar]
- 6.Aubin, J. E. 1998. Advances in the osteoblast lineage. Biochem. Cell Biol. 76:899-910. [PubMed] [Google Scholar]
- 7.Baker, P. J., M. Dixon, R. T. Evans, L. Dufour, E. Johnson, and D. C. Roopenian. 1999. CD4+ T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect. Immun. 67:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonecchi, R., G. Bianchi, P. P. Bordignon, D. D'Ambrosio, R. Lang, A. Borsatti, S. Sozzani, P. Allavena, P. A. Gray, A. Mantovani, and F. Sinigaglia. 1998. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J. Exp. Med. 187:129-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bost, K. L., J. L. Bento, J. K. Ellington, I. Marriott, and M. C. Hudson. 2000. Induction of colony-stimulating factor expression following Staphylococcus or Salmonella interaction with mouse or human osteoblasts. Infect. Immun. 68:5075-5083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bost, K. L., J. L. Bento, C. C. Petty, L. W. Schrum, M. C. Hudson, and I. Marriott. 2001. Monocyte chemoattractant protein-1 expression by osteoblasts following infection with Staphylococcus aureus or Salmonella. J. Interferon Cytokine Res. 21:297-304. [DOI] [PubMed] [Google Scholar]
- 11.Bost, K. L., W. K. Ramp, N. C. Nicholson, J. L. Bento, I. Marriott, and M. C. Hudson. 1999. Staphylococcus aureus infection of mouse or human osteoblasts induces high levels of interleukin-6 and interleukin-12 production. J. Infect. Dis. 180:1912-1920. [DOI] [PubMed] [Google Scholar]
- 12.Bremell, T., S. Lange, R. Holmdahl, C. Ryden, G. K. Hansson, and A. Tarkowski. 1994. Immunopathological features of rat Staphylococcus aureus arthritis. Infect. Immun. 62:2334-2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bresnihan, B. 1999. Pathogenesis of joint damage in rheumatoid arthritis. J. Rheumatol. 26:717-719. [PubMed] [Google Scholar]
- 14.Cunningham, R., A. Cockayne, and H. Humphreys. 1996. Clinical and molecular aspects of the pathogenesis of Staphylococcus aureus bone and joint infections. J. Med. Microbiol. 44:157-164. [DOI] [PubMed] [Google Scholar]
- 15.Ducy, P., T. Schinke, and G. Karsenty. 2000. The osteoblast: a sophisticated fibroblast under central surveillance. Science 289:1501-1504. [DOI] [PubMed] [Google Scholar]
- 16.Ellington, J. K., S. S. Reilly, W. K. Ramp, M. S. Smeltzer, J. F. Kellam, and M. C. Hudson. 1999. Mechanisms of Staphylococcus aureus invasion of cultured osteoblasts. Microb. Pathog. 26:317-323. [DOI] [PubMed] [Google Scholar]
- 17.Finlay, B. B., and P. Cossart. 1997. Exploitation of mammalian host cell functions by bacterial pathogens. Science 276:718-725. [DOI] [PubMed] [Google Scholar]
- 18.Foster, T. J., and M. Hook. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 6:484-488. [DOI] [PubMed] [Google Scholar]
- 19.Gangur, V., F. E. Simons, and K. T. Hayglass. 1998. Human IP-10 selectively promotes dominance of polyclonally activated and environmental antigen-driven IFN-gamma over IL-4 responses. FASEB J. 12:705-713. [DOI] [PubMed] [Google Scholar]
- 20.Graves, D. T., Y. Jiang, and A. J. Valente. 1999. Regulated expression of MCP-1 by osteoblastic cells in vitro and in vivo. Histol. Histopathol. 14:1347-1354. [DOI] [PubMed] [Google Scholar]
- 21.Gundle, R., and J. N. Beresford. 1995. The isolation and culture of cells from explants of human trabecular bone. Calcif. Tissue Int. 56:S8-S10.7719993 [Google Scholar]
- 22.Heine, H., C. J. Kirschning, E. Lien, B. G. Monks, M. Rothe, and D. T. Golenbock. 1999. Cutting edge: cells that carry a null allele for toll-like receptor 2 are capable of responding to endotoxin. J. Immunol. 162:6971-6975. [PubMed] [Google Scholar]
- 23.Horowitz, M. C., D. L. Coleman, P. M. Flood, T. S. Kupper, and R. L. Jilka. 1989. Parathyroid hormone and lipopolysaccharide induce murine osteoblast-like cells to secrete a cytokine indistinguishable from granulocyte-macrophage colony-stimulating factor. J. Clin. Investig. 83:149-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jevon, M., C. Guo, B. Ma, N. Mordan, S. P. Nair, M. Harris, B. Henderson, G. Bentley, and S. Meghji. 1999. Mechanisms of internalization of Staphylococcus aureus by cultured human osteoblasts. Infect. Immun. 67:2677-2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang, Y., and D. T. Graves. 1999. Periodontal pathogens stimulate CC-chemokine production by mononuclear and bone-derived cells. J. Periodontol. 70:1472-1478. [DOI] [PubMed] [Google Scholar]
- 26.Jones, B. D., and S. Falkow. 1996. Salmonellosis: host immune responses and bacterial virulence determinants. Annu. Rev. Immunol. 14:533-561. [DOI] [PubMed] [Google Scholar]
- 27.Jouanguy, E., R. Doffinger, S. Dupuis, A. Pallier, F. Altare, and J. L. Casanova. 1999. IL-12 and IFN-gamma in host defense against mycobacteria and salmonella in mice and men. Curr. Opin. Immunol. 11:346-351. [DOI] [PubMed] [Google Scholar]
- 28.Kawai, T., R. Eisen-Lev, M. Seki, J. W. Eastcott, M. E. Wilson, and M. A. Taubman. 2000. Requirement of B7 costimulation for Th1-mediated inflammatory bone resorption in experimental periodontal disease. J. Immunol. 164:2102-2109. [DOI] [PubMed] [Google Scholar]
- 29.Kikuchi, T., T. Matsuguchi, N. Tsuboi, A. Mitani, S. Tanaka, M. Matsuoka, G. Yamamoto, T. Hishikawa, T. Noguchi, and Y. Yoshikai. 2001. Gene expression of osteoclast differentiation factor is induced by lipopolysaccharide in mouse osteoblasts via Toll-like receptors. J. Immunol. 166:3574-3579. [DOI] [PubMed] [Google Scholar]
- 30.Koehler, L., H. Zeidler, and A. P. Hudson. 1998. Aetiological agents: their molecular biology and phagocyte-host interaction. Baillieres Clin. Rheumatol. 12:589-609. [DOI] [PubMed] [Google Scholar]
- 31.Kondo, A., Y. Koshihara, and A. Togari. 2001. Signal transduction system for interleukin-6 synthesis stimulated by lipopolysaccharide in human osteoblasts. J. Interferon Cytokine Res. 21:943-950. [DOI] [PubMed] [Google Scholar]
- 32.Kong, Y. Y., U. Feige, I. Sarosi, B. Bolon, A. Tafuri, S. Morony, C. Capparelli, J. Li, R. Elliott, S. McCabe, T. Wong, G. Campagnuolo, E. Moran, E. R. Bogoch, G. Van, L. T. Nguyen, P. S. Ohashi, D. L. Lacey, E. Fish, W. J. Boyle, and J. M. Penninger. 1999. Activated T cells regulate bone loss and joint destruction in adjuvant arthritis through osteoprotegerin ligand. Nature 402:304-309. [DOI] [PubMed] [Google Scholar]
- 33.Lalmanach, A. C., and F. Lantier. 1999. Host cytokine response and resistance to Salmonella infection. Microbes Infect. 1:719-726. [DOI] [PubMed] [Google Scholar]
- 34.Lew, D. P., and F. A. Waldvogel. 1997. Osteomyelitis. N. Engl. J. Med. 336:999-1007. [DOI] [PubMed] [Google Scholar]
- 35.Lorenzo, J. 2000. Interactions between immune and bone cells: new insights with many remaining questions. J. Clin. Investig. 106:749-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luster, A. D., and J. V. Ravetch. 1987. Biochemical characterization of a gamma interferon-inducible cytokine (IP-10). J. Exp. Med. 166:1084-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luster, A. D., J. C. Unkeless, and J. V. Ravetch. 1985. Gamma-interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature 315:672-676. [DOI] [PubMed] [Google Scholar]
- 38.Matsuura, K., M. Setoguchi, N. Nasu, Y. Higuchi, S. Yoshida, S. Akizuki, and S. Yamamoto. 1989. Nucleotide and amino acid sequences of the mouse CD14 gene. Nucleic Acids Res. 17:2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nair, S. P., S. Meghji, M. Wilson, K. Reddi, P. White, and B. Henderson. 1996. Bacterially induced bone destruction: mechanisms and misconceptions. Infect. Immun. 64:2371-2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Narumi, S., and T. A. Hamilton. 1991. Inducible expression of murine IP-10 mRNA varies with the state of macrophage inflammatory activity. J. Immunol. 146:3038-3044. [PubMed] [Google Scholar]
- 41.Ohl, M. E., and S. I. Miller. 2001. Salmonella: a model for bacterial pathogenesis. Annu. Rev. Med. 52:259-274. [DOI] [PubMed] [Google Scholar]
- 42.Overturf, G. D. 1999. Infections and immunizations of children with sickle cell disease. Adv. Pediatr. Infect. Dis. 14:191-218. [PubMed] [Google Scholar]
- 43.Ponomaryov, T., A. Peled, I. Petit, R. S. Taichman, L. Habler, J. Sandbank, F. Arenzana-Seisdedos, A. Magerus, A. Caruz, N. Fujii, A. Nagler, M. Lahav, M. Szyper-Kravitz, D. Zipori, and T. Lapidot. 2000. Induction of the chemokine stromal-derived factor-1 following DNA damage improves human stem cell function. J. Clin. Investig. 106:1331-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin, S., J. B. Rottman, P. Myers, N. Kassam, M. Weinblatt, M. Loetscher, A. E. Koch, B. Moser, and C. R. Mackay. 1998. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Investig. 101:746-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qureshi, S. T., L. Lariviere, G. Leveque, S. Clermont, K. J. Moore, P. Gros, and D. Malo. 1999. Endotoxin-tolerant mice have mutations in Toll-like receptor 4 (Tlr4). J. Exp Med. 189:615-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Re, F., and J. L. Strominger. 2001. Toll-like receptor 2 (tlr2) and tlr4 differentially activate human dendritic cells. J. Biol. Chem. 276:37692-37699. [DOI] [PubMed] [Google Scholar]
- 47.Reilly, S. S., M. C. Hudson, J. F. Kellam, and W. K. Ramp. 2000. In vivo internalization of Staphylococcus aureus by embryonic chick osteoblasts. Bone 26:63-70. [DOI] [PubMed] [Google Scholar]
- 48.Reyes-Botella, C., M. J. Montes, M. F. Vallecillo-Capilla, E. G. Olivares, and C. Ruiz. 2000. Expression of molecules involved in antigen presentation and T cell activation (HLA-DR, CD80, CD86, CD44 and CD54) by cultured human osteoblasts. J. Periodontol. 71:614-617. [DOI] [PubMed] [Google Scholar]
- 49.Riancho, J. A., M. T. Zarrabeitia, J. L. Fernandez-Luna, and J. Gonzalez-Macias. 1995. Mechanisms controlling nitric oxide synthesis in osteoblasts. Mol. Cell. Endocrinol. 107:87-92. [DOI] [PubMed] [Google Scholar]
- 50.Rosenshine, I., S. Ruschkowski, V. Foubister, and B. B. Finlay. 1994. Salmonella typhimurium invasion of epithelial cells: role of induced host cell tyrosine protein phosphorylation. Infect. Immun. 62:4969-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sallusto, F., D. Lenig, C. R. Mackay, and A. Lanzavecchia. 1998. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J. Exp. Med. 187:875-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schwandner, R., R. Dziarski, H. Wesche, M. Rothe, and C. J. Kirschning. 1999. Peptidoglycan- and lipoteichoic acid-induced cell activation is mediated by toll-like receptor 2. J. Biol. Chem. 274:17406-17409. [DOI] [PubMed] [Google Scholar]
- 53.Sismey-Durrant, H. J., S. J. Atkinson, R. M. Hopps, and J. K. Heath. 1989. The effect of lipopolysaccharide from bacteroides gingivalis and muramyl dipeptide on osteoblast collagenase release. Calcif. Tissue Int. 44:361-363. [DOI] [PubMed] [Google Scholar]
- 54.Sol, M. A., J. Tkaczuk, J. J. Voigt, M. Durand, M. Sixou, A. Maurette, and M. Thomsen. 1998. Characterization of lymphocyte subpopulations in periapical lesions by flow cytometry. Oral Microbiol. Immunol. 13:253-258. [DOI] [PubMed] [Google Scholar]
- 55.Stashenko, P., S. M. Yu, and C. Y. Wang. 1992. Kinetics of immune cell and bone resorptive responses to endodontic infections. J. Endod. 18:422-426. [DOI] [PubMed] [Google Scholar]
- 56.Takayanagi, H., K. Ogasawara, S. Hida, T. Chiba, S. Murata, K. Sato, A. Takaoka, T. Yokochi, H. Oda, K. Tanaka, K. Nakamura, and T. Taniguchi. 2000. T-cell-mediated regulation of osteoclastogenesis by signalling cross-talk between RANKL and IFN-gamma. Nature 408:600-605. [DOI] [PubMed] [Google Scholar]
- 57.Tamaru, M., Y. Tominaga, K. Yatsunami, and S. Narumi. 1998. Cloning of the murine interferon-inducible protein 10 (IP-10) receptor and its specific expression in lymphoid organs. Biochem. Biophys. Res. Commun. 251:41-48. [DOI] [PubMed] [Google Scholar]
- 58.Taubman, M. A., and T. Kawai. 2001. Involvement of T-lymphocytes in periodontal disease and in direct and indirect induction of bone resorption. Crit. Rev. Oral Biol. Med. 12:125-135. [DOI] [PubMed] [Google Scholar]
- 59.Teitelbaum, S. L. 2000. Bone resorption by osteoclasts. Science 289:1504-1508. [DOI] [PubMed] [Google Scholar]
- 60.Teng, Y. T., H. Nguyen, X. Gao, Y. Y. Kong, R. M. Gorczynski, B. Singh, R. P. Ellen, and J. M. Penninger. 2000. Functional human T-cell immunity and osteoprotegerin ligand control alveolar bone destruction in periodontal infection. J. Clin. Investig. 106:R59-R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trieb, K., T. Lechleitner, S. Lang, R. Windhager, R. Kotz, and S. Dirnhofer. 1998. Evaluation of HLA-DR expression and T-lymphocyte infiltration in osteosarcoma. Pathol. Res. Pract. 194:679-684. [DOI] [PubMed] [Google Scholar]
- 62.Tso, J. Y., X. H. Sun, T. H. Kao, K. S. Reece, and R. Wu. 1985. Isolation and characterization of rat and human glyceraldehyde-3-phosphate dehydrogenase cDNAs: genomic complexity and molecular evolution of the gene. Nucleic Acids Res. 13:2485-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ukai, T., Y. Hara, and I. Kato. 1996. Effects of T cell adoptive transfer into nude mice on alveolar bone resorption induced by endotoxin. J. Periodontal Res. 31:414-422. [DOI] [PubMed] [Google Scholar]
- 64.Vanguri, P., and J. M. Farber. 1990. Identification of CRG-2. An interferon-inducible mRNA predicted to encode a murine monokine. J. Biol. Chem. 265:15049-15057. [PubMed] [Google Scholar]
- 65.Workman, M. R., J. Philpott-Howard, S. Bragman, F. Brito-Babapulle, and A. J. Bellingham. 1996. Emergence of ciprofloxacin resistance during treatment of salmonella osteomyelitis in three patients with sickle cell disease. J. Infect. 32:27-32. [DOI] [PubMed] [Google Scholar]
- 66.Yoshimura, A., E. Lien, R. R. Ingalls, E. Tuomanen, R. Dziarski, and D. Golenbock. 1999. Cutting edge: recognition of Gram-positive bacterial cell wall components by the innate immune system occurs via Toll-like receptor 2. J. Immunol. 163:1-5. [PubMed] [Google Scholar]