Abstract
Previous studies correlated the presence of a 200-kDa protein on the surface of Moraxella catarrhalis with the ability of this organism to agglutinate human erythrocytes (M. Fitzgerald, R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott, FEMS Immunol. Med. Microbiol. 18:209-216, 1997). In the present study, the gene encoding the 200-kDa protein (designated Hag) of M. catarrhalis strain O35E was subjected to nucleotide sequence analysis and then was inactivated by insertional mutagenesis. The isogenic hag mutant was unable to agglutinate human erythrocytes and lost its ability to autoagglutinate but was still attached at wild-type levels to several human epithelial cell lines. The hag mutation also eliminated the ability of this mutant strain to bind human immunoglobulin D. The presence of the Hag protein on the M. catarrhalis cell surface, as well as that of the UspA1 and UspA2 proteins (C. Aebi, I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen, Infect. Immun. 65:4367-4377, 1997), was investigated by transmission electron and cryoimmunoelectron microscopy. Wild-type M. catarrhalis strain O35E possessed a dense layer of surface projections, whereas an isogenic uspA1 uspA2 hag triple mutant version of this strain did not possess any detectable surface projections. Examination of a uspA1 uspA2 double mutant that expressed the Hag protein revealed the presence of a relatively sparse layer of surface projections, similar to those seen on a uspA2 hag mutant that expressed UspA1. In contrast, a uspA1 hag mutant that expressed UspA2 formed a very dense layer of relatively short surface projections. These results indicate that the surface-exposed Hag protein and UspA1 and UspA2 have the potential to interact both with each other and directly with host defense systems.
Moraxella (Branhamella) catarrhalis is an important cause of disease in both the upper and lower respiratory tracts (35, 48). This unencapsulated gram-negative coccobacillus has been shown to express a number of different outer membrane proteins on its cell surface, some of which are antigenically conserved (47, 49). At present, information about the M. catarrhalis gene products that are involved in the ability of this organism to colonize the mucosa of the nasopharynx and survive in this hostile environment is limited at best. Much effort has been expended recently on documenting the human immune response to selected M. catarrhalis surface-exposed proteins (6, 12, 25, 53, 65), providing evidence that these particular gene products are expressed in vivo during otitis media or infections of the bronchial tree. A few of these outer membrane proteins now have a function ascribed to them, mainly with respect to iron acquisition (7, 9, 10, 15, 42, 43).
In contrast, there is relatively little known about other surface proteins of M. catarrhalis that might be involved in the ability of this organism to colonize and survive in the nasopharynx (35). The CD outer membrane protein (33) has been shown to bind middle ear mucin in vitro (51), a function that could be involved in the colonization process or in the development of otitis media. The UspA1 protein has been shown to be an adhesin, at least in vitro (38), whereas both the UspA2 protein (38) and outer membrane protein E (50) have been implicated in serum resistance. Both UspA1 and UspA2, consistent with their functional activities, have been localized to the surface of M. catarrhalis, where they are accessible to antibodies (2, 45).
Scott and colleagues (16, 17) correlated both hemagglutination activity and the expression of a 200-kDa protein by some M. catarrhalis isolates with the presence of a fibrillar surface array. In addition, Sasaki and colleagues reported that the 200-kDa protein expressed by M. catarrhalis was subject to phase variation in vitro (K. Sasaki, L. Myers, S. M. Loosmore, and M. H. Klein, Abstr. 99th Gen. Meet. Am. Soc. Microbiol., abstr. B/D-306, 1999) and determined the nucleotide sequence of the gene encoding this protein (54). In the present study, we used analysis of mutants to show that this protein, designated Hag (hemagglutinin), is involved not only in hemagglutination but also in autoagglutination and the binding of human immunoglobulin D (IgD) by M. catarrhalis strain O35E. In addition, we determined that the Hag protein, together with the UspA1 and UspA2 proteins (3), all form fibrillar projections on the M. catarrhalis cell surface.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains and mutants used in this study are described in Table 1. M. catarrhalis was cultured at 37°C in brain heart infusion (BHI) broth (Difco/Becton Dickinson, Sparks, Md.) or on BHI agar plates in an atmosphere of 95% air-5% CO2. Antimicrobial supplementation for the selection of M. catarrhalis mutants involved the use of chloramphenicol (0.6 μg/ml), Zeocin (Invitrogen, Carlsbad, Calif.) (5 μg/ml), or spectinomycin (15 μg/ml). Mutants were grown without antimicrobial supplementation for biofilm development and for adherence assays.
TABLE 1.
Bacterial strains used in this study
| M. catarrhalis strain | Description or phenotype | Source or reference |
|---|---|---|
| O35E | Wild-type disease isolate | 3 |
| O35E.118CAT | uspA1::cat reporter strain | 39 |
| O35E.2ZEO | Isogenic uspA2 mutant of strain O35E with a Zeocin resistance cartridge inserted into the uspA2 gene | This study |
| O35E.HG | Isogenic hag mutant of strain O35E with a spectinomycin resistance cartridge inserted into the hag gene | This study |
| O35E.ZCS | uspA1 uspA2 hag mutant of strain O35E | This study |
| O35E.ZC | uspA1 uspA2 mutant of strain O35E; expresses Hag | This study |
| O35E.1HG | uspA1 hag mutant of strain O35E; expresses UspA2 | This study |
| O35E.2HG | uspA2 hag mutant of strain O35E; expresses UspA1 | This study |
| O12E | Wild-type disease isolate | 1 |
| 4223 | Wild-type disease isolate | 37 |
| O46E | Wild-type disease isolate | 38 |
| ATCC 25238 | Wild-type strain | American Type Culture Collection |
| ATCC 25240 | Wild-type strain | American Type Culture Collection |
| P44 | Wild-type disease isolate | 34 |
| TTA24 | Wild-type disease isolate | 14 |
| TTA37 | Wild-type disease isolate | Steven Berk |
| E22 | Wild-type disease isolate | 5 |
| V1171 | Wild-type isolate from the nasopharynx of a healthy child | Frederick Henderson |
| ETSU-13 | Serum-resistant wild-type strain | Steven Berk |
| ETSU-25 | Serum-sensitive wild-type strain | Steven Berk |
Growth of biofilms.
The technique described by Budhani and Struthers (8) was used to grow M. catarrhalis in a biofilm. Briefly, a 3-ml portion of an overnight culture was used to inoculate a sterile Sorbarod filter (diameter, 10 mm; length, 20 mm; Ilacon, Kent, United Kingdom) contained within a short piece (length, 3 in.; inside diameter, 3/8 in.) of silicone tubing. After inoculation, sterile BHI broth was dripped onto this Sorbarod filter at a rate of 0.1 ml/min with a multichannel peristaltic pump (Watson Marlow, Wilmington, Mass.). The entire biofilm apparatus was housed in a 37°C environmental room. Cells were routinely harvested after 3 days of growth on the filter.
MAbs and Western blot analysis.
Monoclonal antibody (MAb) 17C7, reactive with both the UspA1 and UspA2 proteins of M. catarrhalis strain O35E (3), and MAb 10F3, reactive with the CopB outer membrane protein of this strain (26), have been described. To obtain a MAb specific for the Hag protein, synthetic peptide DNADGNQVNIADIKKDPNSGSSSNR (Hag-1) was synthesized by the Biopolymers Facility at the University of Texas Southwestern Medical Center and covalently bound to keyhole limpet hemocyanin (KLH; Sigma, St. Louis, Mo.) with glutaraldehyde. The sequence of Hag-1 corresponds to an amino acid sequence in the C-terminal one-third of the Hag protein that is present in all Hag proteins whose open reading frames (ORFs) have been sequenced to date (data not shown). The Hag-1-KLH conjugate was used to immunize mice for hybridoma production as previously described (2); MAb 5D2 was shown by enzyme-linked immunosorbent assay to bind a Hag-1-ovalbumin conjugate and was shown by Western blot analysis to bind a 200-kDa M. catarrhalis antigen. Human IgD κ chain myeloma protein (The Binding Site, San Diego, Calif.) was used as the source of IgD for the IgD binding assays. Western blot analysis was performed using either affinity-purified and radioiodinated goat anti-mouse Ig (38) or horseradish peroxidase-conjugated goat anti-mouse IgG (Jackson ImmunoResearch, West Grove, Pa.) as the secondary antibody to detect mouse MAbs. To detect human IgD, horseradish peroxidase-conjugated goat anti-human IgD (Biosource International, Camarillo, Calif.) was used as the secondary antibody. Horseradish peroxidase-antibody conjugates were detected by chemiluminescence with Western Lightning Chemiluminescence Reagent Plus (New England Nuclear, Boston, Mass.).
TEM.
After M. catarrhalis cells were grown for 3 days on the Sorbarod filter, the BHI growth medium was replaced by transmission electron microscopy (TEM) prefixative consisting of 75 mM lysine monohydrochloride (Sigma), 2% (vol/vol) paraformaldehyde (Electron Microscopy Sciences, Fort Washington, Pa.), and 2.5% (vol/vol) glutaraldehyde (Sigma) in 0.1 M sodium cacodylate buffer (Electron Microscopy Sciences). Prefixative was pumped onto the filter at 0.1 ml/min for 1 h at 37°C. Then, TEM fixative consisting of 2% paraformaldehyde and 2.5% (vol/vol) glutaraldehyde in 0.1 M sodium cacodylate was pumped onto the filters for 2 h at a rate of 0.1 ml/min at 37°C. The filter was then rinsed three times for 10 min in 0.1 M sodium cacodylate buffer at room temperature after which 1% osmium tetroxide in this buffer (Electron Microscopy Sciences) was added and the filter was rocked gently for 90 min. The filter was then washed with distilled water, dehydrated with ethanol, and embedded in Spurr resin (Polysciences, Warrington, Pa.), which was polymerized at 60°C overnight. Sections for TEM were cut at 80 nm with a diamond knife (Micro Star, Huntsville, Tex.) and picked up on copper 200-mesh thin-bar grids (Electron Microscopy Sciences). The grids were stained with uranyl acetate and lead citrate and observed with a JEOL 1200EX II transmission electron microscope.
Cryoimmunoelectron microscopy.
Biofilm-grown M. catarrhalis cells in the Sorbarod filter were pre-fixed as described above except that the prefixative was composed of 2% paraformaldehyde, 0.2% glutaraldehyde, and 75 mM lysine monohydrochloride in phosphate-buffered saline (PBS), pH 7.3. The cells on the filter were then fixed with 2% paraformaldehyde and 0.2% glutaraldehyde in PBS, pH 7.3, at 37°C for 2 h as described above. Subsequently, they were embedded in 10% gelatin as described previously (57) except that the gelatin was not fixed. After centrifugation, the gelatin was solidified on ice and the blocks were prepared for ultramicrotomy and infused with 2.3 M sucrose. Ultrathin sections were obtained and immunolabeled as described previously (58) with minor modifications. In particular, 10% (vol/vol) goat serum was used in the blocking buffer in place of 1% bovine serum albumin and immunolabeling was carried out with a 1:1 dilution of MAb 5D2 or a 1:10 dilution of MAb 17C7 for 2 h and a 1:15 dilution of goat anti-mouse IgG-18-nm colloidal gold (Jackson ImmunoResearch Laboratories) for 1 h. Sections were stained with uranyl acetate and embedded in methyl cellulose according to a modification of the method of Tokuyasu (62) introduced by Griffiths et al. (21). Samples were viewed and photographed with a Zeiss 902 electron microscope.
PCR.
PCR was performed with either XL (Perkin-Elmer Biosystems, Foster City, Calif.) or ExTaq (PanVera, Madison, Wis.) DNA polymerase according to the manufacturers' instructions. Purified chromosomal DNA (Easy-DNA kit; Invitrogen) was used as the template for PCR. Oligonucleotide primers P1 (5′-TTGCCCCATATCTGTACG-3′) and P2 (5′-GGTCATGGTGAAAGAGAATC-3′) were used to amplify a 7-kb product containing the hag gene from strain O35E. Oligonucleotide primers P3 (5′-AGAATGATGATGCCTACGAG-3′) and P2 were used to amplify the hag gene from strain O12E.
Nucleotide sequence analysis.
PCR products were sequenced with a model 373A or model 377 automated DNA sequencer (Perkin-Elmer Biosystems). DNA sequence information was analyzed by using the MacVector analysis package (version 6.5; Oxford Molecular Group, Campbell, Calif.).
Construction of isogenic mutants.
Strain O35E.118CAT, an isogenic uspA1 mutant version of M. catarrhalis strain O35E, has been described (39). Isogenic uspA2 mutants were produced by using oligonucleotide primers 5′-CGGGATCCTTCTCCCCCTAAAAATCGCTGT-3′ and 5′-AGGGATCCCGCTGTATGCCGCTACTCGCAGCT-3′ (BamHI sites are underlined) for the PCR-based amplification of a 2.6-kb fragment containing an incomplete uspA2 ORF from wild-type M. catarrhalis strain P44; this fragment was cloned into pCR2.1 (Invitrogen). This uspA2 sequence was then subcloned as an EcoRI fragment into pBluescript KS(+) (Stratagene, La Jolla, Calif.). A 0.4-kb BglII fragment was deleted from the middle of the uspA2 sequence, and a 0.5-kb Zeocin resistance cassette was ligated into this site to create plasmid pELU244ZEO. This plasmid was electroporated into M. catarrhalis strain O35E to produce uspA2 mutant O35E.2ZEO by allelic exchange. uspA1 uspA2 double mutant O35E.ZC was constructed by using plasmid pELU1CAT (39) to electroporate uspA2 mutant O35E.2ZEO and by identifying a transformant resistant to both chloramphenicol and Zeocin.
With the working assumption that the 200-kDa protein of M. catarrhalis strain 4223 (Sasaki et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol., 1999) was likely the same protein described phenotypically by Fitzgerald and coworkers (16), the nucleotide sequence of the gene from M. catarrhalis strain 4223 encoding the 200-kDa protein (54) was used to design oligonucleotide primers for the PCR-based amplification of the hag gene. Isogenic hag mutants were constructed by using oligonucleotide primers 5′-ATTCTAGAGCTCAGGGTGATGCCTCGATTGCC-3′ and 5′-ATTCTAGATGGAAGAAGCGGATACCTTGTTC-3′ (XbaI sites are underlined) together with M. catarrhalis strain P44 chromosomal DNA to amplify a 5.5-kb fragment from within the hag ORF, which was subsequently cloned into the XbaI site in pUC19 (New England Biolabs, Beverly, Mass.). A spectinomycin resistance cartridge (63) was then ligated into the EcoRV site in the hag fragment to make plasmid pELHGSPEC. uspA1 uspA2 hag triple mutant O35E.ZCS was constructed by electroporating uspA1 uspA2 mutant O35E.ZC with pELHGSPEC and identifying a transformant resistant to chloramphenicol, Zeocin, and spectinomycin. The other hag mutants used in this study (Table 1) were constructed by electroporating wild-type and mutant strains of M. catarrhalis with pELHGSPEC.
Hemagglutination assays.
Overnight 4-ml cultures of M. catarrhalis were centrifuged at 7,500 × g for 8 min and resuspended in PBS to a density of 300 Klett units with a Klett-Summerson colorimeter (Klett Mfg. Co., New York, N.Y.). A 50-μl portion of this suspension and serial twofold dilutions of this suspension were added in triplicate to a 96-well U-bottom Costar polypropylene plate (Fisher Scientific Co., Pittsburgh, Pa.). Citrated human blood (Rockland, Gilbertsville, Pa.) was centrifuged at 1,000 × g, and a 2% (vol/vol) suspension of erythrocytes in PBS was prepared. A 50-μl portion of the erythrocyte suspension was then added to each well, and the microtiter plate was gently agitated on a Vortex mixer for 30 s. Hemagglutination was recorded photographically after 15 min.
Autoagglutination assays.
M. catarrhalis cells scraped from the surface of BHI agar plates were suspended in 1 ml of PBS (pH 7.3). Portions of this suspension were added to 4 ml of PBS to attain a density of 400 Klett units in a glass tube. Autoagglutination was measured as the decrease in Klett units over time.
Adherence assays.
The ability of M. catarrhalis to attach to Chang human conjunctival epithelial cells in vitro was measured as described previously (38).
RESULTS
Characterization of the M. catarrhalis strain O35E hag gene and its encoded protein product.
The hag gene was amplified by PCR from M. catarrhalis strain O35E chromosomal DNA with oligonucleotide primers designed from the sequence of the gene encoding the 200-kDa surface protein from M. catarrhalis strain 4223 (54). The hag ORF contained 5,895 nucleotides (nt) (GenBank accession no. AY077637). The hag gene from strain O35E has at least two putative translational start sites, separated by 45 nt, and encodes predicted proteins with molecular masses of 201,566 and 199,700 Da. Inverted repeats that might function as transcriptional terminators were located 15 and 269 nt 3′ from the end of the hag ORF. The N-terminal amino acid sequence of the predicted protein has properties consistent with the presence of a signal peptide: a short hydrophilic sequence followed by a longer hydrophobic region which contains a putative signal peptidase I cleavage site (i.e., AYA) at residues 64 to 66. It should be noted that the 5′ end of this ORF contains a region with six consecutive G residues, similar to the nine consecutive G residues observed in the 5′ end of the ORF encoding the 200-kDa protein from M. catarrhalis strain 4223 (Sasaki et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol., 1999). The deduced amino acid sequence of the Hag protein from strain O35E has several regions predicted to form coiled-coil structures (data not shown) and has homology with several bacterial surface proteins that are members of the autotransporter family (27, 28). In particular, the C-terminal domain of Hag has a predicted β-sheet structure very similar to that found in the well-characterized Hia autotransporter of Haemophilus influenzae (60, 61).
Effect of the hag mutation on hemagglutination ability.
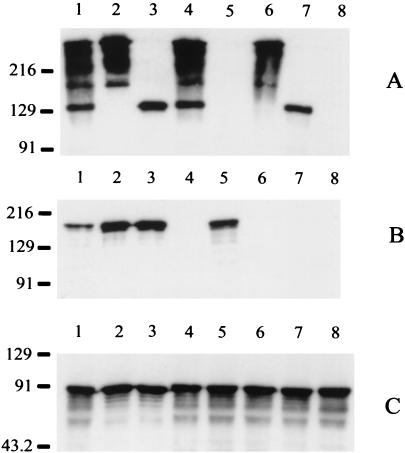
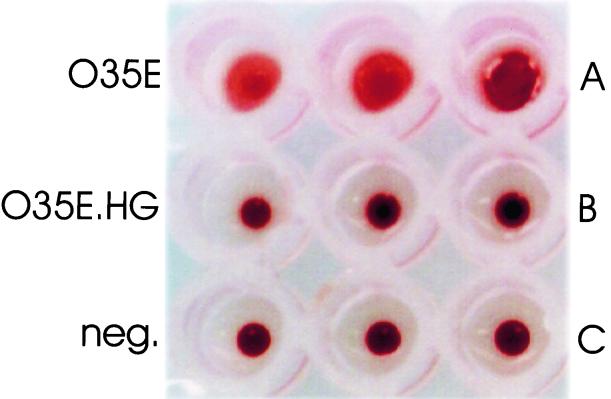
To determine whether the 200-kDa protein described by Scott and coworkers (46) as being associated with the hemagglutination ability of some M. catarrhalis strains was encoded by the hag gene, we constructed an isogenic hag mutant version of M. catarrhalis strain O35E. The hag mutant, O35E.HG (Fig. 1B, lane 4), did not express any detectable Hag protein, whereas the wild-type parent strain, O35E (Fig. 1B, lane 1), expressed a readily detectable 200-kDa antigen that bound Hag-specific MAb 5D2. This hag mutant (Fig. 1A, lane 4) also expressed wild-type levels of both UspA1 and UspA2. The wild-type parent strain (Fig. 2A) caused hemagglutination of human erythrocytes. In contrast, hag mutant O35E.HG did not cause hemagglutination (Fig. 2B).
FIG. 1.
Expression of Hag, UspA1, and UspA2 by wild-type and mutant strains of M. catarrhalis O35E. Whole-cell lysates were probed in Western blot analysis with UspA1- and UspA2-reactive MAb 17C7 (A), with Hag-specific MAb 5D2 (B), and with CopB-specific MAb 10F3 (C). Lanes: 1, wild-type parent strain; 2, uspA1 mutant O35E.118CAT; 3, uspA2 mutant O35E.2ZEO; 4, hag mutant O35E.HG; 5, uspA1 uspA2 double mutant O35E.ZC; 6, uspA1 hag double mutant O35E.1HG; 7, uspA2 hag double mutant O35E.2HG; 8, uspA1 uspA2 hag triple mutant O35E.ZCS. In this gel system, as described previously (13), the UspA1 protein migrates as a 130-kDa band whereas the UspA2 protein migrates as a series of bands near the top of the gel. Both UspA1 and UspA2 bind MAb 17C7. The CopB protein was used as an internal control for standardizing antigen loads. Molecular size position markers (in kilodaltons) are shown on the left.
FIG. 2.
Hemagglutination ability of wild-type and mutant strains of M. catarrhalis O35E. Cells of the wild-type parent strain, O35E (A), the hag mutant O35E.HG (B) , and PBS (negative control) (C) were mixed in triplicate with citrated human red blood cells and incubated at room temperature for 15 min.
Autoagglutination ability of the hag mutant.
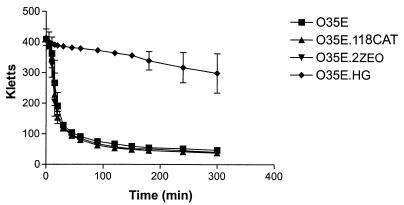
When dense bacterial suspensions were prepared, it was observed that the wild-type parent strain settled out of suspension more rapidly than did the hag mutant. To further investigate this phenomenon, we determined the autoagglutination characteristics of both the wild-type parent strain and relevant mutants with altered expression of the Hag, UspA1, and UspA2 surface proteins. The wild-type parent strain, O35E, uspA1 mutant O35E.118CAT, and uspA2 mutant O35E.2ZEO (Fig. 3) all exhibited the same rate and extent of autoagglutination in PBS. In contrast, hag mutant O35E.HG exhibited little or no tendency to autoagglutinate even after 5 h in suspension (Fig. 3).
FIG. 3.
Autoagglutination ability of wild-type and mutant strains of M. catarrhalis O35E. Cells grown on BHI agar plates overnight were suspended in PBS, and the change in turbidity over time was measured.
Effect of the hag mutation on attachment ability of M. catarrhalis strain O35E.
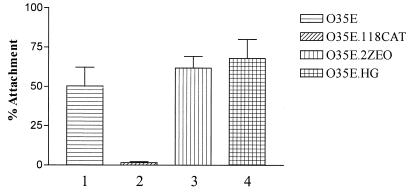
Previous studies from this laboratory had indicated that M. catarrhalis readily attaches to Chang conjunctival epithelial cells in vitro and that the UspA1 protein is responsible for the observed attachment ability (2, 38). Because the Hag protein was involved in both hemagglutination and autoagglutination and therefore had the potential to interact directly with the surfaces of eukaryotic cells, we first tested the ability of the hag mutant to attach to Chang cells. Wild-type strain O35E attached to these human cells at readily detectable levels (Fig. 4, bar 1), whereas, as expected, the uspA1 mutant, O35E.118CAT, had little or no ability to attach to these same cells (Fig. 4, bar 2). In addition, the uspA2 mutant, O35E.2ZEO, attached to the Chang cells at wild-type levels (Fig. 4, column 3), consistent with previously published results obtained with an independently isolated uspA2 mutant (2). The presence of the hag mutation had no detectable deleterious effect on the attachment ability of M. catarrhalis strain O35E (Fig. 4, column 4). Similarly, this hag mutant attached to several other human epithelial cell lines in vitro at levels similar or identical to those obtained with the wild-type parent strain (data not shown). These cell lines included HEp-2 cells (ATCC CCL-23), 16HBE14o- bronchial epithelial cells (22), and NCI-H292 epithelial cells (ATCC CRL 1848) derived from a lung mucoepidermoid carcinoma.
FIG. 4.
Attachment of wild-type and mutant strains of M. catarrhalis to Chang conjunctival epithelial cells in vitro. Attachment was measured as the percentage of the initial inoculum adherent to the Chang cells.
The Hag protein has IgD-binding activity.
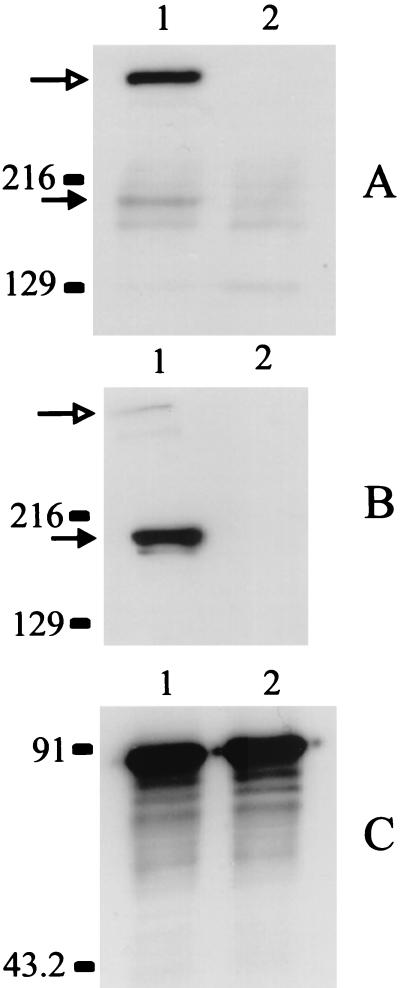
While the studies described above were in progress, Forsgren and colleagues (19) identified a 200-kDa protein, designated Mid, from M. catarrhalis strain Bc5 that possessed IgD-binding activity. To determine whether the 200-kDa protein encoded by the hag gene of M. catarrhalis strain O35E was capable of binding IgD, we tested both the wild-type parent strain and the hag mutant, O35E.HG, in a Western blot assay for IgD-binding activity. When a whole-cell lysate of the wild-type O35E strain was incubated with human IgD, the majority of the IgD-binding activity was associated with a band (Fig. 5A, lane 1) that just entered the separating gel, together with a minor band (Fig. 5A, lane 1) that migrated to a point just below the 216-kDa standard. These two different binding activities were also described by Forsgren and coworkers in their studies of the Mid protein (19). In contrast, there was no IgD-binding activity detected in the whole-cell lysate of the hag mutant (Fig. 5A, lane 2) other than some minor reactive bands that also appeared to be expressed by the wild-type parent strain. Western blot analysis of the whole-cell lysate of the wild-type parent strain (Fig. 5B, lane 1) with Hag-reactive MAb 5D2 indicated that this MAb bound an antigen that migrated to the same position as did the minor IgD-binding activity expressed by this strain (Fig. 5B, lane 1). In addition, this MAb also yielded weak reactivity with an antigen of the wild-type strain that migrated to the same position (i.e., near the top of the separating gel) (Fig. 5B, lane 1) as did the major IgD-binding activity present in the wild-type parent strain (Fig. 5A, lane 1). CopB-specific MAb 10F3 (Fig. 5C) was used to ensure that equal protein loads were used in these Western blot experiments.
FIG. 5.
Binding of human IgD by wild-type and mutant strains of M. catarrhalis. Whole-cell lysates of wild-type strain O35E (lane 1) and hag mutant O35E.HG (lane 2) were probed in a Western blot analysis with human IgD (A), Hag-reactive MAb 5D2 (B), and CopB-specific MAb 10F3 (C). In lane 1 (A and B), the open arrow indicates the position of the form of Hag that just barely enters the separating gel whereas the solid arrow indicates the position of the form that has an apparent molecular weight of approximately 200,000. Molecular size position markers (in kilodaltons) are shown on the left.
Conservation of a Hag antigenic determinant.
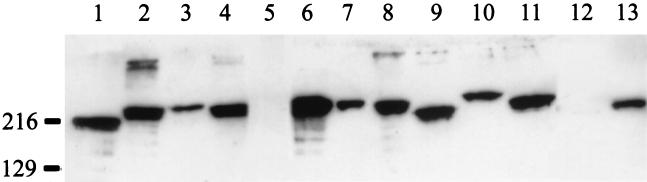
Western blot analysis of 12 additional M. catarrhalis isolates revealed that 10 of these 12 strains expressed a Hag protein that was readily detectable with MAb 5D2 (Fig. 6, lanes 2 to 13). The apparent molecular weights of these different Hag proteins varied somewhat from strain to strain. In addition, the very large form of the Hag protein which barely entered the separating gel was readily apparent with a few of these strains (Fig. 6, lanes 2, 4, 8, and 9). The lack of a detectable Hag protein with M. catarrhalis strains ATCC 25240 and ETSU-13 (Fig. 6, lanes 5 and 12, respectively) indicated that these two strains lacked a hag gene, possessed a nonfunctional hag gene, or expressed a Hag protein that lacked the epitope bound by MAb 5D2. To discriminate among these possibilities, we used PCR to amplify the 5′ end of each strain's putative hag ORF; this is the region which contains the poly(G) tract. Nucleotide sequence analysis revealed that this region contained 8 G residues in strain ATCC 25240 and 11 G residues in strain ETSU-13. In both cases, these numbers of G residues would result in premature termination of translation of the hag ORF, in the same manner as that postulated to occur in a spontaneous M. catarrhalis strain 4223 mutant that had lost the ability to express the 200-kDa protein as the result of a change from nine to eight G residues in its poly(G) tract (Sasaki et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol., 1999).
FIG. 6.
Reactivity of 12 M. catarrhalis strains with Hag-specific MAb 5D2. Whole-cell lysates of the following M. catarrhalis wild-type isolates were probed in a Western blot analysis with Hag-reactive MAb 5D2: O35E (lane 1), O12E (lane 2), O46E (lane 3), ATCC 25238 (lane 4), ATCC 25240 (lane 5), P44 (lane 6), TTA24 (lane 7), TTA37 (lane 8), E22 (lane 9), V1171 (lane 10), 4223 (lane 11), ETSU-13 (lane 12), and ETSU-25 (lane 13). Molecular size position markers (in kilodaltons) are shown on the left.
We also performed nucleotide sequence analysis of the hag gene from M. catarrhalis strain O12E, whose encoded protein product was reactive with MAb 5D2 (Fig. 6, lane 2). The hag ORF from strain O12E contained 6,945 nt (GenBank accession no. AY977638). The deduced amino acid sequence of the Hag protein from M. catarrhalis strain O35E was 60 to 76% identical to the Hag protein from strain O12E, the 200-kDa protein from strain 4223, and the Mid protein from strain Bc5. The first 60 amino acids of these four proteins had 96 to 100% identity, with the C-terminal 400 amino acids having 99% identity. All four proteins also possessed IAIGXXXXXXXXXXIAIG amino acid repeat motifs, which tended to be clustered at the beginning and end of these macromolecules and which appear to be present in several other large, surface-expressed bacterial proteins included in the autotransporter family (29).
Construction of mutants deficient in expression of surface-exposed proteins.
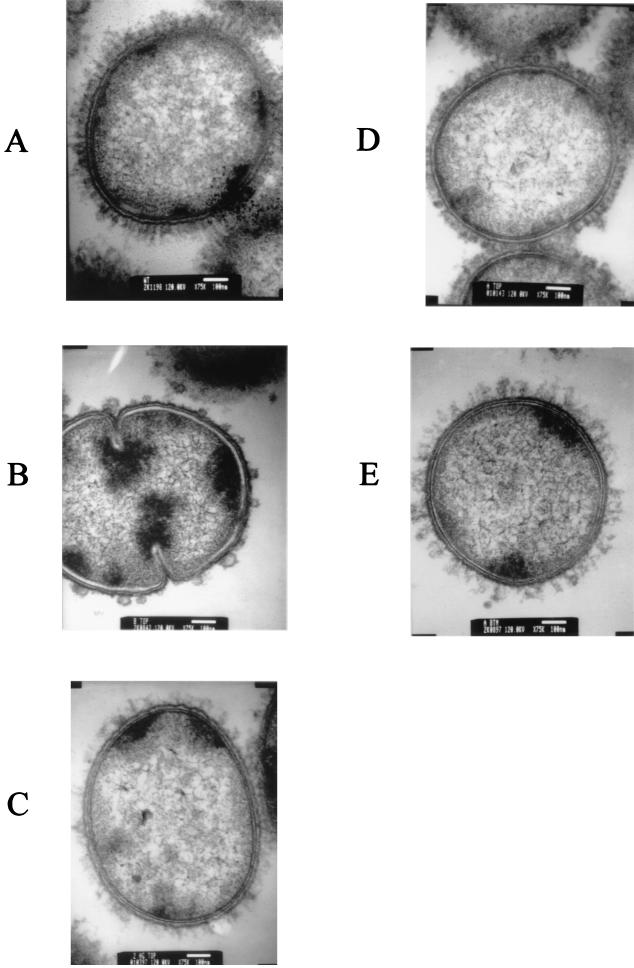
Previous studies from this laboratory indicated that both UspA1 and UspA2 are present on the bacterial cell surface (2), and it was apparent from the functional activity of the Hag protein (e.g., hemagglutination) that it was also surface exposed. In preliminary experiments examining the growth of M. catarrhalis in biofilms, we were able to use TEM of biofilm-derived cells to detect the presence of a dense layer of projections extending from the cell surface of wild-type strain O35E (Fig. 7A). To determine whether the Hag, UspA1, and UspA2 proteins were present in this surface array on the wild-type parent strain, we first constructed an M. catarrhalis strain O35E mutant that was unable to express these three proteins. This uspA1 uspA2 hag triple mutant (Fig. 1, lane 8) expressed none of these proteins at a detectable level, whereas the wild-type parent strain expressed all three macromolecules, as determined by Western blot analysis of whole-cell lysates (Fig. 1, lane 1). TEM of biofilm-derived cells of this uspA1 uspA2 hag mutant (Fig. 7B) revealed that it appeared to possess no surface projections except for some bleb-like structures.
FIG. 7.
Detection of projections on the surfaces of wild-type and mutant strains of M. catarrhalis by TEM. Each strain was grown as a biofilm for 3 days before being processed for TEM, as described in Materials and Methods. (A) Wild-type parent strain O35E; (B) uspA1 uspA2 hag triple mutant O35E.ZCS; (C) uspA2 hag double mutant O35E.2HG, expressing only the UspA1 protein; (D) uspA1 hag double mutant O35E.1HG, expressing only the UspA2 protein; (E) uspA1 uspA2 double mutant O35E.ZC, expressing only the Hag protein.
To determine the individual contributions of the Hag, UspA1, and UspA2 proteins to the wild-type surface layer, we constructed strain O35E mutants that expressed each of these proteins in the absence of the other two. The uspA2 hag double mutant (Fig. 1, lane 7) expressed only UspA1, and cells of this mutant (Fig. 7C) exhibited relatively long, thin projections that were sparsely distributed on the cell. The uspA1 hag double mutant (Fig. 1, lane 6), which expressed UspA2, formed much shorter projections (Fig. 7D), which were very densely distributed on the cell surface. The uspA1 uspA2 double mutant (Fig. 1, lane 5), which expressed only the Hag protein, formed projected structures (Fig. 7E), which resembled those formed by the uspA2 hag mutant that expressed only UspA1.
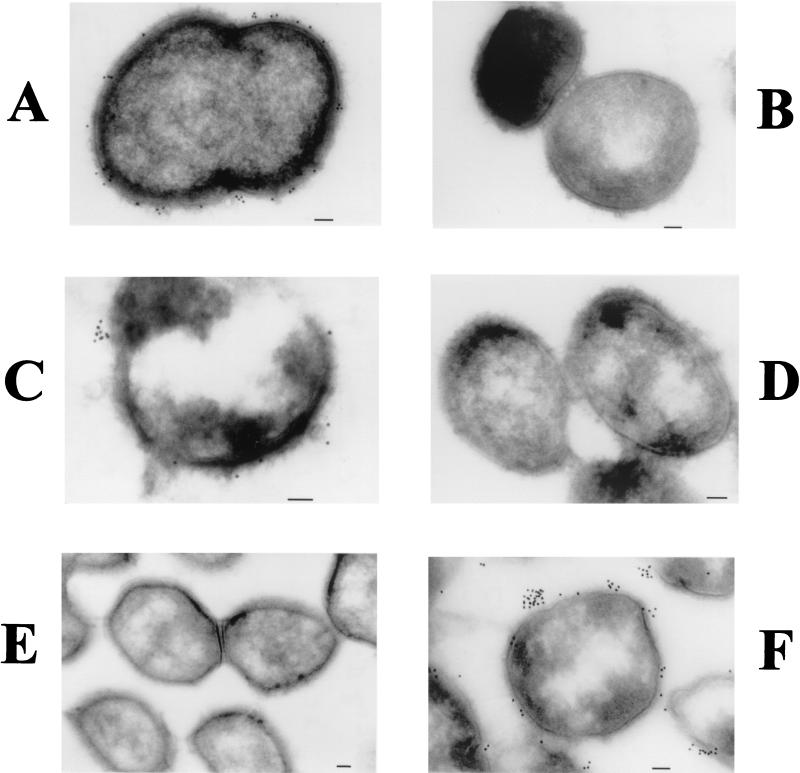
Cryoimmunoelectron microscopy was used to confirm that all three of these proteins were expressed on the M. catarrhalis cell surface. The uspA2 hag double mutant, which expressed only the UspA1 protein, bound the UspA1- and UspA2-reactive MAb 17C7 (Fig. 8A) and did not bind the Hag-specific MAb 5D2 (Fig. 8B). The uspA1 hag double mutant, which expressed only UspA2, also bound MAb 17C7 (Fig. 8C) and did not bind MAb 5D2 (Fig. 8D). Finally, the uspA1 uspA2 double mutant, which expressed only the Hag protein, bound the Hag-specific MAb 5D2 (Fig. 8F) but did not react with MAb 17C7 (Fig. 8E). In all cases, MAb-mediated binding of the antibody-conjugated gold particles was localized almost exclusively to the surface of the bacterial cell or to the area immediately exterior to the cell surface.
FIG. 8.
Use of cryoimmunoelectron microscopy to detect UspA1, UspA2, and Hag on the M. catarrhalis cell surface. Biofilm-derived cells of uspA2 hag double mutant O35E.2HG, expressing only the UspA1 protein (A and B), uspA1 hag double mutant O35E.1HG, expressing only the UspA2 protein (C and D), and uspA1 uspA2 double mutant O35E.ZC, expressing only the Hag protein (E and F) were probed with UspA1- and UspA2-reactive MAb 17C7 (A, C, and E) and with Hag-specific MAb 5D2 (B, D, and F) prior to incubation with gold particle-conjugated goat anti-mouse IgG. Bars (all panels), 100 nm.
DISCUSSION
The presence of filamentous projections on the surface of M. catarrhalis was documented over 3 decades ago by Wistreich and Baker (64), who described the presence of putative fimbriae on the surface of a strain of Neisseria (Moraxella) catarrhalis. The ability of some M. catarrhalis strains to hemagglutinate different types of erythrocytes was also described by Wistreich and Baker (64) and was studied in some detail by several other laboratories (4, 36, 52). Some of these early workers differed on whether hemagglutination ability could be correlated with fimbriation (4, 52) or with the ability to attach to eukaryotic cells (36, 52). The more recent studies of Scott and coworkers (16-18, 46) showed that the hemagglutination ability of different strains of M. catarrhalis was associated with the expression of a 200-kDa protein which was present on the bacterial cell surface.
Our previous studies had indicated that both UspA1 and UspA2 were exposed on the surface of M. catarrhalis (2, 14) and that both of these proteins had regions that were likely to form coiled coils (14), which could project from the cell surface. The reported association of the 200-kDa protein with a fibrillar layer on some strains of M. catarrhalis (17) prompted us to investigate whether the UspA1 and UspA2 proteins could also be involved in the formation of this fibrillar layer. Examination by TEM of a triple mutant lacking the ability to express the Hag, UspA1, and UspA2 proteins (Fig. 7B) revealed that there were no detectable fibrillar projections on the surface of this strain. We then examined the surface phenotype of double mutants which could express only one of each of these three proteins. These studies revealed that a uspA2 hag mutant with a functional uspA1 gene (Fig. 7C) and a uspA1 uspA2 mutant with a functional hag gene (Fig. 7E) both expressed fibrillar structures that were sparsely distributed on the bacterial cell surface. In contrast, the uspA1 hag mutant, which could express UspA2 (Fig. 7D), had a much denser layer of shorter projections on its cell surface. Close inspection of the wild-type parent strain (Fig. 7A) reveals the presence of two layers of projections. It is of interest to note that Wistreich and Baker (64) reported that the single fimbriate strain of M. catarrhalis used in their study expressed two fimbrial types, with one being much longer than the other.
A recent report from Hoiczyk et al. (32), which appeared while our study was in progress, indicated that a uspA1 uspA2 mutant version of strain O35E that had been constructed previously by our laboratory (2) lacked filamentous projections. There are two possible explanations for the reported absence of filamentous projections on this particular uspA1 uspA2 mutant. The first is that the sparsely distributed surface projections associated with expression of the hag gene may simply have been destroyed in the preparation of their samples for TEM. The second possibility would involve a mutation in the hag gene. Sasaki and coworkers (Abstr. 99th Gen. Meet. Am. Soc. Microbiol., 1999) reported the presence of nine consecutive G residues near the 5′ end of the ORF encoding the 200-kDa protein of M. catarrhalis strain 4223. A spontaneous change in the number of G residues from nine to eight, likely as the result of slipped-strand mispairing (30), caused lack of expression of the 200-kDa protein. The latter possibility seems less likely to have occurred in uspA1 uspA2 mutant O35E.12 (2), however, in view of the fact that the corresponding region of the hag gene of strain O35E has only six G residues and this shorter poly(G) tract would be less likely to undergo slipped-strand mispairing.
Inactivation of the hag gene in M. catarrhalis strain O35E resulted in the loss of both hemagglutination (Fig. 2) and autoagglutination (Fig. 3) ability. This result confirmed that the Hag protein is responsible for hemagglutination by strain O35E. In addition, the propensity of this strain to form large aggregates (i.e., autoagglutination) was eliminated by the hag mutation. Interestingly, loss of the ability to express Hag was not associated with a decreased ability to attach to several human epithelial cell lines, including Chang conjunctival epithelial cells (Fig. 4) and HEp-2 cells. This finding confirms an earlier report by Scott and colleagues (18) which indicated that the hemagglutination activity of M. catarrhalis isolates appeared to be independent of their ability to attach to HEp-2 cells in vitro. Whether the Hag protein could be involved in the attachment of M. catarrhalis to other cell types or to respiratory tract mucin (51) remains to be determined.
The lack of autoagglutination by the hag mutant (Fig. 3) mimics that described for a spontaneous mutant or variant version of M. catarrhalis strain 4223 that was isolated by Murphy and colleagues (37). These workers selected a nonclumping variant of strain 4223 by sequential passage in broth. This variant lacked detectable expression of a 200-kDa protein and also had reduced expression of the high-molecular-weight outer membrane protein (i.e., UspA2) as well as altered surface accessibility of some but not all of the surface epitopes of its outer membrane protein CD and its lipooligosaccharide molecule (37). It is likely that the 200-kDa protein missing from this variant is Hag, although this needs to be confirmed by Western blot analysis. The existence of a spontaneous frameshift mutant of strain 4223 that lacked the ability to express the 200-kDa protein (Sasaki et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol., 1999) raises the possibility that the nonclumping variant isolated by broth passage (37) may have had the same type of mutation in the hag ORF. The other alterations in the phenotype of this nonclumping variant could have been the effect of the lack of expression of the 200-kDa protein on the surface architecture of the outer membrane. Alternatively, this variant of strain 4223 may have possessed additional genetic changes.
It is of interest that the majority of the IgD-binding activity associated with the Hag protein (Fig. 5A) did not migrate to the same position in the separating gel as did the Hag antigen which bound the Hag-specific MAb 5D2 (Fig. 5B). Most of the MAb-reactive Hag protein migrated to a position just beneath the 216-kDa marker, whereas the IgD-binding activity migrated much more slowly, with an apparent molecular mass well in excess of 216 kDa. This behavior of the Hag protein in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) is similar to that described for UspA1 and UspA2, both of which form very large aggregates or complexes when analyzed by SDS-PAGE (14). It should also be noted that the IgD-binding activity of the Mid protein, described by Forsgren et al. (19), had two different forms in SDS-PAGE, with one form migrating near the 200-kDa marker and the other form barely entering the separating gel. It appears from our data that IgD preferentially binds to the multimeric form of the Hag protein from strain O35E; the structural basis for this recognition pattern remains to be determined.
The biologic significance of two (i.e., hemagglutination and autoagglutination) of the three different phenotypes associated with expression of the Hag protein by M. catarrhalis strain O35E has been determined for a number of different organisms. Hemagglutination in gram-negative bacteria is often associated with expression of pili or nonpilus adhesive proteins that promote attachment to and colonization of host mucosal surfaces (40, 41, 44, 55). The M. catarrhalis hag mutant did not display any decrease in its ability to attach to a number of human cell lines in vitro, although it is possible that lack of Hag expression could affect attachment to cell types not included in the present study. Autoagglutination, often caused either by certain types of pili or by outer membrane proteins, has been used as a marker of virulence or attachment ability for several gram-negative pathogens including such diverse organisms as Vibrio cholerae (13), Yersinia enterocolitica (56), and Fusobacterium nucleatum (24). The lack of a relevant animal model for testing the virulence of M. catarrhalis precludes determination of whether the hag mutant is truly less virulent than its wild-type parent strain.
The biologic relevance of IgD-binding activity is more difficult to ascertain. It has been known for many years that M. catarrhalis readily binds soluble human IgD (20), and Forsgren and colleagues have reported recently that the Mid protein binds IgD-expressing B cells (19). Soluble IgD is present in the nasopharynges of healthy children (59), thus providing the opportunity for this Ig to be bound by M. catarrhalis. Whether the interaction between IgD and the Hag protein might somehow augment the ability of M. catarrhalis to colonize the nasopharynx or cause otitis media remains to be determined.
The N-terminal and C-terminal regions of the 200-kDa protein from strain 4223 (54), the Hag proteins of strains O35E and O12E, and the Mid protein of strain Bc5 (19) have nearly perfect identity, with significant amino acid sequence differences being found in the more-central regions of these proteins. This situation is similar to that described individually for the UspA1 and UspA2 proteins of M. catarrhalis, where the N and C termini of each type of macromolecule are virtually identical among different strains (14). If the four 200-kDa proteins described immediately above are truly all Hag proteins, it appears that there is a clustering of amino acid sequence polymorphisms between the highly conserved N and C termini. This is similar to the mosaic structures of other surface-exposed proteins from different pathogens including Neisseria gonorrhoeae (23) and Neisseria meningitidis (31), where mosaic genes have been proposed to result from horizontal genetic exchange. The fact that M. catarrhalis can be transformed in vitro (11) makes this possibility feasible.
The IgD-binding activity of the Hag protein described in this study makes it likely that Hag and the Mid protein described by Forsgren et al. (19) are the same macromolecule with strain-specific amino acid sequence differences. Definitive proof that Hag and Mid are the same protein is dependent on the demonstration that an isogenic mid mutant version of M. catarrhalis strain Bc5 has lost its abilities to bind human IgD and hemagglutinate human erythrocytes.
Acknowledgments
This study was supported by U.S. Public Health Service grant AI36344 to E.J.H.
We thank John D. Nelson, Timothy F. Murphy, Steven L. Berk, Frederick W. Henderson, and Paul Roy for providing the M. catarrhalis strains used in this study. We also thank both Dennis Belotto and Marilyn Levy for expert technical assistance in performing TEM and cryoimmunoelectron microscopy, respectively.
Editor: A. D. O'Brien
REFERENCES
- 1.Aebi, C., L. D. Cope, J. L. Latimer, S. E. Thomas, C. A. Slaughter, G. H. McCracken, Jr., and E. J. Hansen. 1998. Mapping of a protective epitope of the CopB outer membrane protein of Moraxella catarrhalis. Infect. Immun. 66:540-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aebi, C., E. R. Lafontaine, L. D. Cope, J. L. Latimer, S. R. Lumbley, G. H. McCracken, Jr., and E. J. Hansen. 1998. Phenotypic effect of isogenic uspA1 and uspA2 mutations on Moraxella catarrhalis O35E. Infect. Immun. 66:3113-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aebi, C., I. Maciver, J. L. Latimer, L. D. Cope, M. K. Stevens, S. E. Thomas, G. H. McCracken, Jr., and E. J. Hansen. 1997. A protective epitope of Moraxella catarrhalis is encoded by two different genes. Infect. Immun. 65:4367-4377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed, K., N. Rikitomi, and K. Matsumoto. 1992. Fimbriation, hemagglutination and adherence properties of fresh clinical isolates of Branhamella catarrhalis. Microbiol. Immunol. 36:1009-1017. [DOI] [PubMed] [Google Scholar]
- 5.Beaulieu, D., M. Ouellette, M. G. Bergeron, and P. H. Roy. 1988. Characterization of a plasmid isolated from Branhamella catarrhalis and detection of plasmid sequences within the genome of a B. catarrhalis strain. Plasmid 20:158-162. [DOI] [PubMed] [Google Scholar]
- 6.Bhushan, R., C. Kirkham, S. Sethi, and T. F. Murphy. 1997. Antigenic characterization and analysis of the human immune response to outer membrane protein E of Branhamella catarrhalis. Infect. Immun. 65:2668-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonnah, R. A., H. Wong, S. M. Loosmore, and A. B. Schryvers. 1999. Characterization of Moraxella (Branhamella) catarrhalis lbpB, lbpA, and lactoferrin receptor orf3 isogenic mutants. Infect. Immun. 67:1517-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Budhani, R. K., and J. K. Struthers. 1998. Interaction of Streptococcus pneumoniae and Moraxella catarrhalis: investigation of the indirect pathogenic role of beta-lactamase-producing moraxellae by use of a continuous-culture biofilm system. Antimicrob. Agents Chemother. 42:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Campagnari, A. A., T. F. Ducey, and C. A. Rebmann. 1996. Outer membrane protein B1, an iron-repressible protein conserved in the outer membrane of Moraxella (Branhamella) catarrhalis, binds human transferrin. Infect. Immun. 64:3920-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Campagnari, A. A., K. L. Shanks, and D. W. Dyer. 1994. Growth of Moraxella catarrhalis with human transferrin and lactoferrin: expression of iron-repressible proteins without siderophore production. Infect. Immun. 62:4909-4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Catlin, B. W., and L. S. Cunningham. 1964. Genetic transformation of Neisseria catarrhalis by deoxyribonucleate preparations having different average base compositions. J. Gen. Microbiol. 37:341-352. [DOI] [PubMed] [Google Scholar]
- 12.Chen, D., V. Barniak, K. R. VanDerMeid, and J. C. McMichael. 1999. The levels and bactericidal capacity of antibodies directed against the UspA1 and UspA2 outer membrane proteins of Moraxella (Branhamella) catarrhalis in adults and children. Infect. Immun. 67:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang, S. L., R. K. Taylor, M. Koomey, and J. J. Mekalanos. 1995. Single amino acid substitutions in the N-terminus of Vibrio cholerae TcpA affect colonization, autoagglutination, and serum resistance. Mol. Microbiol. 17:1133-1142. [DOI] [PubMed] [Google Scholar]
- 14.Cope, L. D., E. R. Lafontaine, C. A. Slaughter, C. A. Hasemann, Jr., C. Aebi, F. W. Henderson, and G. H. McCracken, Jr. 1999. Characterization of the Moraxella catarrhalis uspA1 and uspA2 genes and their encoded products. J. Bacteriol. 181:4026-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, R. P., Q. J. Wang, Y.-P. Yang, A. B. Schryvers, P. Chong, M. H. Klein, and S. M. Loosmore. 1998. Cloning and expression of the Moraxella catarrhalis lactoferrin receptor genes. Infect. Immun. 66:3656-3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fitzgerald, M., R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott. 1997. A 200 kDa protein is associated with haemagglutinating isolates of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 18:209-216. [DOI] [PubMed] [Google Scholar]
- 17.Fitzgerald, M., R. Mulcahy, S. Murphy, C. Keane, D. Coakley, and T. Scott. 1999. Transmission electron microscopy studies of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 23:57-66. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald, M., S. Murphy, R. Mulcahy, C. Keane, D. Coakley, and T. Scott. 1999. Tissue culture adherence and haemagglutination characteristics of Moraxella (Branhamella) catarrhalis. FEMS Immunol. Med. Microbiol. 24:105-114. [DOI] [PubMed] [Google Scholar]
- 19.Forsgren, A., M. Brant, A. Mollenkvist, A. Muyombwe, H. Janson, N. Woin, and K. Riesbeck. 2001. Isolation and characterization of a novel IgD-binding protein from Moraxella catarrhalis. J. Immunol. 167:2112-2120. [DOI] [PubMed] [Google Scholar]
- 20.Forsgren, A., and A. O. Grubb. 1979. Many bacterial species bind human IgD. J. Immunol. 122:1468-1472. [PubMed] [Google Scholar]
- 21.Griffiths, G., A. McDowall, R. Back, and J. Dubochet. 1984. On the preparation of cryosections for immunocytochemistry. J. Ultrastruct. Res. 89:65-78. [DOI] [PubMed] [Google Scholar]
- 22.Gruenert, D. C., C. B. Basbaum, M. J. Welsh, M. Li, W. E. Finkbeiner, and J. A. Nadel. 1988. Characterization of human tracheal epithelial cells transformed by an origin-defective simian virus 40. Proc. Natl. Acad. Sci. USA 85:5951-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halter, R., J. Pohlner, and T. F. Meyer. 1989. Mosaic-like organization of IgA protease genes in Neisseria gonorrhoeae generated by horizontal genetic exchange in vivo. EMBO J. 8:2737-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Han, Y. W., W. Shi, G. T. Huang, H. S. Kinder, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harabuchi, Y., H. Murakata, M. Goh, H. Kodama, A. Kataura, H. Faden, and T. F. Murphy. 1998. Serum antibodies specific to CD outer membrane protein of Moraxella catarrhalis, P6 outer membrane protein of non-typeable Haemophilus influenzae and capsular polysaccharides of Streptococcus pneumoniae in children with otitis media with effusion. Acta Otolaryngol. 118:826-832. [DOI] [PubMed] [Google Scholar]
- 26.Helminen, M. E., I. Maciver, J. L. Latimer, L. D. Cope, G. H. McCracken, Jr., and E. J. Hansen. 1993. A major outer membrane protein of Moraxella catarrhalis is a target for antibodies that enhance pulmonary clearance of the pathogen in an animal model. Infect. Immun. 61:2003-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henderson, I. R., R. Cappello, and J. P. Nataro. 2000. Autotransporter proteins, evolution and redefining protein secretion. Trends Microbiol. 8:529-532. [DOI] [PubMed] [Google Scholar]
- 28.Henderson, I. R., and J. P. Nataro. 2001. Virulence functions of autotransporter proteins. Infect. Immun. 69:1231-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson, I. R., F. Navarro-Garcia, and J. P. Nataro. 1998. The great escape: structure and function of the autotransporter proteins. Trends Microbiol. 6:370-378. [DOI] [PubMed] [Google Scholar]
- 30.Henderson, I. R., P. Owen, and J. P. Nataro. 1999. Molecular switches—the ON and OFF of bacterial phase variation. Mol. Microbiol. 33:919-932. [DOI] [PubMed] [Google Scholar]
- 31.Hobbs, M. M., A. Seiler, M. Achtman, and J. G. Cannon. 1994. Microevolution within a clonal population of pathogenic bacteria: recombination, gene duplication and horizontal genetic exchange in the opa gene family of Neisseria meningitidis. Mol. Microbiol. 12:171-180. [DOI] [PubMed] [Google Scholar]
- 32.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsiao, C. B., S. Sethi, and T. F. Murphy. 1995. Outer membrane protein CD of Branhamella catarrhalis—sequence conservation in strains recovered from the human respiratory tract. Microb. Pathog. 19:215-225. [DOI] [PubMed] [Google Scholar]
- 34.Jordan, K. L., S. H. Berk, and S. L. Berk. 1990. A comparison of serum bactericidal activity and phenotypic characteristics of bacteremic, pneumonia-causing strains, and colonizing strains of Branhamella catarrhalis. Am. J. Med. 88:28S-32S. [DOI] [PubMed] [Google Scholar]
- 35.Karalus, R., and A. Campagnari. 2000. Moraxella catarrhalis: a review of an important human mucosal pathogen. Microbes Infect. 2:547-559. [DOI] [PubMed] [Google Scholar]
- 36.Kellens, J., M. Persoons, M. Vaneechoutte, F. van Tiel, and E. Stobberingh. 1995. Evidence for lectin-mediated adherence of Moraxella catarrhalis. Infection 23:37-41. [DOI] [PubMed] [Google Scholar]
- 37.Kyd, J. M., A. W. Cripps, and T. F. Murphy. 1998. Outer-membrane antigen expression by Moraxella (Branhamella) catarrhalis influences pulmonary clearance. J. Med. Microbiol. 47:159-168. [DOI] [PubMed] [Google Scholar]
- 38.Lafontaine, E. R., L. D. Cope, C. Aebi, J. L. Latimer, G. H. McCracken, Jr., and E. J. Hansen. 2000. The UspA1 protein and a second type of UspA2 protein mediate adherence of Moraxella catarrhalis to human epithelial cells in vitro. J. Bacteriol. 182:1364-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lafontaine, E. R., N. J. Wagner, and E. J. Hansen. 2001. Expression of the Moraxella catarrhalis UspA1 protein undergoes phase variation and is regulated at the transcriptional level. J. Bacteriol. 183:1540-1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li, X., D. E. Johnson, and H. L. Mobley. 1999. Requirement of MrpH for mannose-resistant Proteus-like fimbria-mediated hemagglutination by Proteus mirabilis. Infect. Immun. 67:2822-2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Locht, C., P. Bertin, F. D. Menozzi, and G. Renauld. 1993. The filamentous haemagglutinin, a multifaceted adhesin produced by virulent Bordetella spp. Mol. Microbiol. 9:653-660. [DOI] [PubMed] [Google Scholar]
- 42.Luke, N. R., and A. A. Campagnari. 1999. Construction and characterization of Moraxella catarrhalis mutants defective in expression of transferrin receptors. Infect. Immun. 67:5815-5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luke, N. R., T. A. Russo, N. Luther, and A. A. Campagnari. 1999. Use of an isogenic mutant constructed in Moraxella catarrhalis to identify a protective epitope of outer membrane B1 defined by monoclonal antibody 11C6. Infect. Immun. 67:681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marsh, J. W., and R. K. Taylor. 1999. Genetic and transcriptional analyses of the Vibrio cholerae mannose-sensitive hemagglutinin type 4 pilus gene locus. J. Bacteriol. 181:1110-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McMichael, J. C., M. J. Fiske, R. A. Fredenburg, D. N. Chakravarti, K. R. VanDerMeid, V. Barniak, J. Caplan, E. Bortell, S. Baker, R. Arumugham, and D. Chen. 1998. Isolation and characterization of two proteins from Moraxella catarrhalis that bear a common epitope. Infect. Immun. 66:4374-4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murphy, S., M. Fitzgerald, R. Mulcahy, C. Keane, D. Coakley, and T. Scott. 1997. Studies on haemagglutination and serum resistance status of strains of Moraxella catarrhalis isolated from the elderly. Gerontology 43:277-282. [DOI] [PubMed] [Google Scholar]
- 47.Murphy, T. F. 1996. Branhamella catarrhalis: epidemiology, surface antigenic structure, and immune response. Microbiol. Rev. 60:267-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murphy, T. F. 2000. Bacterial otitis media: pathogenetic considerations. Pediatr. Infect. Dis. J. 19:S9-S15. [DOI] [PubMed] [Google Scholar]
- 49.Murphy, T. F., and L. C. Bartos. 1989. Surface-exposed and antigenically conserved determinants of outer membrane proteins of Branhamella catarrhalis. Infect. Immun. 57:2938-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murphy, T. F., A. L. Brauer, N. Yuskiw, and T. J. Hiltke. 2000. Antigenic structure of outer membrane protein E of Moraxella catarrhalis and construction and characterization of mutants. Infect. Immun. 68:6250-6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reddy, M. S., T. F. Murphy, H. S. Faden, and J. M. Bernstein. 1997. Middle ear mucin glycoprotein; purification and interaction with nontypeable Haemophilus influenzae and Moraxella catarrhalis. Otolaryngol. Head Neck Surg. 116:175-180. [DOI] [PubMed] [Google Scholar]
- 52.Rikitomi, N., B. Andersson, K. Matsumoto, R. Lindstedt, and C. Svanborg. 1991. Mechanism of adherence of Moraxella (Branhamella) catarrhalis. Scand. J. Infect. Dis. 23:559-567. [DOI] [PubMed] [Google Scholar]
- 53.Samukawa, T., N. Yamanaka, S. Hollingshead, T. F. Murphy, and H. Faden. 2000. Immune response to surface protein A of Streptococcus pneumoniae and to high-molecular-weight outer membrane protein A of Moraxella catarrhalis in children with acute otitis media. J. Infect. Dis. 181:1842-1845. [DOI] [PubMed] [Google Scholar]
- 54.Sasaki, K., R. E. Harkness, and M. H. Klein. September 1998. Nucleic acids encoding high molecular weight major outer membrane protein of Moraxella. U.S. patent 5808024.
- 55.Scheuerpflug, I., T. Rudel, R. Ryll, J. Pandit, and T. F. Meyer. 1999. Roles of PilC and PilE proteins in pilus-mediated adherence of Neisseria gonorrhoeae and Neisseria meningitidis to human erythrocytes and endothelial and epithelial cells. Infect. Immun. 67:834-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skurnik, M., I. Bolin, H. Heikkinen, S. Piha, and H. Wolf-Watz. 1984. Virulence plasmid-associated agglutination in Yersinia spp. J. Bacteriol. 158:1031-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slot, J. W., and H. J. Geuze. 1998. Localization of macromolecular components by application of the immunogold technique on cryosectioned bacteria. Methods Microbiol. 20:211-236. [Google Scholar]
- 58.Slot, J. W., H. J. Geuze, S. Gigengack, G. E. Lienhard, and D. E. James. 1991. Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113:123-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sorensen, C. H., and P. L. Larsen. 1988. IgD in nasopharyngeal secretions and tonsils from otitis-prone children. Clin. Exp. Immunol. 73:149-154. [PMC free article] [PubMed] [Google Scholar]
- 60.St. Geme, J. W., III, and D. Cutter. 2000. The Haemophilus influenzae Hia adhesin is an autotransporter protein that remains uncleaved at the C terminus and fully cell associated. J. Bacteriol. 182:6005-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.St. Geme, J. W., III, D. Cutter, and S. J. Barenkamp. 1996. Characterization of the genetic locus encoding Haemophilus influenzae type b surface fibrils. J. Bacteriol. 178:6281-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tokuyasu, K. T. 1980. Immunochemistry on ultrathin frozen sections. Histochem. J. 12:381-403. [DOI] [PubMed] [Google Scholar]
- 63.Whitby, P. W., D. J. Morton, and T. L. Stull. 1998. Construction of antibiotic resistance cassettes with multiple paired restriction sites for insertional mutagenesis of Haemophilus influenzae. FEMS Microbiol. Lett. 158:57-60. [DOI] [PubMed] [Google Scholar]
- 64.Wistreich, G. A., and R. F. Baker. 1971. The presence of fimbriae (pili) in three species of Neisseria. J. Gen. Microbiol. 65:167-173. [DOI] [PubMed] [Google Scholar]
- 65.Yu, R. H., R. A. Bonnah, S. Ainsworth, and A. B. Schryvers. 1999. Analysis of the immunological responses to transferrin and lactoferrin receptor proteins from Moraxella catarrhalis. Infect. Immun. 67:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]