Abstract
Combinatorial cloning and expression library analysis were used to isolate human antibody Fab fragments specific for the capsular polysaccharide of Streptococcus pneumoniae serotype 23F. Thirty 23F-specific Fabs were isolated from seven vaccinated donors, and the sequences of the heavy (H)- and light (L)-chain variable regions were determined. All individuals utilized either the Vκ A23 L chain, the Vκ L6 L chain, or both chains in forming the 23F-specific combining site. Vκ A23 L chains paired primarily with VH3-23 H chains. Vκ L6 L chains were more promiscuous in heavy-chain usage between individuals. Both H and L chains were mutated, primarily in the complementarity-determining regions, compared to their closest germ line counterpart, suggesting a recall response that has undergone affinity maturation. H-chain isotypes were reflective of those found in the serum. Shared somatic modifications demonstrated that immunoglobulin G2 (IgG2) and IgA antibodies arose from the same somatically matured B cell. Our results indicate that the response to the serotype 23F pneumococcal capsular polysaccharide is oligoclonal within the individual, with one or two paratope families accounting for the majority of expressed antibody. We also determined that, in spite of the combinatorial diversity available to the immune system, the 23F-specific response is highly restricted at the population level, with the same two L-chain-determined paratope families recurring in all individuals. Lastly, analysis of the isolated Fabs indicate all have undergone extensive somatic mutation, as well as class switch, maturational events that presumably require the participation of T cells.
Streptococcus pneumoniae is a significant human pathogen causing pneumonia, bacteremia, meningitis, and otitis media. The pathogenic pneumococci are surrounded by a complex capsule composed of polymeric sugars, C polysaccharide, peptidoglycan, and surface proteins. The pneumococcal capsular polysaccharides (PPS), are heterogeneous in structure, with at least 90 different serotypes occurring within the species S. pneumoniae. PPS epitopes are immunogenic in adults and elicit antibodies that protect against infection. A vaccine containing capsular polysaccharides from 23 pneumococcal serotypes (23-valent) is available and is currently recommended for persons over 65 years of age and for other adults considered to be at increased risk of developing pneumococcal disease. Purified capsular polysaccharides do not, however, induce a protective antibody response in infants, who comprise one of the primary populations at risk. Consequently, a 7-valent polysaccharide-protein conjugate vaccine has been developed and shown to be efficacious in infants and has recently been licensed for use in this age group.
Capsular polysaccharides are defined as TI-2 (T-cell-independent) antigens based on their repetitive structure and lack of immunogenicity in xid mice and human infants. The serum response to these polysaccharides in adults is oligoclonal and restricted in isotype, with immunoglobulin G2 (IgG2) and IgA antibodies predominating after vaccination or infection. Little is known about differing immunoglobulin gene usage in antibodies specific for different PPS serotypes or in antibodies specific for the same serotype in different individuals, due primarily to the difficulty in establishing human hybridomas. Combinatorial cloning circumvents this limitation and provides a means to analyze antibody repertoires without the necessity of generating hybridomas.
In this report we examine the expressed repertoire of human antibodies specific for the capsular polysaccharide of S. pneumoniae serotype 23F. Heavy (H)- and light (L)-chain sequences are reported for 30 PPS 23F-specific Fabs isolated from seven individuals. We demonstrate that the majority of individuals use the same H- and L-chain pairs to form PPS 23F-specific paratopes. We confirm the oligoclonality of PPS 23F-specific response at the level of immunoglobulin gene expression within the individual. Lastly, we show this response to be complex in terms of somatic mutation and class switch, maturational events thought to require T-cell participation.
MATERIALS AND METHODS
Subjects.
Adult volunteers were randomly assigned to receive either the licensed 23-valent polysaccharide vaccine (Pnu-Immune; Wyeth-Lederle) or a 9-valent polysaccharide-protein conjugate vaccine consisting of PPS from serotypes 1, 4, 5, 6B, 9V, 14, 18C, 19F, and 23F conjugated to the mutant diphtheria toxin CRM197 (Wyeth-Lederle). Blood was collected on the day of vaccination and at 30 days after vaccination to determine serum antibody response. A 100-ml blood sample was also collected 7 days after vaccination for the isolation of mononuclear cells (MNC). Human subject protocols were reviewed and approved by the Institutional Review Boards at both Children's Hospital Oakland and St. Louis University School of Medicine.
Affinity selection of cells.
The enrichment of PPS-specific B cells has been previously described in detail (17). Briefly, MNC were isolated from the 7 day postvaccination blood sample by using Ficoll-Hypaque. An aliquot (106 cells) was placed into culture for 7 days in 1 ml of RPMI 1640 supplemented with 5% fetal calf serum, and the supernatant was assayed for PPS 23F-specific antibody production. PPS 23F was biotinylated as previously described and used to “arm” avidin-coated paramagnetic beads (Immunotech, Inc., Marseilles, France). These PPS 23F-coated beads were washed and then added to 2 × 107 MNC (preabsorbed with avidin-coated magnetic beads), and the mixture was incubated on ice for 30 min. C-polysaccharide (C-PS) (10 μg/ml), a bacterial product that copurifies with all serotype-specific polysaccharides, was included in the incubation buffer to inhibit the binding of C-PS specific cells. PPS 23F-binding cells were then isolated with a magnet. Positively selected cells were washed twice with cold phosphate-buffered saline-0.5% bovine serum albumin and used for RNA extraction.
Production of Fab expression libraries.
The procedures for the construction of Fab libraries has been previously described in detail (3, 17, 19,). Briefly, total RNA was prepared from affinity-isolated cells (RNeasy; Qiagen, Valencia, Calif.) and cDNA prepared by using the Thermoscript RT-RCR System (Gibco-BRL, Carlsbad, Calif.) according to the manufacturer's instructions. cDNA was used as a template in the PCR to generate H-chain Fd fragments and total kappa L chains for insertion into the expression vector pComb 3H (3). The primer sets used to generate the H and L fragments were the same as those listed elsewhere (17), with the exclusion of the lambda L-chain primers and the IgM downstream primers. L-chain fragments were inserted into the SacI/XbaI site of the pComb 3H vector, and the resulting L-chain library was electroporated into XL1-Blue E. coli cells. An aliquot was plated to determine the transformation efficiency, and the balance was expanded for 8 h. Plasmid DNA was purified from the expanded culture and digested with XhoI and SpeI, and the purified H-chain Fd fragments were ligated into the XhoI/SpeI site of L-chain library plasmid DNA. The Fd×L library was electroporated into XL1-Blue cells, plated at low density on Luria broth-carbenicillin plates, and grown overnight, and individual colonies were selected for analysis.
Identification of PPS 23F-specific Fabs.
Individual transfected E. coli colonies were selected, mastered onto an LB-carbenicillin agar plate, and grown in 1-ml overnight cultures under antibiotic selection. Bacteria were pelleted by centrifugation, resuspended in 100 μl of lysis buffer (PBS plus protease inhibitor cocktail [Complete; Roche Molecular Biochemicals, Indianapolis, Ind.]), and rapidly frozen and thawed three times by using liquid nitrogen, and the cellular debris was pelleted by centrifugation. Next, 50 μl of the lysate was assayed for 23F-specific binding activity by the facilitated radioantigen-binding assay (RABA) described below. Clones that scored positive (2× background) for PPS 23F binding were restreaked for single-colony isolation from the master plate, individual clones picked and processed as described above to verify binding, and the VH- and VL-chain DNA sequence of positive clones was determined.
Sequencing and sequence analysis.
Plasmids containing H- and L-chain genes were submitted to Davis Sequencing, LLC (Davis, Calif.), for VH- and VL-chain sequence determination. The L-chain sequencing primer LSEQ (5′-GCTTCCGGCTCGTATGTTGTGTGG-3′) and H-chain sequencing primer HSEQ (5′-GCAGCCGCTGGATTGTTATTACTC-3′) both bind to the vector. Initial sequence analysis utilized the NCBI IgBlast server (http://www.ncbi.nlm.nih.gov/igblast/) to identify candidate germ line gene (2). Subsequent analysis, alignments, and translations were performed by using MacVector (Accelrys, Inc., Princeton, N.J.). The kappa V region gene nomenclature is as described by Schable and Zachau (20). H-chain V region gene nomenclature is described in the ImMunoGeneTics database (13, 18). The complementarity-determining regions (CDRs) were as defined previously (12).
Fab purification.
Representatives of each of the two dominant L-chain families were chosen for large-scale purification and binding analysis. The H-chain CH1 domain of each of these Fabs were engineered to contain a C-terminal polyhistidine region as previously described (17). These were grown in 1-liter cultures, induced with IPTG (isopropyl-β-d-thiogalactopyranoside), and periplasmic proteins extracted with B-Per (Pierce Chemical Co., St. Louis, Mo.). Fab protein in the crude extracts was then purified by means of metal-chelate chromatography (Ni-NTA; Qiagen).
Isolation and protein sequence determination of serum antibody.
PPS 23F-specific antibody was purified from donor 023 as described by Lucas et al. (15). Briefly, sera collected 30 days postvaccination were purified by a combination of affinity chromatography on PPS 23F-coupled Sepharose and immunoprecipitation with PPS 23F. Purified antibody was separated into H and L chains by sodium dodecyl sulfate-polyacrylamide gel electrophoresis under reducing conditions, transferred onto polyvinylidene difluoride membranes, stained with Coomassie blue, washed, and dried, and H and L protein bands were excised and sent to the Protein Structure Laboratory of the University of California, Davis, for N-terminal amino acid sequence determination.
Antigen-binding and Fab concentration assays.
The ability of Fabs and serum samples to bind PPS 23F was determined by a modified RABA. The preparation of radiolabeled PPS and the RABA has been described elsewhere (15). Serum anti-PPS 23F levels were calculated by comparison to the reference serum 89-SF as previously described (15). In the analyses of Fab samples, 5 μg of affinity-purified goat anti-human kappa antisera (Biosource International, Camarillo, Calif.)/ml was included in the reaction mixture to increase avidity and facilitate precipitation. Fab concentration was determined by a capture enzyme-linked immunosorbent assay in which goat anti-human Fd (The Binding Site, Birmingham, United Kingdom) or goat anti-IgA (Sigma, St. Louis, Mo.) immobilized on a microtiter plate, captures Fab, which is then detected by alkaline phosphatase-labeled goat anti-human kappa L chain (Biosource International). This assay is standardized with a purified Fab standard whose concentration was calculated from UV absorbance at 280 nm.
Accession numbers.
Clone (VL/VH) accession numbers were as follows: 023.102 (AF485420/AF485421), 023.115 (AF485422/AF485423), 023.125 (AF485424/AF485425), 027.064 (AF485426/AF485427), 027.242 (AF485428/AF485429), 027.304 (AF485430/AF485431), 027.343 (AF485432/AF485433), 018.P6G5 (AF485434/AF485435), 025.P1C6 (AF485436/AF485437), 025.P1H10 (AF485438/AF485439), 025.P4E12 (AF485440/AF485441), 025.P5D3 (AF485442/AF485443), 025.P7A6 (AF485444/AF485445), 025.P7F5 (AF485446/AF485447), 002.P3B11 (AF485448/AF485449), 002.P7F3 (AF485450/AF485451), 002.P8E11 (AF485454/AF485455), 008.1F9 (AF485456/AF485457), 008.3A2 (AF485458/AF485459), 008.3C12 (AF485460/AF485461), 008.3D7 (AF485462/AF485463), 008.4A3 (AF485464/AF485465), 008.4D1 (AF485466/AF485467), 008.4B7 (AF485468/AF485469), 008.4B12 (AF485470/AF485471), 008.2E7 (AF485472/AF485473), 008.2F7 (AF485474/AF485475), 014.7A2 (AF485476/AF485477), 014.6G9 (AF485478/AF485479), and 014.2B6 (AF485480/AF485481)
RESULTS
Isolation of recombinant PPS 23F-specific Fabs.
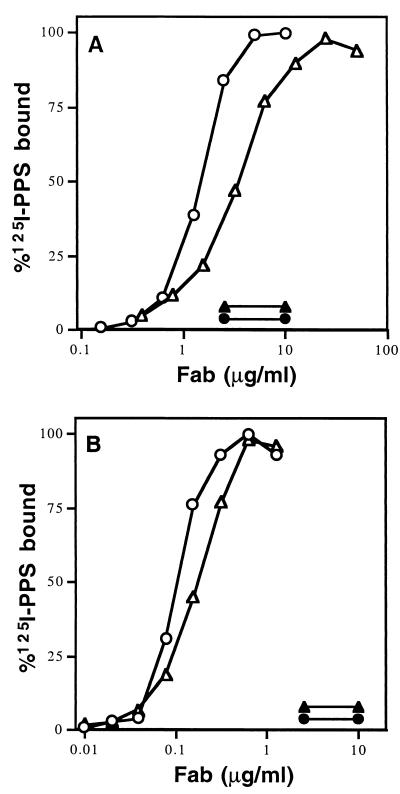
A total of 30 adults, aged 24 to 45 years, were immunized with either the licensed 23-valent polysaccharide vaccine Pnu-Immune or a 9-valent conjugate vaccine consisting of serotype 1, 4, 5, 6B, 9V, 14, 18C, 19F, and 23F PPS individually conjugated to the mutant diphtheria toxin CRM197. MNC were isolated from each individual 7 days after vaccination. An aliquot of cells was placed into culture to verify specific antibody secretion, and PPS 23F-specific cells isolated from the remaining MNC with PPS 23F-armed magnetic beads. Based primarily on the production of 23F-specific antibody in vitro and the magnitude of serum antibody response after vaccination, nine donors were selected as cloning candidates (Table 1). From these, Fd-chain, kappa L-chain, and Fd×L Fab expression libraries were constructed as described in the Materials and Methods. A total of 300 to 1,200 individual clones from each expression library were assayed for the production of PPS 23F-specific Fabs. Positive clones (those precipitating twice that of the negative control) were restreaked for single-colony isolation and recloned, and the binding and specificity were verified. H- and L-chain sequences were determined for each positive clone. A total of 49 PPS 23F-specific Fabs were isolated from seven of the nine donors assayed, 30 of which were unique in L-chain sequence, H-chain sequence, or both (Table 2). None of the isolated Fabs precipitated radiolabeled PPS 14, and none were inhibited by C-PS (10 μg/ml), indicating specificity for PPS 23F. Inspection of the sequences revealed that paratopes utilizing either the Vκ L6 or Vκ A23 L-chain genes accounted for most of the Fabs isolated from all donors. A representative Fab from each of these major families was produced in quantity and purified, and the specificity and relative affinity for PPS 23F were determined (Fig. 1). Both the Vκ A23 Fab (027.242) and the Vκ L6 Fab (023.102) bound to radiolabeled PPS 23F in a specific and concentration-dependent manner under both monovalent and polyvalent conditions.
TABLE 1.
Characteristics of MNC donors
| Donor | Vaccinea | Serum anti-PPS23F titerb (μg/ml)
|
Serum isotype | No. of Fabs/no. of clonesc | No. of unique Fabsd | |
|---|---|---|---|---|---|---|
| Pre | Post | |||||
| 001 | Conjugate | 0.9 | 112.5 | IgG2/IgA | 0/800 | 0 |
| 002 | Conjugate | 0.17 | 16.9 | IgG2/IgA | 13/800 | 3 |
| 004 | Conjugate | 1.43 | 10.95 | IgG2/IgA | 0/500 | 0 |
| 008 | Conjugate | 2.6 | 34.3 | IgG2/IgA | 11/400 | 10 |
| 014 | Conjugate | 1.34 | 15.6 | IgG2/IgA | 3/800 | 3 |
| 018 | PPS | 11.9 | 22.07 | IgG2/IgA | 3/1200 | 1 |
| 023 | PPS | 11.0 | 75.8 | IgG2/IgA | 8/300 | 3 |
| 025 | PPS | 0.3 | 10.98 | IgG2/IgA | 7/700 | 6 |
| 027 | PPS | 5.6 | 24.32 | IgG2/IgA | 4/500 | 4 |
Conjugate vaccine (PPS types 1, 4, 5, 6B, 9V, 14, 18C, 19F, and 23F conjugated to CRM197) or PPS vaccine (23-valent PPS vaccine) was administered 7 days prior to the collection of MNC.
Pre- and postvaccination PPS 23F-specific serum titers were determined for sera collected before and 30 days after vaccination.
The number of PPS 23F-specific Fabs isolated divided by the number of individual colonies screened.
The number of Fabs unique in L-chain sequence, H-chain sequence, or both.
TABLE 2.
Characteristics of the unique PPS 23F-specific Fabs
| Donor | Clone | Vκ | % Identitya | JL | L3 (aa)b | VH | % Identitya | Isotype | JH | H3 (aa)b |
|---|---|---|---|---|---|---|---|---|---|---|
| 002 | 3B11 | L6 | 98.2 | JK4 | 10 | VH3-64 | 96.1 | IgG2 | JH6 | 14 |
| 002 | 7F3 | L6 | 100.0 | JK1 | 10 | VH3-64 | 96.1 | IgG2 | JH6 | 14 |
| 002 | 8E11 | L6 | 100.0 | JK1 | 10 | VH3-64 | 95.7 | IgG2 | JH6 | 14 |
| 008 | 1F9 | A23 | 95.8 | JK2 | 9 | VH3-23 | 90.7 | IgG1 | JK4 | 6 |
| 008 | 2E7 | A23 | 96.1 | JK2 | 9 | VH3-23 | 92.1 | IgG2 | JH4 | 6 |
| 008 | 2F7 | L6 | 96.7 | JK4 | 7 | VH3-48 | 93.5 | IgG2 | JH2 | 16 |
| 008 | 3A2 | L6 | 95.3 | JK4 | 7 | VH3-48 | 93.5 | IgA2 | JH2 | 15 |
| 008 | 3C12 | A23 | 95.4 | JK3 | 9 | VH3-23 | 93.2 | IgG2 | JH1 | 6 |
| 008 | 3D7 | A23 | 96.5 | JK3 | 9 | VH3-23 | 91.1 | IgG2 | JH6 | 6 |
| 008 | 4A3 | L6 | 96.7 | JK4 | 7 | VH3-48 | 91.7 | IgG2 | JH2 | 16 |
| 008 | 4B12 | A23 | 95.5 | JK2 | 9 | VH3-23 | 94.3 | IgG2 | JH1 | 6 |
| 008 | 4B7 | L5 | 93.7 | JK2 | 9 | VH4-4 | 92.1* | IgG2 | JH5 | 13 |
| 008 | 4D1 | L6 | 96.6 | JK4 | 7 | VH3-48 | 93.2 | IgG2 | JH2 | 16 |
| 014 | 2B6 | L6 | 98.2 | JK4 | 10 | VH3-23 | 94.6 | IgG2 | JH4 | 13 |
| 014 | 6G9 | L6 | 98.2 | JK4 | 10 | VH3-23 | 94.3 | IgA1 | JH4 | 13 |
| 014 | 7A2 | L6 | 97.1 | JK2 | 11 | VH3-30 | 95.7 | IgG2 | JH4 | 11 |
| 018 | 6G5 | A23 | 96.5 | JK2 | 9 | VH3-7 | 90.1* | IgG2 | JH4 | 8 |
| 023 | 102 | L6 | 98.2 | JK1 | 11 | VH3-30 | 95.3 | IgG2 | JH4 | 11 |
| 023 | 115 | A2 | 96.9 | JK1 | 8 | VH4-59 | 93.1 | IgA2 | JH5 | 7 |
| 023 | 125 | L6 | 96.4 | JK1 | 11 | VH3-30 | 95.3 | IgG2 | JH4 | 11 |
| 025 | 1C6 | A23 | 97.7 | JK1 | 9 | VH3-23 | 95.6 | IgA2 | JH5 | 3 |
| 025 | 1H10 | L6 | 97.9 | JK3 | 11 | VH3-30 | 93.1 | IgG2 | JH4 | 15 |
| 025 | 4E12 | A23 | 93.8 | JK1 | 9 | VH3-23 | 94.9 | IgA2 | JH5 | 3 |
| 025 | 5D3 | L6 | 97.9 | JK3 | 11 | VH3-30 | 88.2 | IgA1 | JH4 | 15 |
| 025 | 7A6 | L6 | 97.5 | JK3 | 11 | VH3-30 | 93.1 | IgA2 | JH4 | 15 |
| 025 | 7F5 | A23 | 97.6 | JK1 | 9 | VH3-23 | 95.7 | IgA2 | JH5 | 3 |
| 027 | 064 | A23 | 95.8 | JK2 | 9 | VH3-23 | 93.0* | IgG2 | JH5 | 6 |
| 027 | 242 | A23 | 95.1 | JK2 | 9 | VH3-23 | 89.6 | IgA2 | JH5 | 6 |
| 027 | 304 | A23 | 95.1 | JK2 | 9 | VH3-23 | 89.6 | IgA1 | JH5 | 6 |
| 027 | 343 | A23 | 94.1 | JK2 | 9 | VH3-23 | 84.9 | IgA2 | JH5 | 6 |
Percent nucleotide identy over the entire V region compared to the corresponding germ line gene.
The number of residues comprising the Vκ (L3) and VH (H3) CDR3 regions. Fabs that contain in-frame insertions in the VH region (two residues in CDR1 of 008.4B7, two residues in framework 1 in 018.6G5, and one residue in CDR2 of 027.064) that were not considered in calculating the percent homology are marked by an asterisk.
FIG. 1.
RABA analysis of PPS binding by representative Fabs 023.102 and 027.242 under monovalent (A) and polyvalent (B) conditions. Symbols: ○, Fab 023.102 binding PPS 23F; •, Fab 023.102 binding PPS 14; ▵, Fab 027.242 binding PPS 23F; ▴, Fab 027.242 binding PPS 14.
Two L-chain V genes dominate the response to PPS 23F.
All individuals we examined utilized either the Vκ A23 L chain, the Vκ L6 L chain, or both in forming 23F-specific combining sites. Vκ A23 L chains were, with a single exception, paired with VH3-23 H chains to form the paratope. Vκ L6 L chains were more promiscuous in H-chain pairing between individuals but utilized a single H chain within any single individual. The VH3-23 gene has previously been demonstrated to be expressed at high frequency (25%) in randomly selected IgM+ B cells (6). Normal IgM+ B cells express Vκ A23 (1%) and Vκ L6 (6%) at frequencies in line with their expected occurrence in the genome (9).
In addition to these two recurring L-chain families, Fabs were isolated in which a Vκ L5 L chain was paired with a VH4-4 H chain (008.4B7), and a Vκ A2 L chain was paired with a VH4-59 H chain (023.115). We have previously reported two additional PPS 23F-specific Fabs, one utilizing Vκ A23/VH3-23 gene products, and one that utilizes Vκ A2 paired with VH3-11 (17).
Vκ A23 Fab family.
Four of the seven individuals we analyzed utilized Vκ A23-derived L chains in forming PPS 23F-specific binding domains. All Vκ A23 L chains were rearranged with a 9-amino-acid CDR3 and were mutated (as compared to the germ line) in the CDRs (Tables 3 and 4). In some individuals (donor 027, for example) the isolated L chains appeared to derive from a single rearrangement event, as indicated by the pattern of mutation and shared Jκ segment usage. In others (i.e., donor 008) sequences indicative of at least two rearrangement events were isolated. In several cases identical substitutions were found in different individuals (Asp28 to Asn28 in CDR1, Asn53 to Lys53 in CDR2, and Met89 to Thr89 in CDR3, for example), suggesting antigen-driven selection for these particular residues at these locations during affinity maturation. Alternately, these residues may indicate the use of a previously undescribed allele of Vκ A23. The Vκ A23 L chain isolated from donor 018 differed in its mutation pattern from the others in that substitutions were primarily in CDR3, with an almost germ line configuration in CDRs 1 and 2. Notably, this was the only Vκ A23 L chain isolated paired with an H chain other than VH3-23.
TABLE 3.
Vκ A23 Fab family: L chainsa
| Fab | Sequence of L chain
|
Jκ | VH | ||
|---|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | |||
| Vκ A23 | RSSQSLVHSDGNTYLS | KISNRFS | MQATQFP | ||
| 008.1F9 | T-------RN-Q-F-- | Q--K--- | T------ YT | Jκ2 | VH3-23 |
| 008.3C12 | ---------N-D---T | G--K--- | ------- FT | Jκ3 | VH3-23 |
| 008.3D7 | ---------N-D---T | E--K--- | --TS--- FT | Jκ3 | VH3-23 |
| 008.4B12 | --------RT-E-F-- | K----V- | T------ YT | Jκ2 | VH3-23 |
| 008.2E7 | --------GN-E-F-- | K------ | T----Y- YT | Jκ2 | VH3-23 |
| 025.1C6 | S------R-------- | E--K--- | ------- WT | Jκ1 | VH3-23 |
| 025.4E12 | S-G---------I--N | R----L- | ---S-Y- WT | Jκ1 | VH3-23 |
| 025.7F5 | ---------NRD---- | E--K--- | ------- WT | Jκ1 | VH3-23 |
| 027.064 | ---------N------ | QV-D--- | T------ YT | Jκ2 | VH3-23 |
| 027.242 | --------RN------ | E--K-L- | T------ YT | Jκ2 | VH3-23 |
| 027.304 | ------------A--- | Q--K-V- | T------ YT | Jκ2 | VH3-23 |
| 027.343 | ------------A--N | Q--K-V- | T------ YT | Jκ2 | VH3-23 |
| 018.6G5 | ---------------- | -----L- | -HG--L- RT | Jκ2 | VH3-7 |
CDR residues and gene usage of the A23 family of PPS 23F-specific Fabs. Germ line positions where identical substitutions were isolated from different individuals are shown in boldface. Fab 027.064 has a single residue insertion between Ser52 and Gly52a in the H-chain CDR2; a space was inserted at the appropriate location of the germ line sequence for purposes of alignment (see asterisks, Table 4). Donors 008 received the conjugate vaccine. Donors 018, 025, and 027 received the 23-valent PPS vaccine.
TABLE 4.
Vκ A23 Fab family: H chainsa
| Fab | Sequence of H chain
|
JH | Isotype | ||
|---|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | |||
| VH3-23 | SYAMS | AIS*GSGGSTYYADSVKG | |||
| 008.1F9 | -HT-T | SLN*EP-DV-R------- | VHGSAY | JH4 | IgG1 |
| 008.3C12 | NNT-T | S-N*E--DR-H------- | VHGSAH | JH1 | IgG2 |
| 008.3D7 | DNT-T | S-I*DGD-PP-------- | VHGVPD | JH6 | IgG2 |
| 008.4B12 | INT-S | S--*E--DR-R------- | VHGSAH | JH1 | IgG2 |
| 008.2E7 | -HT-T | S-N*EN-DT-H------- | VHGDAY | JH4 | IgG2 |
| 025.1C6 | -D--- | G--*---TI-------R- | STY | JH5 | IgA2 |
| 025.4E12 | -D--- | G--*---TI-------R- | STF | JH5 | IgA2 |
| 025.7F5 | ND--- | G-G*---DV--------- | STF | JH5 | IgA2 |
| 027.064 | A-T-T | S-GD-G--K--------- | VHGVPV | JH5 | IgG2 |
| 027.242 | R-T-T | S-V*D-NNRP-------- | VHGEAY | JH5 | IgA2 |
| 027.304 | R-T-T | S-V*DGNNRP-------- | VHGEAY | JH5 | IgA1 |
| 027.343 | R-T-T | S-V*NGNNRP-------- | VHGEPS | JH5 | IgA2 |
| VH3-7 | SYWMS | NIK*QDGSEKYYVDSVKG | |||
| 018.6G5 | -S--G | ---*H----QS------- | DWYRTFDY | JH4 | IgG2 |
See Table 3, footnote a.
The VH3-23-derived H chains paired with the A23 L chains were extensively mutated throughout CDRs 1 and 2 (Tables 3 and 4). Sequences from donor 027 probably represent at least two rearrangement events. In addition to a different pattern of substitutions, clone 027.064 has a single amino acid insertion in CDR2 that differentiates it from the other VH3-23-derived chains isolated from that individual. All isolates from donor 025 appear to be derivative of a single rearrangement event. Variations in CDR3 and JH usage indicates at least three rearrangements events occurred in donor 008. As in the L chains, there were identical substitutions seen in VH3-23 H chains from different individuals (Ala33 to Thr33 in CDR1, Ser35 to Thr35 in CDR1, and Ala50 to Ser50 in CDR2, for example) again implying antigen-driven selection. The VH3-23 locus is more heterogeneous than that of Vκ A23, however, and the assignment of such shared differences to mutation as opposed to common allele usage is somewhat less certain.
L6 Fab family.
The paratope family defined by the Vκ L6 gene is much more diverse, both in VH gene usage and L-chain CDR3 structure (Tables 5 and 6). Five of the seven donors utilized the Vκ L6 L chain in forming PPS 23F specific paratopes. The most common pairing (three of five donors) was a Vκ L6 L chain with an 11-amino-acid CDR3 paired with a VH3-30-derived H chain. The unusually long CDR3 results from the addition of six nontemplated bases at the Vκ-Jκ junction. Donor 008 utilizes an equally uncommon 7-amino-acid L6 CDR3 (apparently generated by the deletion of the first two residues of the J region during rearrangement) paired with a VH3-48 H chain. Substitutions shared between individuals in L-chain CDRs 1 and 2 are less common than in the Vκ A23 chains. Residues 95a and 95b, the nontemplated additions, do tend to be conserved between individuals that utilize extended CDR3s, however. Although there is no conservation in Jκ region usage between individuals, residue 96, the only varying residue contributed by the Jκ region to CDR3, has been mutated in all cases, most commonly to Ala in the 11-amino-acid CDR3s. These facts, taken together, suggest that the majority of antigen-specific selection pressure on the L chains contribution to the paratope is exerted on the CDR3 loop.
TABLE 5.
Vκ L6 Fab family: L chainsa
| Fab | Sequence of L chain
|
Jκ | VH | ||
|---|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | |||
| Vκ L6 | RASQSVSSYLA | DASNRAT | QQRSNWP | ||
| 014.7A2 | K------RS-- | ------- | -H-N--- PG AT | Jκ2 | VH3-30 |
| 023.102 | ------TN--- | G------ | ---D--- PD AT | Jκ1 | VH3-30 |
| 023.125 | ------G-Q-- | ------- | ---K--- PD GT | Jκ1 | VH3-30 |
| 025.1H10 | -------R--- | ------- | -H-G--- PG AT | Jκ3 | VH3-30 |
| 025.5D3 | -------R--- | ------- | -H----- PG AT | Jκ3 | VH3-30 |
| 002.3B11 | ------G-F-- | ------- | ------- P LT | Jκ4 | VH3-64 |
| 002.7F3 | ----------- | ------- | ------- P VT | Jκ4 | VH3-64 |
| 002.P8E11 | ----------- | ------- | ------- P VA | Jκ4 | VH3-64 |
| 014.6G9 | -------I--- | ------P | ---L--- P LT | Jκ4 | VH3-23 |
| 008.2F7 | ---E---T--- | ---H--- | ---TD-G | Jκ4 | VH3-48 |
| 008.3A2 | ---E---TF-- | ---D--- | ---T--G | Jκ4 | VH3-48 |
| 008.4A3 | ---E---T--- | ---D--- | ---T--G | Jκ4 | VH3-48 |
| 008.4D1 | -----IN---- | ---Y--- | ------G | Jκ4 | VH3-48 |
CDR residues and gene usage of the L6 family of PPS 23F-specific Fabs. Germ line positions where identical substitutions were isolated from different individuals are shown in boldface. Grouping is by L-chain CDR3 length. Fabs isolated from donor 008 have 7-amino-acid L-chain CDR3 region resulting from the deletion of the first two residues of Jκ4. Donors 002, 008, and 014 received the conjugate vaccine. Donors 023 and 025 received the 23-valent PPS vaccine.
TABLE 6.
Vκ L6 Fab family: H chainsa
| Fab | Sequence of H chain
|
JH | Isotype | ||
|---|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | |||
| VH3-30 | SYGMH | VISYDGSNKYYADSVKG | |||
| 014.7A2 | -F-V- | -V-AG-TTT-------D | EGSGSKWCFDY | JH4 | IgG2 |
| 023.102 | R---- | -V-S--RTT-------- | EGGDNKFSFDY | JH4 | IgG2 |
| 025.1H10 | N---- | -V-SH-NT--------- | EGHGSDMTWRADFDY | JH4 | IgG2 |
| 025.5D3 | N---- | -LGA--TTT-------- | EGHASGMTWRADFDY | JH4 | IgA1 |
| 025.7A6 | H---- | -V-SG-ET--------- | EGHSSGMTWRADFDY | JH4 | IgA2 |
| VH3-64 | SYAMH | AISSNGGSTYYADSVKG | |||
| 002.7F3 | G-S-- | --N---D-----V---- | DIYDVSSAYYGMDV | JH6 | IgG2 |
| VH3-48 | SYSMN | YISSSSSTIYYADSVKG | |||
| 008.3A2 | T--A- | ---GD-G---------- | SK QQVSGGFYWYFDL | JH2 | IgA2 |
| 008.4A3 | -N-V- | ----T-DI--------- | SRGQQSPLGFYWYFDL | JH2 | IgG2 |
| 008.4D1 | -N-V- | ---NTG-I----N---- | SKQQQSTAGFYWYFDL | JH2 | IgG2 |
| 008.2F7 | TN-V- | ----T-TI--------- | SKAQQATAGFYWYFDL | JH2 | IgG2 |
| VH3-23 | SYAMS | AISGSGGSTYYADSVKG | |||
| 014.6G9 | N---- | SVF--ADN-H------- | VLSPGGSNMVWDY | JH4 | IgA1 |
| 014.2B6 | N---- | SVF--ADN-H------- | VLSPGGSNMVWDY | JH4 | IgG2 |
See Table 5, footnote a.
H-chain usage in the Vκ L6 family is more diverse than in the Vκ A23 Fabs (Tables 5 and 6). There is a conservation of VH and JH gene usage, however, in Vκ L6 Fabs of a given CDR3 length. All Fabs (from three different donors) that utilized a Vκ L6 L chain with an 11-amino-acid CDR3 used VH3-30 and JH4 to generate the paratope. All but 1 of the 10-amino-acid CDR3 Vκ L6 Fabs utilized VH3-64 and JH6, and all of the 7-amino-acid CDR3 Fabs used VH3-48 and JH2. All VH chains were mutated in the CDRs, and there were identical substitutions at several residues in H-chain CDR2 in the VH3-30 Fabs (Ile51 to Val51, Asn56 to Thr56, and Lys57 to Thr57, for example). The VH3-30 locus is highly polymorphic, however (13), and although none of the shared substitutions appears to correspond to any of the known alleles, the possibility of an undescribed allele cannot be excluded.
Postvaccination sera obtained from donor 023 appeared to contain a single predominant clonotype when analyzed by isoelectric focusing (data not shown). Affinity-purified antibody from this donor was reduced and separated into H and L chains, and the separated chains were submitted for N-terminal protein sequence determination. Data was obtained for residues 1 to 21 of the light chain. The determined sequence (EIVLTQSPATLSLSPGEXATL) was 100% identical to Vκ L6, Vκ L20, and Vκ A11 and therefore consistent with the predominant L chain isolated from this individual by combinatorial cloning. Our attempts to obtain sequence data for the H chain were not successful.
In addition to the Vκ A23 and Vκ L6 Fabs isolated from all donors, a Fab was isolated that used a Vκ L5 L chain paired with VH4-4, and one that used a Vκ A2 L chain paired with VH4-59 (Tables 7 and 8). Both were mutated in the CDRs of both H and L chains. The H chain of Fab 008.4B7 (Vκ L5/VH4-4) had a two-residue insertion into CDR1. The A2 L chain of Fab 023.115 is noteworthy in that the Lys30-to-Arg30 and Tyr34-to-His34 substitutions in CDR1, as well as the deletion of Pro95 in CDR3, are shared with our previously reported A2-utilizing PPS 23F specific Fab (17).
TABLE 7.
Non-A23/L6 Fabs: L chainsa
| Fab | Sequence of L chain
|
Jκ | ||
|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | ||
| Vκ L5 | RASQGISSWLA | AASSLQS | QQANSFP | |
| 008.4B7 | -----V-Q--G | DVF--HG | --TK--- HT | Jκ2 |
| Vκ A2 | KSSQSLLHSDGKTYLY | EVSNRFS | MQSIQLP | |
| 023.115 | R----------R--FH | ------- | ---M-Y RT | Jκ1 |
H- and L-chain CDRs and isotypes of the two non-A23/L6 Fabs isolated in this study. Fab 008.4B7 has a two-residue insertion in H-chain CDR1; a space was inserted at the appropriate location of the germ line sequence for purposes of alignment (as indicated by asterisks, see Table 8). Donor 008 received the conjugate vaccine. Donor 023 received the 23-valent PPS vaccine.
TABLE 8.
Non-A23/L6 Fabs: H chainsa
| Fab | Sequence of H chain
|
JH | Isotype | ||
|---|---|---|---|---|---|
| CDR1 | CDR2 | CDR3 | |||
| VH4-4 | S**YYWS | RIYTSGSTNYNPSLKS | |||
| 008.4B7 | NSNF--- | H--A--T--------- | GAYASGNYYPPGE | JH5 | IgG2 |
| VH4-59 | SYYWS | YIYYSGSTNYNPSLKS | |||
| 023.115 | DHS-- | -FHF--T--------- | GSAMMGS | JH5 | IgA2 |
See Table 7, footnote a.
The H-chain isotypes of the isolated Fabs were IgG2 and IgA (with one exception) reflecting what is usually found in the serum (Tables 3 to 6). There were several examples of different isotypes associated with the same H-chain V region sequence. Fabs 027.304 (IgA1) and 027.343 (IgA2) are similar in their CDRs (Tables 3 and 4) and clearly share the same postrearrangement somatic maturational history. An IgG2 Fab from this same individual (027.064) shares some substitutions but has an insertion in CDR2 and may have arisen from a separate rearrangement event. The three VH3-30 isolates from donor 025 differ in isotype (Tables 5 and 6) but share significant homology in CDR3, as well as some substitutions in CDR2, and probably arose from the same initial rearrangement. The two VH3-23 H chains paired with L6 from donor 014 (IgA1 and IgG2) are identical in their CDRs and undoubtedly arose from the same rearrangement event.
DISCUSSION
The molecular and genetic diversity inherent in an antibody response to a particular antigen is difficult to study in detail. Mouse models are unsuitable for such analysis since members of genetically identical inbred strains are not expected to produce the variability observed in outbred populations. Repertoire analysis is intrinsically difficult in humans. Stable antigen-specific human hybridomas are difficult to establish, and in only a few cases has it been possible to generate hybridomas specific for a single antigen from multiple donors (1, 4). Protein sequencing of purified serum antibodies has provided repertoire information for some specificities (22, 23) but is technically challenging, labor-intensive, and expensive when applied at a population level. Idiotypic analysis can be applied to large numbers of donors (14) but gives more limited information and is severely compromised by the necessity of generating and characterizing specific reagents for each response being studied.
Reverse transcription-PCR is a convenient and standardized methodology for deriving sequence information concerning expressed immunoglobulin genes. The two relevant approaches for repertoire analysis are to either amplify and express the products of single antigen-specific cells or to construct combinatorial expression libraries from donor-derived cells that are then screened for the production of specific Fabs. Single-cell PCR (8) retains native pairing, but the necessity of identifying antigen-specific cells and the multitude of manipulations that must be carried out on each cell, a small fraction of which are antigen specific even in enriched populations, together with the necessity of protein expression and specificity determinations for each isolated Fab, make it unsuitable for the analysis of repertoires where, by definition, the response of multiple individuals must be examined. The combinatorial approach we have taken in cloning and screening expression libraries is more adaptable to repertoire analysis but suffers from the fact that native H- and L-chain pairing is lost. It is not the case, however, that de novo specific paratopes are created by this methodology. In the few cases in which immunoglobulin protein sequence is available from purified serum antibody, gene usage inferred from protein sequence corresponds to that identified by combinatorial Fabs specific for the same antigen. Protein sequence obtained from purified Haemophilus influenzae type B capsular polysaccharide (Hib PS) antibodies (22) identify the same canonical H and L chains as are obtained from combinatorial cloning (19). In the current study, amino-terminal L-chain sequence (21 residues) obtained from purified donor 023 PPS 23F-specific antibody was identical to Vκ L6, Vκ L20 (the distal homolog of L6), and Vκ A11, a closely related Vκ III V gene. Most Fabs isolated from this donor were members of the Vκ L6 family. The relatively high frequency of Fabs in these libraries, as well as the isolation of intraclonal variants of the same H and L chains, also suggests that these chains are representative of those involved in forming specific paratopes in vivo. In addition to the polysaccharide antigens cited above, others (7) have shown that influenza virus hemagglutinin-specific Fabs isolated by combinatorial cloning recapitulate naturally occurring H- and L-chain pairs. Fabs isolated by cloning procedures also reflect serum antibodies of the donor in both H- and L-chain isotype (17, 19; the present study). Together, these findings strongly suggest that Fabs obtained by combinatorial cloning are representative of the paratopes actually used in the response. The information that is lost by using a combinatorial approach relates primarily to the effect individual mutations have on binding affinities by specific H- and L-chain pairs. Although laborious, this information can be inferred from H×L recombination or “back crossing” experiments where all relevant chain pairs from a given individual are recreated. Although the somatic history of the individual chain pairs is lost, combinatorial cloning allows the investigator to determine the repertoire of H- and L-chain gene products which combine to form antigen-specific paratopes. When library complexity is limited by antigen-specific cell selection prior to mRNA extraction, combinatorial cloning provides a powerful methodology for repertoire analysis in humans.
Our analysis of the PPS 23F-specific repertoire has revealed several points. We have demonstrated that the 23F-specific repertoire utilized by the individual is limited. Only one of the donors we examined (008) used more than two paratope families in response to vaccination. This finding concurs with serological analysis that has shown the 23F specific-response to be oligoclonal (15), with at most two or three clonotypes accounting for the majority of the antibody in serum. Oligoclonality is believed to be a characteristic of antibody responses to polysaccharide antigens in general and has been demonstrated serologically for several other polysaccharide antigens including Hib PS (11), PPS 6B, and PPS 14 (15). A more surprising finding is our determination that most individuals use the same H- and L-chain pairings in forming PPS 23F-specific antibodies. These results do not necessarily follow from oligoclonality itself and, given the combinatorial diversity available to the immune system, it is quite remarkable that such a limited diversity is seen between individuals. A similar recurrence in gene usage is seen in the response to Hib PS, where most individuals utilize an A2 L chain paired with a VH3-23 H chain to form Hib PS-specific Fabs (16). It has been suggested that the high-molecular-weight capsular polysaccharides, which are polymers of smaller repeating subunits, lack epitope diversity and that this results in the observed oligoclonality of the response. Our findings suggest not only a paucity of antigenic epitopes associated with the PPS 23F polysaccharide but also that, for any given epitope, only a few members of the germ line repertoire are normally used to form the corresponding paratope.
The genetic elements utilized by the Fabs isolated in this study have clearly undergone extensive somatic modification. These mutations are preferentially found in the CDR loops and, together with the observation that identical substitutions occur in different individuals, strongly suggest an antigen-driven mechanism of somatic mutation. Studies of type 2 TI antigen responses in mice (5) have also determined that identical substitutions arise in independently derived clones and are directly linked to increases in affinity. Such “affinity maturation” is thought to be T cell dependent. Somatically modified antibodies have been reported for both Hib PS and other PPS antigens (see below), and our findings suggest a role for T cells in the response to the 23F polysaccharide as well. The presence at 7 days of such extensively mutated paratopes implies that our findings after vaccination reflects a recall response generated from preexisting memory B cells. We found no significant correlation between the degree of mutation and the T-cell-dependent or T-cell-independent form of the vaccine. A possible explanation for our findings is that these individuals had previously encountered the antigen through natural exposure and that the 23F polysaccharide was originally encountered in a T-cell-dependent form, perhaps as consequence of being associated with the intact bacteria.
We have previously reported the isolation of two PPS 23F-specific Fabs from a single donor immunized as part of a separate study (17). One of these (23F.2) utilized an Vκ A2 L chain paired with a VH3-11 H chain to form the paratope. As mentioned above, the 23F.2 L chain shares two substitutions in CDR1, as well as the shortened CDR3, with the Vκ A2 isolate in this study. The Vκ A23/VH3-23 Fab previously reported (23F.1) is also highly homologous to the members of that family presented here. The 23F.1 L chain shares a substitution in CDR1 and all three substitutions in CDR2 with 027.242 L chain reported in this study. The 23F.1 H chain and the 027.064 H chain in this study shares two substitutions in CDR1, as well as two substitutions and the insertion of an Asp residue between Ser52 and Gly53 in CDR2. In all cases sufficient differences exist between the isolates to rule out laboratory cross-contamination. The only other reported human antibody specific for PPS 23F utilized a VH3-48 H chain that shares two CDR2 substitutions with the 008.2F7 H-chain sequence we report here (4). No L-chain sequence for this hybridoma was reported. The isolation of the same gene products in different studies and from different laboratories and with similar substitutions supports our conclusion that this antibody response arises from a limited V gene repertoire and is subjected to antigen-driven somatic modification.
There is a substantial body of evidence suggesting that antibody responses to capsular polysaccharides undergo the same maturational processes as do responses directed toward protein determinants. In humans, non-Vκ A2 Hib PS-specific antibodies are mutated (10, 21, 22), and even the “canonical” Vκ A2 antibodies often undergo some degree of somatic modification. PPS-specific paratopes isolated either by combinatorial cloning (17) or traditional hybridoma technology (4) also show evidence of somatic mutation. Partial protein sequence obtained from purified PPS 6B antibodies show differences from the most likely germ line V genes, and the reported V gene sequences of a PPS 6B-specific human hybridoma are mutated in the V region of both the L and H chain (23). In murine models, PS antigens induce germinal center formation (24), affinity maturation, and class switch (5). These maturational events are generally thought to be T cell dependent. Although PPS antigens do not undergo intracellular processing and major histocompatibility complex-associated display as normally required for T-cell activation, the evidence supports a substantial role for T cells (or possibly NK cells) acting in a regulatory manner. Taken together, these findings suggest that the inability to respond to “T-cell-independent” polysaccharide antigens early in life may actually result from a maturational delay in the T-cell compartment of the immune system.
Our isolation of Fabs whose H chains are the same IgG2/IgA isotypes as PPS 23F-specific serum antibody supports our conclusion that these combinatorial Fabs are representative of those actually used in the response. Interestingly, our findings suggest both that PPS-specific secretory IgA and serum IgG derive from the same original somatically matured B cell and that class switching can occur independent of concomitant somatic mutation. The two VH3-23 H chains paired with L6 from donor 014 (IgA1 and IgG2) are identical in their CDRs, share several silent mutations, and undoubtedly arose from the same rearrangement event. The VH regions of Fabs 027.304 (IgA1) and 027.343 (IgA2) are both significantly mutated compared to the germ line but highly homologous to each other and clearly share the same postrearrangement somatic maturational history. Our data do not allow us to determine whether somatic mutation occurred prior to the switch from IgM to IgG or IgA, nor can we rule out further mutational events affecting only the L chain. It does appear, however, that little further somatic modification of the VH region takes place between the time of isotype differentiation and our sampling point at 7 days postvaccination.
Our determination of gene family usage may have been biased by the selection criteria we employed in selecting donors for library construction and by certain aspects of the cloning methodology. We preferentially selected donors whose MNC produced PPS 23F-specific antibody in vitro and who demonstrated a significant rise in serum antibody titer after vaccination. This may have introduced a bias toward individuals utilizing higher-affinity paratopes that would be reflected in restricted gene usage in the isolated Fabs. We have also restricted our analysis to kappa L chain Fabs. This was based on our observation that the 23F-specific response is highly skewed toward antibodies utilizing this L chain (15). There are, however, individuals that have lambda antibodies in their serum, and we are currently assessing the contribution of this L-chain isotype to the 23F-specific repertoire in those individuals. Also, the cloning methodology we employ utilizes restriction enzymes whose recognition sequences, although very rare in human germ line V genes, may arise as a result of somatic mutation, leading to the elimination of these gene products from a given library. This usually can be detected during library construction and a solution engineered, but the loss of these sequences from analysis cannot be entirely ruled out. It should also be pointed out that our analysis was restricted to specific B cells circulating 7 days after vaccination. B cells present in other compartments (bone marrow or spleen) or that circulate at other time points might utilize other gene products to make PPS 23F-specific antibodies.
Our findings represent one of the most extensive analyses, at the molecular level, of a specific antibody response in humans. The somatic history of the response to the “T-cell-independent” PPS 23F capsular polysaccharide provides compelling evidence of T-cell participation in the maturation of the adult repertoire. Given the combinatorial diversity available to the immune system, it is remarkable to find not only the same H- and L-chain pairing in different individuals, but the sharing of postrearrangement somatic modification as well. The two L-chain defined paratope families we describe suggest the existence of two distinct antigenic determinants associated with the polysaccharide. If so, this would imply that for each antigenic epitope a limited number of L- and H-chain genes can pair to form the specific paratope.
Acknowledgments
This work was supported by Public Health Service grants AI47136, AI25008, and AI045250 from the National Institute of Allergy and Infectious Diseases.
We thank Adam P. O'Connor and Mistique C. Felton for technical assistance and Betty M. Ho for critically reading the manuscript.
Editor: D. L. Burns
REFERENCES
- 1.Adderson, E. E., J. M. Johnston, P. G. Shackelford, and W. L. Carroll. 1992. Development of the human antibody repertoire. Pediatr. Res. 32:257-263. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbas, C. F., A. S. Kang, R. A. Lerner, and S. J. Benkovic. 1991. Assembly of combinatorial antibody libraries on phage surfaces: the gene III site. Proc. Natl. Acad. Sci. USA 88:7978-7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baxendale, H. E., Z. Davis, H. N. White, M. B. Spellerberg, F. K. Stevenson, and D. Goldblatt. 2000. Immunogenetic analysis of the immune response to pneumococcal polysaccharide. Eur. J. Immunol. 30:1214-1223. [DOI] [PubMed] [Google Scholar]
- 5.Boswell, C. M., and K. E. Stein. 1996. Avidity maturation, repertoire shift, and strain differences in antibodies to bacterial levan, a type 2 thymus-independent polysaccharide antigen. J. Immunol. 157:1996-2005. [PubMed] [Google Scholar]
- 6.Brezinschek, H. P., S. J. Foster, R. I. Brezinschek, T. Dorner, R. Domiati-Saad, and P. E. Lipsky. 1997. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5+/IgM+ and CD5-IgM+ B cells. J. Clin. Investig. 99:2488-2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caton, A. J., and H. Koprowski. 1990. Influenza virus hemagglutinin-specific antibodies isolated from a combinatorial expression library are closely related to the immune response of the donor. Proc. Natl. Acad. Sci. USA 87:6450-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coronella, J. A., P. Telleman, T. D. Truong, F. Ylera, and R. P. Junghans. 2000. Amplification of IgG VH and VL (Fab) from single human plasma cells and B cells. Nucleic Acids Res. 28:E85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foster, S. J., H. P. Brezinschek, R. I. Brezinschek, and P. E. Lipsky. 1997. Molecular mechanisms and selective influences that shape the kappa gene repertoire of IgM+ B cells. J. Clin. Investig. 199:1614-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hougs, L., L. Juul, H. J. Ditzel, C. Heilmann, A. Svejgaard, and T. Barington. 1999. The first dose of a Haemophilus influenzae type b conjugate vaccine reactivates memory B cells: evidence for extensive clonal selection, intraclonal affinity maturation, and multiple isotype switches to IgA2. J. Immunol. 162:224-237. [PubMed] [Google Scholar]
- 11.Insel, R. A., A. Kittelberger, and P. Anderson. 1985. Isoelectric focusing of human antibody to the Haemophilus influenzae b capsular polysaccharide: restricted and identical spectrotypes in adults. J. Immunol. 135:2810-2816. [PubMed] [Google Scholar]
- 12.Kabat, E., T. Wu, H. Perry, K. Gottesman, and C. Foeller. 1991. Sequence of proteins of immunological interest, 5 ed. U.S. Department of Health and Human Services, National Institutes of Health, Bethesda, Md.
- 13.Lefranc, M. P., V. Giudicelli, C. Ginestoux, J. Bodmer, W. Muller, R. Bontrop, M. Lemaitre, A. Malik, V. Barbie, and D. Chaume. 1999. IMGT, the international ImMunoGeneTics database. Nucleic Acids Res. 27:209-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lucas, A. H., R. J. Langley, D. M. Granoff, M. H. Nahm, M. Y. Kitamura, and M. G. Scott. 1991. An idiotypic marker associated with a germ-line encoded kappa light chain variable region that predominates the vaccine-induced human antibody response to the Haemophilus influenzae b polysaccharide. J. Clin. Investig. 88:1811-1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lucas, A. H., D. M. Granoff, R. E. Mandrell, C. C. Connolly, A. S. Shan, and D. C. Powers. 1997. Oligoclonality of serum immunoglobulin G antibody responses to Streptococcus pneumoniae capsular polysaccharide serotypes 6B, 14, and 23F. Infect. Immun. 65:5103-5109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lucas, A. H., and D. C. Reason. 1999. Polysaccharide vaccines as probes of antibody repertoires in man. Immunol. Rev. 171:89-104. [DOI] [PubMed] [Google Scholar]
- 17.Lucas, A. H., K. D. Moulton, V. R. Tang, and D. C. Reason. 2001. Combinatorial library cloning of human antibodies to Streptococcus pneumoniae capsular polysaccharides: variable region primary structures and evidence for somatic mutation of Fab fragments specific for capsular serotypes 6B, 14, and 23F. Infect. Immun. 69:853-8564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuda, F., K. Ishii, P. Bourvagnet, K. Kuma, H. Hayashida, T. Miyata, and T. Honjo. 1998. The complete nucleotide sequence of the human immunoglobulin heavy chain variable region locus. J. Exp. Med. 188:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reason, D. C., T. C. Wagner, and A. H. Lucas. 1997. Human Fab fragments specific for the Haemophilus influenzae b polysaccharide isolated from a bacteriophage combinatorial library use variable region gene combinations and express an idiotype that mirrors in vivo expression. Infect. Immun. 65:261-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schable, K. F., and H. G. Zachau. 1993. The variable genes of the human immunoglobulin kappa locus. Biol. Chem. Hoppe-Seyler 374:1001-1022. [PubMed] [Google Scholar]
- 21.Scott, M. G., D. L. Crimmins, D. W. McCourt, G. Chung, K. F. Schable, R. Thiebe, E. M. Quenzel, H. G. Zachau, and M. H. Nahm. 1991. Clonal characterization of the human IgG antibody repertoire to Haemophilus influenzae type b polysaccharide. IV. The less frequently expressed VL are heterogeneous. J. Immunol. 147:4007-4013. [PubMed] [Google Scholar]
- 22.Scott, M. G., H. G. Zachau, and M. H. Nahm. 1992. The human antibody V region repertoire to the type B capsular polysaccharide of Haemophilus influenzae. Int. Rev. Immunol. 9:45-55. [DOI] [PubMed] [Google Scholar]
- 23.Sun, Y., M. K. Park, J. Kim, B. Diamond, A. Solomon, and M. H. Nahm. 1999. Repertoire of human antibodies against the polysaccharide capsule of Streptococcus pneumoniae serotype 6B. Infect. Immun. 67:1172-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sverremark, E., and C. Fernandez. 1998. Role of T cells and germinal center formation in the generation of immune responses to the thymus-independent carbohydrate dextran B512. J. Immunol. 161:4646-4651. [PubMed] [Google Scholar]