Abstract
Neisseria meningitidis strains belonging to the hypervirulent lineage ET-37 and several unrelated strains are extremely UV sensitive. The phenotype is consequent to the presence of a nonfunctional recBET-37 allele carrying multiple missense mutations. Phenotypic analysis has been performed with congenic meningococcal strains harboring either the wild-type recB allele or the recBET-37 allele. Congenic recBET-37 meningococci, in addition to being sensitive to UV, were defective both in repair of DNA lesions induced by UV treatment and, partially, in recombination-mediated transformation. Consistently, the wild-type, but not the recBET-37, allele was able to complement the Escherichia coli recB21 mutation to UV resistance and proficiency in recombination. recBET-37 meningococci did not exhibit higher frequencies of spontaneous mutation to rifampin resistance than recB-proficient strains. However, mutation rates were enhanced following UV treatment, a phenomenon not observed in the recB-proficient counterpart. Interestingly, the results of PCR-based assays demonstrated that the presence of the recBET-37 allele considerably increased the frequency of recombination at the pilin loci. The main conclusion that can be drawn is that the presence of the defective recBET-37 allele in N. meningitidis isolates causes an increase in genetic diversity, due to an ineffective RecBCD-dependent DNA repair and recombination pathway, and an increase in pilin antigenic variation.
Neisseria meningitidis, a microorganism naturally competent for transformation, is a paradigm of genetic variation. A continuous horizontal flow of genetic material affects the population's genetic structure, which is characterized by an occasional outgrowth of clones responsible for epidemics and outbreak of disease in an otherwise panmictic contest (19). There is also ample circumstantial evidence for horizontal exchange of genes between meningococci and nonpathogenic Neisseria species (1, 19). In addition to being able to transform naturally, meningococci have evolved sophisticated mechanisms for optimizing adaptation to unpredictable challenges in a new environment: antigenic and phase variation.
Phase variation randomly alters the expression of over 30 surface-associated components, including capsule, pilus, and surface sugar biosynthesis proteins involved in lipooligosaccharide metabolism and outer membrane proteins acting as porins, adhesins, or invasins or involved in iron uptake (1, 19, 30), through slippage-like mechanisms (3, 20, 26, 27). This mechanism ensures that at least a fraction of a bacterial population expresses a phenotype that is compatible with a productive infection cycle.
Antigenic variation allows (i) the optimization of adhesion and invasion processes through the fine-tuning of intermolecular interactions with available host receptors and, at a later stage, (ii) escape from the immune response (27). In the case of the pilin genes, variation is caused by intragenic recombination involving the transfer of variable sequences (minicassettes) from a repertoire of nonexpressed or silent loci (pilS) to the expression locus (pilE). Such rearrangements appear to be nonreciprocal and were originally interpreted as gene conversion events. The phenomenon of pilin antigenic variation in Neisseria gonorrhoeae has been studied extensively. It occurs at a high frequency of >10−2 pilE variants per total number of pilE genes (32), and it involves both homologous recombination activities and specific factors. The Sma/Cla DNA repeat, located at the 3′ ends of all pilin loci, is required for efficient variation (37) and is the target for multiple site-specific binding proteins (37, 38). The RecA and the RecF-like pathways of recombination are essential for pilin antigenic (and phase) variation (10, 18). In contrast, the RecBCD pathway is involved in DNA transformation and DNA repair but is not required for pilin antigenic variation (18). However, the frequency of pilin phenotypic variation was considerably higher in a recD mutant than in a congenic RecD-proficient strain (5).
In this study we demonstrate that N. meningitidis strains belonging to the hypervirulent lineage ET-37 and several unrelated strains are extremely UV sensitive (UVS). This phenotype is associated with a biochemical lesion at the level of the RecBCD (exonuclease V [Exo V]) pathway as a consequence of the presence of a nonfunctional recBET-37 allele that has suffered several missense mutations. In Escherichia coli, RecBCD is the primary pathway for DNA recombination and repair (11). The products of genes recB and recC, essential for homologous recombination following conjugation or transduction, are responsible for the repair of chromosomal double-strand breaks. The RecBCD enzyme is a potent exonuclease that inhibits recombination by destroying linear DNA. The RecD ejection model predicts that the RecBCD enzyme enters double-strand DNA at one end and travels destructively along the DNA until it encounters the recombination hot spot sequence chi. chi weakens the affinity of RecD to the complex, and the resulting RecBC heterodimer is deficient for exonuclease activity and proficient as a recombinagenic helicase (21, 33).
To investigate the role of the RecBCD pathway in genetic variation in meningococci and the biological relevance of the biochemical lesion of the RecBCD pathway, phenotypic analysis of congenic strains harboring either the wild-type recB or the recBET-37 allele was performed.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Meningococcal strains used in this study are listed in Table 1. Invasive strains were derived from a collection of strains isolated during outbreaks of epidemic disease which have occurred in different places in Italy during the last 20 years. The strains' serogroups, serotypes, and assigned hypervirulent lineages are shown in Table 1. A total of 24 commensal strains were included in this study. These strains were sampled from the nasopharynxes of different healthy subjects at the times of their military enlistment in the course of a routine screening program for surveillance of meningococcal disease. The commensal strains used in this study were isolated in different geographical areas in Italy and France. Two strains, BL9513 and BF9513, were isolated, respectively, from the cerebrospinal fluid and the nasopharynx of a single sick subject in France.
TABLE 1.
Typing and UV sensitivities of clinical isolates of N. meningitidis
| Strain | Serogroup and/or serotypea | Lineageb | Clinical specimen(s)c | UV sensitivityd | Sourcee |
|---|---|---|---|---|---|
| BF2 | B | ET-37 | NP | S | i |
| BF3 | B | ET-37 | NP | S | i |
| BF9 | B | Other | NP | R | i |
| BF13 | B | Other | NP | R | i |
| BL847 | B:14:P1.12 | Lineage 3 | CSF | R | ii |
| BL851 | B:4:P1.13 | Lineage 3 | CSF | R | ii |
| BL857 | B:21:P1.7,16 | Other | CSF | R | ii |
| BL858 | B:15,21:P1.15 | Other | CSF | R | ii |
| BL859 | B:4:P1.13 | Lineage 3 | CSF | R | ii |
| BS843 | B:4:P1.13 | Lineage 3 | BL | R | ii |
| BS845 | B:4:P1.13 | Lineage 3 | BL | R | ii |
| BS849 | B:4:P1.13 | Lineage 3 | BL | R | ii |
| BL855 | B | ET-37 | CSF | S | ii |
| B1940 | B:NT:P1.3,6,15 | Other | CSF | R | iii |
| BL9513 | B:4:P1.4 | Lineage 3 | CSF and BL | R | iv |
| BL892 | B:4:P1.4 | Lineage 3 | CSF | R | iv |
| BL942 | B:1:NST | Lineage 3 | CSF | R | iv |
| BL911 | B:NT:P1.9 | Other | CSF | R | iv |
| BL915 | B:NT:P1.5 | Other | CSF | S | iv |
| BL951 | B:NT:P1.1 | Lineage 3 | CSF | R | iv |
| BL899 | B:4:P1.2,5 | ET-5 | CSF | R | iv |
| BL932 | B:4:P1.1 | Other | CSF | R | iv |
| BL937 | B:4:NST | ET-5 | CSF | R | iv |
| BL947 | B:1:NST | Other | CSF | R | iv |
| BL897 | B | Other | CSF | R | iv |
| BF10 | B | Other | NP | S | v |
| BF17 | B | Other | NP | R | v |
| BF18 | B | Other | NP | S | v |
| BF21 | B | Other | NP | S | v |
| BF65 | B | Other | NP | S | v |
| BF16 | B | Other | NP | R | v |
| BF23 | B | ET-5 | NP | R | v |
| BF40 | B | ET-37 | NP | S | v |
| BF52 | B | ET-37 | NP | S | v |
| BF8960 | B | Other | NP | R | vi |
| BF8961 | B | Other | NP | S | vi |
| BF8964 | B | Other | NP | R | vi |
| BF8969 | B | Other | NP | R | vi |
| BF9216 | B | Other | NP | R | vi |
| BF5425 | B | Other | NP | R | vi |
| BF32B | B | Lineage 3 | NP | R | vii |
| BF37B | B | Other | NP | R | vii |
| BF43B | B | Other | NP | R | vii |
| BF6L | B | Other | NP | R | vii |
| BF8L | B | Other | NP | R | vii |
| BF57L | B | Other | NP | R | vii |
| 205900 | A | IV-1 | CSF and BL | R | viii |
| 93/4286 | C | ET-37 | CSF and BL | S | viii |
| NGP165 | B | ET-37 | CSF and BL | S | viii |
| BZ169 | B | ET-5 | CSF and BL | R | viii |
| H44/76 | B | ET-5 | CSF and BL | R | viii |
| MC58 | B | ET-5 | CSF and BL | R | viii |
| NGF26 | B | Other | CSF and BL | R | viii |
| 1000 | B | Other | CSF and BL | R | viii |
| NGE31 | B | Other | CSF and BL | R | viii |
| NGH15 | B | Other | CSF and BL | R | viii |
| ZF15 | Z | Other | NP | R | v |
| XL929 | X | Other | CSF and BL | S | iv |
| XF47A | X | Other | NP | S | v |
| YL896 | Y | Other | CSF | R | iv |
| CF5C | C | Other | NP | R | v |
NT, not serotypeable; NST, not subserotypeable.
The assignment of the isolates to hypervirulent lineages was determined as previously described (28).
NP, nasopharynx swab; CSF, cerebrospinal fluid; BL, blood.
Dilutions of bacteria grown to an OD550 of 0.5 were plated on GC agar plates and irradiated with a UV254 germicidal lamp. The dose was 90 J/m2. Survival fractions of resistant (R) and sensitive (S) strains were, respectively, >10−3 and <10−5.
i, II policlinico, Università di Napoli, Naples, Italy; ii, Istituto Superiore di Sanità, Rome, Italy; iii, Bayerische Julius-Maximilians Universität, Würzburg, Germany; iv, Institut Pasteur, Paris, France; v, Hôpital d'Instruction des Armée, Brest Naval, France; vi, Institut de Médecine Tropicale du Service de Santé dès Armées, Marseille Armées, France; vii, Laboratory of Microbiology, Università di Lecce, Lecce, Italy; viii, Chiron S.p.A, Siena, Italy.
All meningococcal strains were cultured on chocolate agar (Becton Dickinson) or on GC agar or broth (Difco) supplemented with 1% (vol/vol) Polyvitox (Biomerieux) at 37°C in 5% CO2.
E. coli strain DH5α [F−φ80d lacZΔM15 endA1 recA1 hsdR17 supE44 thi-1 λ−gyrA96 Δ(lacZYA-argF) U169] was used in cloning procedures. Strains GY4709 (E. coli K-12 KS468F−lacMS286 φ80 dII lacBK1) and derivative strain GY5873 (recB21), harboring a nontandem duplication of two partially deleted lactose operons, were used in the Lac+ papillation assay (43). The E. coli strains were grown in Luria-Bertani broth or in McConkey plates with 40 g of MacConkey agar base (Difco) plus 1% lactose per liter-50 mg of 2,3,5-triphenyltetrazolium chloride per liter. To allow plasmid selection, Luria-Bertani medium was supplemented with ampicillin (100 μg ml−1) and/or tetracycline (10 μg ml−1).
Transformation of meningococci.
Transformations were performed as previously described (8) by using 1 μg of chromosomal DNA. Transformants were selected on GC agar medium supplemented with rifampin (36 μg ml−1).
DNA procedures.
High-molecular-weight genomic DNAs from the different N. meningitidis strains were prepared as previously described (3). DNA fragments were isolated through acrylamide slab gels and recovered by electroelution as described previously (29). Coding regions of the genes recA, uvrA, uvrB, uvrC, uvrD, rep, and recB were amplified from chromosomal DNA of strain BL859 by using PCR with the following primer pairs: 5′-CCGAATCCTCCGGCAAAACCACCC-3′ and 5′-CCGATCTTCATCCGGATTTGGTTGATG-3′ (recA), 5′-GCTCGTGGTGGTAACAGGATTGTCGGG-3′ and 5′-CAAAAGGCGCAGATAGTCGTGGATTTC-3′ (uvrA), 5′-GAACATATCGAGCAGATGCGCCTTTCC-3′ and 5′-GCTCATTAAATCGTCGACTTGGGTGGC-3′ (uvrB), 5′-GCAAAGTCTTATACGTCGGCAAAGC-3′ and 5′-GGTGTGGCTGATGTCGAAGCATTC-3′ (uvrC), 5′-GTGCTGACCACGCGCATCGCATGGC-3′ and 5′-GTTGGTGTCTTGGAACTCGTCAACGAG-3′ (uvrD), 5′-TGCTCGTCCTTGCCGGTGCAGGCAGCG-3′ and 5′-CGCGGTGGAGCGGTAGTTTTGCTCCAG-3′ (rep), and 5′-CCAAACTTGGAGACGACACGCTTTCAGACG-3′ and 5′-CCTTTTTTCAGTCCGGCTTCGAGTTTGTC-3′ (recB). The lengths of the PCR products were 400 bp (recA), 248 bp (uvrA), 955 bp (uvrB), 1,129 bp (uvrC), 561 bp (uvrD), 755 bp (rep), and 687 bp (recB). The amplification reactions consisted of 30 cycles including 1 min of denaturation at 94°C, 1 min of annealing at 55°C, and 1 to 2 min of extension at 72°C. They were carried out in a PerkinElmer Cetus model 480 DNA thermal cycler. Labeling of the DNA fragments to be used as probes in Southern blot and library screening procedures was performed by random priming with the Klenow fragment of E. coli DNA polymerase I, [α-32P]dATP, and [α-32P]dGTP (3,000 Ci mmol−1) by standard protocols (29)
DNA sequencing reactions were carried out by the dideoxy chain termination procedure using a TAQence cycle sequencing kit from U.S. Biochemicals (distributed by Amersham Life Science) according to the instructions of the manufacturers.
Processing of the DNA sequences was performed with the software GeneJockey Sequence Processor (published and distributed by Biosoft). The CDD (conserved domain database) and CD-Search service at the National Center for Biotechnology Information was used to identify the conserved domains present in protein sequences.
Plasmids and cloning procedures.
The chromosomal DNAs derived from the meningococcal strains MC58 and BF2 were restricted by SalI and used to construct two genomic libraries in SalI-restricted pBR322. The screening was performed by colony hybridization with the recB probe above described and led to isolation of recombinant plasmids, pNMRecBwt and pNMRecBET-37, containing genomic 4,548-bp-long SalI DNA fragments from MC58 and BF2, respectively.
UV survival assays.
Meningococcal cells were grown to late logarithmic phase in GC medium supplemented with Vitox. When the cultures reached an an optical density at 550 nm (OD550) of 1.0, 10-μl serial dilutions were spotted onto petri dishes containing GC agar plus Vitox supplement. Irradiation was achieved by opening the plates under a SpectrolineR model ENF-260C/F 254-nm-wavelength UV (UV254) germicidal lamp (Spectronics Corporation, Westbury, N.Y.) at a distance of 15 cm for different times in the absence of daylight illumination. As a control, dilutions from the same cultures were spotted onto nonirradiated plates. UV fluences were determined with a UV254 light-sensitive photodiode (type S1336-18BQ; Hamamatsu). Exposure to the germicidal lamp never resulted in an increase of temperature of the GC agar medium of more than 0.1°C, as measured with a platinum probe PT 100 (RS Components) that was placed at the surface of the medium.
Determination of spontaneous and UV-induced mutation frequencies.
Meningococcal cells grown to an OD550 of 1.0 were collected by centrifugation and gently resuspended in GC medium to concentrate them. About 2 × 1010 bacteria were plated on selective GC agar medium supplemented with rifampin (36 μg ml−1). To determine the UV-induced mutation frequencies, thin layers of bacterial suspensions (2 ml, about 2 × 1010 cells) were placed in empty petri dishes and exposed to the UV254 germicidal lamp at different fluences, in the absence of daylight illumination, before being plated on selective GC agar medium with rifampin. Serial dilutions were treated in parallel and plated on GC agar to determine the numbers of viable CFU. Frequencies were determined by dividing the number of resistant CFU ml−1 by the total number of viable CFU ml−1.
Marker repair assay.
A rifampin-resistant derivative strain of MC58, MC58Rif, was isolated on selective medium. The chromosomal DNA from this strain was used to transform MC58-1 to rifampin resistance, resulting in MC58-1Rif. MC58Rif and MC58-1Rif were grown to late logarithmic phase (OD550 = 1.0). Then cells were collected by centrifugation and gently resuspended in GC medium to concentrate them. Thin layers of bacterial suspensions were placed into open petri dishes and were irradiated with the UV254 germicidal lamp at different fluences. Irradiated and nonirradiated bacteria were incubated at 37°C for 20 min to allow DNA repair before DNA extraction. One microgram of DNA was used to transform the MC58 recipient strain to rifampin resistance.
Pilin variation assay.
Recombinant pilin genes were detected in DNAs extracted from liquid growing meningococci by a PCR-based assay. The primers 5′-GCCATCGTCGGCATTTTGGCGGCAGTC-3′ (pil1) and 5′-GCGTCGCGGCAGGTTGACGGCAGGTGCTTGG-3′ (pil2) amplified a 444-bp fragment corresponding to part of the coding region of the pilE gene (GenBank accession number NMB0018). They map in the conserved N-terminal peptide region (with primer pil1) and in the conserved Cys2 C-terminal region (with primer pil2) in mature pilin (25). The primers pil1 and pil3 (5′-TGGTTTTCTTGTTGCCGGTGTTGTC-3′) were designed to detect recombinant pilE-pilS7 gene products. The sequence of pil3 was specific to the pilS7 large cassette (accession number NMB0025). The PCRs were carried out using different amounts (1 pg to 100 ng) of genomic DNA as a template under the same conditions described in the “DNA procedures” section. PCR products were detected either directly on ethidium bromide-stained agarose gels or by Southern blot hybridization.
Nucleotide sequence accession number.
The GenBank accession number for the nucleotide sequence of the recB region from BF2 is AF495855.
RESULTS
Effects of UV treatment on the survival of clinical isolates of N. meningitidis.
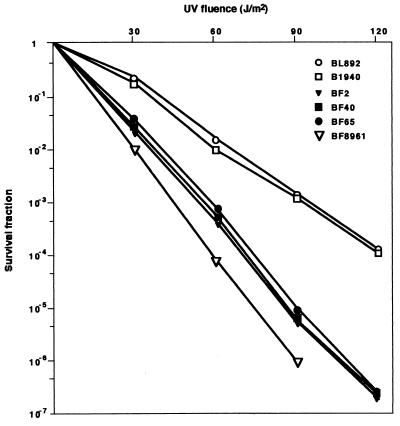
We analyzed the effect of UV254 treatment on the survival of clinical isolates of N. meningitidis. The meningococcal strains were isolated in different regions of Italy and France over the last 10 years from either patients suffering from meningitis or sepsis (invasive strains) or healthy subjects (commensal strains) (Table 1). Genetic relationships among 50 clinical isolates and 10 reference strains (28) are shown in Table 1. Bacteria were irradiated in the dark to prevent photoreactivation repair activity (9). The strains exhibited different UV sensitivities and were arbitrarily classified in two groups, UV-resistant (UVR) and UVS strains. Phylogenetic analysis indicated that all isolates or strains belonging to the ET-37 lineage (7 out of 7) and some strains belonging to other unrelated lineages (8 out of 54) were UVS (Table 1). Examples of results of the analysis of two UVR strains and two unrelated and two ET-37 UVS strains are shown in Fig. 1.
FIG. 1.
UV254 radiation sensitivities of various N. meningitidis strains. Strains BL892, B1940, BF2, BF40, BF65, and BF8961 were grown to late logarithmic phase. Serial dilutions (10 μl) were spotted onto agar plates containing GC agar plus Vitox supplement. Irradiation was achieved by opening the plates under a UV254 germicidal lamp at a distance of 15 cm for different times in the absence of daylight illumination as detailed in Materials and Methods. Survival fractions are reported on the ordinate. UV fluence values are indicated on the abscissa.
Molecular and genetic analyses of UVR and UVS meningococcal isolates.
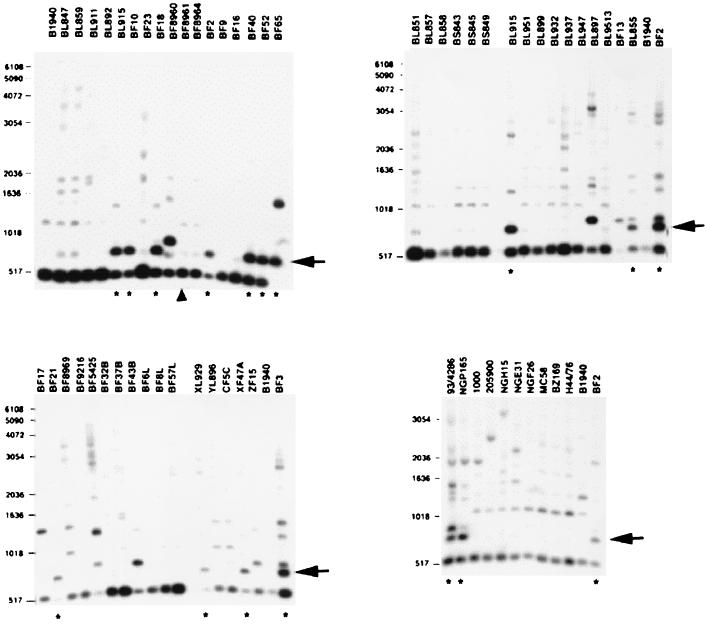
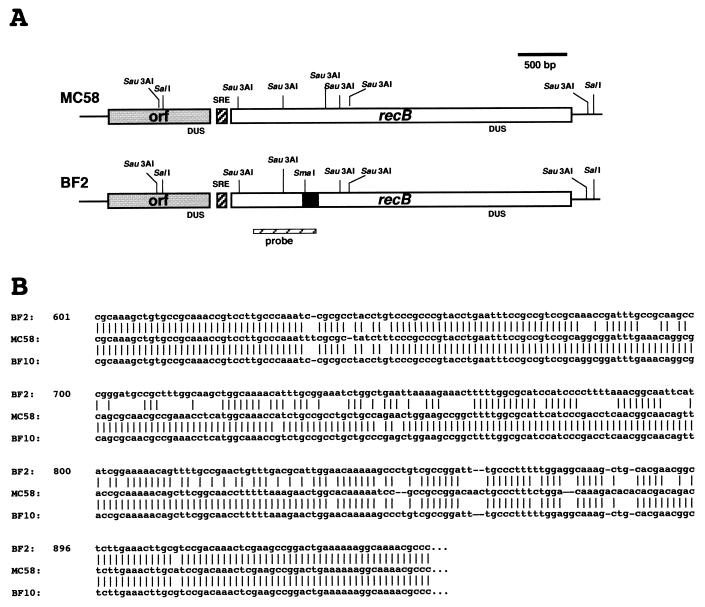
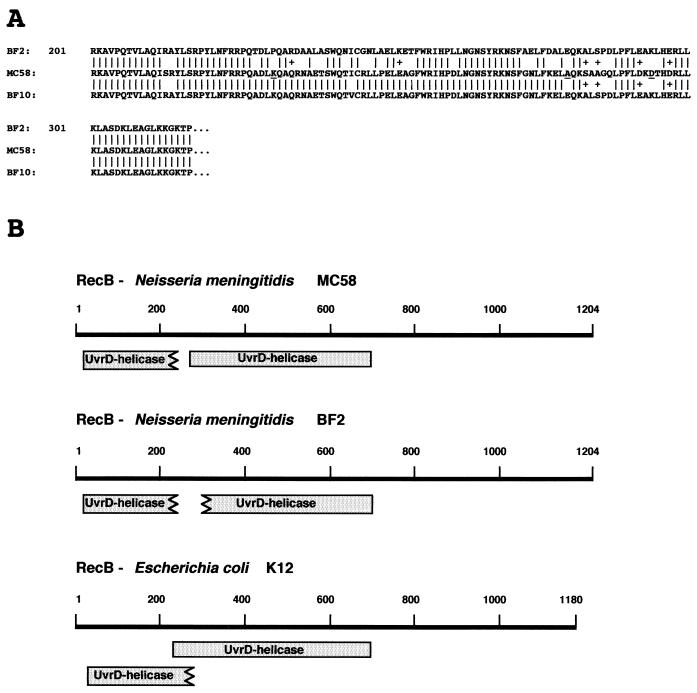
In E. coli, lesions in several genes for DNA recombination and/or repair pathways increase the sensitivity of the cells to UV treatment. With the aim of defining the molecular basis of the UV phenotype in meningococci, we developed a strategy based on DNA analysis in an attempt to identify restriction fragment length polymorphisms (RFLP) in DNA recombination and/or repair genes possibly associated with the UV phenotype. Sau3AI-generated polymorphisms in uvrA, uvrB, uvrC, uvrD, recA, recB, and rep genes were detected by Southern analysis of the genomic DNAs derived from the 50 clinical isolates or from the 10 reference strains. Data for the recB allele are presented in Fig. 2. All UVS strains, but not the UVS BF8961 strain (Fig. 1), shared a common restriction pattern with the recB-specific probe, producing a diagnostic 644-bp-long fragment (Fig. 2). The chromosomal region containing the recB gene was therefore isolated from the UVS ET-37 isolate BF2 and from the UVR ET-5 strain MC58 by screening genomic libraries with a recB-specific probe (see Materials and Methods). Two recombinant plasmids, pNMRecBwt and pNMRecBET-37, containing genomic 4,548-bp-long SalI DNA fragments from MC58 and BF2, respectively, were isolated. Nucleotide sequence analysis of the DNA fragment from the BF2 strain (Fig. 3) revealed that the chromosome of BF2 contains an entire, in-frame recB gene mapping downstream from a conserved hypothetical open reading frame (Fig. 3A). In both the ET-5 strain MC58 (36) and BF2, the predicted recB coding region is 3,615 bp long and the gene is preceded by a “nemis” (neisserial miniature DNA insertion sequence) (17). However, alignment of the nucleotide sequences of the BF2 and MC58 recB genes showed that numerous insertions, deletions, and substitutions have occurred in the coding region spanning nucleotides 637 to 885 (Fig. 3B). The sequences of this region from strains Z2491, belonging to serogroup A (23), and BL859, a lineage 3 isolate, were identical to the MC58 sequence (data not shown). Alignment of the deduced amino acid sequences from BF2 and MC58 evidenced that the recB gene from the ET-37 isolate suffered a number of missense mutations (Fig. 4A). In particular, three mutations affected amino acid residues that are evolutionarily conserved in RecB proteins from different microorganisms. These amino acid substitutions were predicted to destroy the UvrD-like type II helicase domain (Fig. 4B).
FIG. 2.
RFLP analysis in the recB genomic region by Southern blot hybridization. Chromosomal DNAs were extracted from different meningococcal strains (Table 1) and treated with Sau3AI. The digested DNAs were analyzed by Southern blotting using a 32P-labeled recB-specific probe (Fig. 3A) (Materials and Methods). Arrows on the right indicate a restriction fragment common to all UVS strains, which are marked by stars below the panels. The triangle marks the UVS strain BF8961. The relative lengths of migration of DNA molecular markers and their weights are shown on the left.
FIG. 3.
Physical and genetic map and partial nucleotide sequence of the recB gene from BF2, MC58, and BF10. (A) Physical and genetic map of the genomic recB region from strains MC58 and BF2. The black box within the recB gene of BF2 indicates the region that contains many missense mutations. The map of the recB region from MC58 was deduced on the basis of the available sequencing data (36). The homologous region was sequenced from strain BF2 (accession number AF495855). The position of the recB-specific probe used in the Southern blot experiments (Fig. 2) is also indicated below the map. Abbreviations: DUS, DNA uptake sequence; SRE, neisserial small repetitive element; orf, open reading frame. (B) Alignment of partial nucleotide sequences of the recB coding region from strains BF2, MC58, and BF10. The sequences of the recB coding region containing missense mutations in BF2 and BF10 (from nucleotides 601 to 951) are presented.
FIG. 4.
Partial primary-structure and functional domains of the RecB proteins from BF2, MC58, and BF10. (A) Alignment of deduced partial amino acid sequences of the RecB proteins from BF2, MC58, and BF10. Vertical bars indicate identical amino acids; pluses indicate synonymous substitutions. Amino acids that are evolutionarily conserved in the RecB proteins from many bacteria are underlined in the RecB sequences of MC58. (B) Location of UvrD-like type II helicase domains in RecB proteins from N. meningitidis strains MC58 and BF2 and from E. coli K-12. Domains were identified by computer program analysis (CDD at the National Center for Biotechnology Information [http://www.ncbi.nlm.nih.gov:80/Structure/cdd/cdd.shtml]) designed to identify the conserved domains present in a protein sequence (shaded boxes). The lengths of the RecB proteins from MC58 and BF2 are identical (1,204 amino acids); E. coli K-12 RecB has 1,180 residues.
The nucleotide sequences of this part of the recB coding region were determined in the UVS strains BL915, BF10, BF18, BF21, XL929, and XF47A, phylogenetically unrelated to the ET-37 lineage. The analysis demonstrated that the nucleotide sequences in strains BL915, BF18, XL929, and XF47A were identical to that of BF2 (data not shown). Strains BF10 and BF21 shared a common mosaic allele. The nucleotide sequence of the allele was identical to that of BF2 upstream of nucleotide 630, similar to that of MC58 between nucleotides 631 and 846 and again identical to that of BF2 downstream from nucleotide 847 (Fig. 3B and data not shown). As a consequence of these rearrangements, two out of the three conserved amino acid residues were mutated (Fig. 4A).
The UVS strain BF8961 contained a wild-type recB allele, suggesting an alternative mechanism responsible for the UV phenotype. Southern blot analysis of RFLP demonstrated the presence of an allele in the rep locus restricted to this strain (data not shown). Preliminary sequencing data indicated the presence of mutations affecting the primary structure of the Rep protein, encoding a UvrD-like type II helicase involved in DNA replication and, possibly, in DNA recombination and repair (41). Functional characterization of these mutations is under investigation.
The ability of the recB genes derived from MC58 and BF2 to promote recombination in E. coli was tested. The assay measured the ability of the meningococcal recB alleles to complement the genetic defect of the E. coli recB21 mutation. The plasmids pNMRecBwt and pNMRecBET-37 were used to transform strains GY4709 (recB proficient) and GY5873 (recB21) harboring a nontandem duplication of two partially deleted lactose operons. The assay, which measures the ability to form Lac+ recombinants, requires intact recA, recB, and recC gene products (43). The mean numbers of Lac+ papillae were similar in RecB-proficient strains harboring either pNMRecBwt or a control vector (Table 2). Intriguingly, pNMRecBET-37 decreased the recombination frequencies about threefold, possibly as a result of the interference of the defective meningococcal RecBET-37 protein on the activity of endogenous RecB. pNMRecBwt but not pNMRecBET-37 was able to restore recombination proficiency in the E. coli recB21 strain almost to wild-type levels.
TABLE 2.
Effects of expression of meningococcal recBwt or recBET-37 on homologous recombination in E. coli
| Strain (relevant genotype) | Plasmid | Mean no. of Lac+ papillae (10−7 bacteria)a |
|---|---|---|
| GY4709 (recB+) | pBR322 | 272 (13) |
| pNMRecBwt | 348 (37) | |
| pNMRecBET-37 | 127 (26) | |
| GY5873 (recB21) | pBR322 | 33 (5) |
| pNMRecBwt | 221 (18) | |
| pNMRecBET-37 | 45 (7) |
Values are means of results from at least five independent experiments. Standard errors are reported in parentheses.
Phenotypic analysis of the recBET-37 allele.
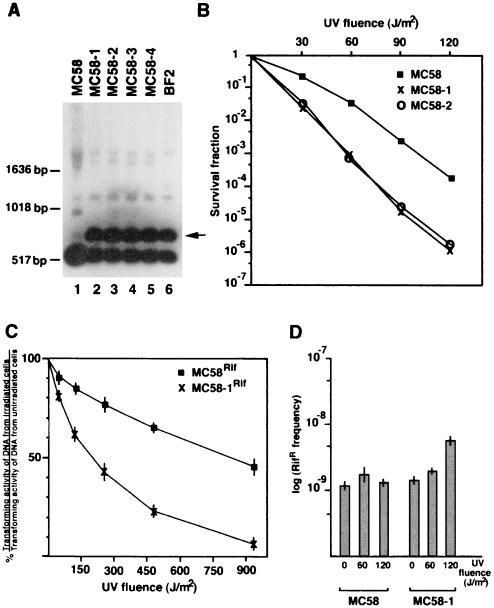
To obtain direct evidence that the mutated recB allele in the ET-37 strains and isolates (recBET-37) was responsible for UV sensitivity, we constructed congenic strains harboring either wild-type recB or the recBET-37 allele by transformation and homologous recombination. The 4,548-bp-long SalI DNA fragment derived from pNMRecBET-37 was used to transform the UVR strain MC58. UVS transformants were isolated with a frequency of about 0.5% and four of them, MC58-1, MC58-2, MC58-3, and MC58-4, were characterized by Southern blotting. In all recombinant strains the recBET-37 allele replaced the functional recB allele (Fig. 5A).
FIG. 5.
Effects of substitution of the recBET-37 allele for the wild-type recB allele in N. meningitidis strain MC58 on UV254 radiation sensitivity, DNA repair of UV-induced damages, and mutability. (A) Southern blot experiment demonstrating allelic replacement. Genomic DNAs were extracted from the UVR strain MC58 (lane 1); from the UVS strains MC58-1, MC58-2, MC58-3, and MC58-4 (transformed with the recBET-37 allele) (lanes 2 to 5, respectively); and from BF2 (source of the recBET-37 allele) (lane 6) and were treated with Sau3AI. The digested DNAs were analyzed by Southern blotting using the 32P-labeled recB-specific probe (Fig. 3A). The arrow on the right indicates the diagnostic restriction fragment of the allelic replacement. The relative lengths of migration of DNA molecular markers and their weights are shown on the left. (B) UV254 radiation sensitivity of meningococcal strains MC58, MC58-1, and MC58-2. (C) Repair of UV-induced damages assayed in a marker repair experiment. Rifampin-resistant strains MC58Rif and MC58-1Rif were grown to late logarithmic phase and irradiated with a UV254 germicidal lamp at different fluences (indicated on the abscissa), as detailed in Materials and Methods. Then the DNA was extracted from irradiated bacteria and nonirradiated controls after a postirradiation period of 20 min at 37°C under nongrowth conditions to allow DNA repair to occur. The DNA was used to transform MC58 to rifampin resistance. Relative transforming activities from irradiated and nonirradiated cells are reported on the ordinate. (D) Spontaneous and UV-induced mutability to rifampin resistance in MC58 and MC58-1. Mutation frequencies were determined in nonirradiated or in UV254-irradiated bacteria at different fluences as described in Materials and Methods. Values are reported as logs of frequencies of mutation to rifampin resistance. Error bars are indicated.
Phenotypes associated with the recBET-37 allele, including UV sensitivity, DNA repair of UV lesions, mutation rates, transformation frequencies, and recombination at the pilin loci, were analyzed (Fig. 5B to D, Fig. 6, and Table 3). The survival fractions after UV treatment at different fluences in MC58-1 and MC58-2 were considerably lower (about 2 orders of magnitude at 120 J/m2) than in the parental recB-proficient strain MC58 (Fig. 5B). The ability to repair lesions in DNA was tested in a “marker repair” experiment. The extent of repair was estimated by determining the recovery of the transforming activity of DNA extracted from UV-treated bacteria after a postirradiation incubation period to allow DNA repair to occur (see Materials and Methods). Figure 5C shows that marker repair activity was dramatically reduced in the presence of the recBET-37 allele. The recBET-37 allele did not affect the spontaneous mutability determined as frequencies of the appearance of rifampin-resistant colonies on selective medium (Fig. 5D). Mutation frequencies were also determined after exposure to UV radiation. In MC58, this treatment did not result in an increase of mutation frequencies to rifampin resistance (Fig. 5D). The absence of UV-induced mutagenesis was consistent with the lack of an SOS-like system in Neisseriaceae (2). However, unexpectedly, the frequencies of resistant clones moderately increased in MC58-1 with increasing UV fluences.
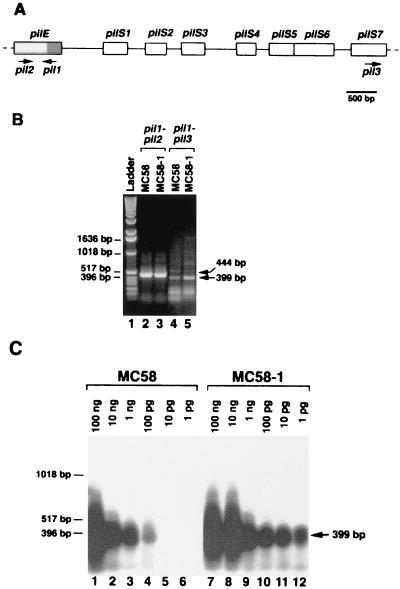
FIG. 6.
Effect of the allelic substitution of recBET-37 on recombination at the pilin loci in MC58. (A) Map of the pilin gene cluster in the chromosome of MC58, as derived from the sequencing data (accession numbers NMB0018 for pilE and NMB0019 to NMB0025 for pilS1 to pilS7). Arrows indicate oligonucleotides used as primers (pil1, pil2, and pil3) in PCR results shown in panel B. The shaded box within pilE represents the region encoding the conserved N-terminal region in mature pilins. (B) Ten nanograms of chromosomal DNA derived from strain MC58 or MC58-1 was used as the template in PCRs with the primers pil1 and pil2 or pil1 and pil3. pil1 and pil2 amplified part of the coding region of pilE; pil1 and pil3 were designed to detect recombination products between pilE and pilS7. The PCRs were performed with 1% agarose gel, and results were analyzed. Arrows on the right indicate specific PCR products and their relative sizes (444 and 399 bp). The relative lengths of migration of DNA molecular markers (lane 1) and their weights are shown on the left. (C) Different amounts (1 pg to 100 ng) of chromosomal DNAs derived from strains MC58 and MC58-1 were used as templates in PCRs with the primers pil1 and pil3. The recombination pilE-pilS7 molecules were analyzed by Southern blotting using the purified 399-bp-long PCR product shown in panel B, labeled with 32P, as a probe. The sizes of the DNA molecular weight markers are shown on the left.
TABLE 3.
Frequencies of natural transformation in congenic recBwt and recBET-37 meningococci
| DNA donor strain (Neisseria sp.) | Frequency of transformation (10−7)a to rifampin resistance of recipient strain:
|
Relative transformation frequency (MC58-1/MC58) (%) | |
|---|---|---|---|
| MC58 | MC58-1 | ||
| MC58Rif (N. meningitidis) | 4,420 | 902 | 20.4 |
| NL21Rif (N. lactamica) | 575 | 80 | 13.9 |
| NL56Rif (N. lactamica) | 768 | 252 | 32.8 |
| NL104Rif (N. lactamica) | 1,040 | 392 | 37.7 |
| NF3264Rif (N. flava) | 356 | 66 | 18.5 |
| NS3260Rif (N. subflava) | 513 | 128 | 24.9 |
| NP462Rif (N. polysaccharea) | 620 | 53 | 8.5 |
| NM405Rif (N. mucosa) | 133 | 9 | 6.8 |
| NC415Rif (N. cinerea) | 138 | 17.6 | 12.7 |
| NS408Rif (N. sicca) | 85 | 1.7 | 2.0 |
Transformation frequencies were measured by transforming MC58 or MC58-1 with DNA derived from rifampin-resistant variants of Neisseria strains. All these variants were obtained by selection on GC agar plates containing rifampin (36 μg/ml). Values indicate numbers of transformants/numbers of viable cells and are means of results from three independent experiments.
In N. gonorrhoeae the RecBCD pathway is involved DNA transformation. Disruption of the gonococcal recB, recC, or recD gene by transcriptionally nonpolar ermC′ gene cassettes resulted in a significant decrease in DNA transformation efficiency (18). Therefore, we determined the frequencies of transformation of congenic MC58 and MC58-1 meningococci in DNAs from different rifampin-resistant strains of Neisseria spp. (Table 3). We found that the presence of the recBET-37 allele in MC58-1 resulted in decreases in transformation efficiencies, ranging from 2 to 37.7% of the respective values measured in MC58. The difference in transformability between the two strains was generally higher when the transforming activity of DNA was lower in MC-58. For instance, the DNA from the N. sicca strain NS408Rif transformed MC58 to rifampin resistance at a frequency about 50-fold lower than that of the DNA from MC58Rif; MC58-1 was transformed by this DNA at a frequency of 2% with respect to the frequency for MC58.
To analyze the effect of the allelic substitution of recBET-37 for the wild-type allele on antigenic variation at the pilin loci, a PCR-based assay was developed (Fig. 6). The nucleotide sequence of the active pilE gene and of the silent pilS cassette cluster (pilS1 to pilS8) mapping immediately upstream of pilE is available in the data bank of strain MC58. On the basis of the sequence data, oligonucleotides were designed to detect recombinant gene products between the expression pilE locus and the silent large pilS7 cassette (Fig. 6A). Primers pil1 and pil2 amplified the pilE coding region in the DNA templates from both the MC58 and MC58-1 strains (Fig. 6B, lanes 2 and 3). With the oligonucleotides pil1 and pil3 as primers, a recombinant pilE-pilS7 gene product of the expected size was detectable in the two strains (Fig. 6B, lanes 4 and 5). To approximately estimate the amount of the recombinant pilin gene in the DNA templates, PCRs were carried out at different genomic DNA concentrations, ranging from 1 pg to 100 ng. The specific PCR products were detected by Southern blot hybridization (Fig. 6C). The data demonstrated quantitative differences between the two strains. In fact, the recombinant pilin gene product could be amplified from the MC58 genomic DNA only at concentrations above 100 pg/50 μl of reaction volume. On the contrary, the specific PCR product was detectable also when 1 pg of DNA from MC58-1 per 50 μl was used, suggesting that the presence of the defective recBET-37 allele dramatically enhanced (by about 2 orders of magnitude) the recombination frequency at the pilin loci.
DISCUSSION
In this study we have identified and characterized a defective recB allele (comprising a cluster of missense mutations in a critical region of the protein) in clinical isolates of N. meningitidis. The allele has been termed recBET-37 because of its prevalence in meningococci belonging to the hypervirulent ET-37 complex, although it is also common to other strains phylogenetically unrelated to this complex (Fig. 2 to 4 and Table 1). This commonality suggests that these clones diverged from each other after the mutations occurred or that the recBET-37 allele was acquired by horizontal gene transfer, as implied by the occurrence of a mosaic recB allele in two UVS strains.
Because the recB gene encodes a protein that is part of the multifunctional RecBCD enzyme, which controls the primary pathway for DNA recombination and repair in many bacteria, an intriguing question raised and partly addressed by this paper is which selective advantage is conferred by the defective allele to the meningococci.
Congenic meningococci harboring the recBET-37 allele are extremely sensitive to UV radiation (Fig. 5B) and are unable to repair lesions in DNA efficiently (Fig. 5C). These traits have been previously associated with mutations designed to destroy the activity of the RecB protein in E. coli (4, 11, 12), as well as in N. gonorrhoeae. In gonococci, insertional inactivation of the recB, recC, or recD gene by transcriptionally nonpolar ermC′ gene cassettes resulted in reduced levels of resistance to both UV and ionizing radiation and in a growth defect (18). Consistently with a current model about the function of the RecBCD pathway (12), recBET-37 meningococci did not exhibit higher spontaneous mutation frequencies than those of recB-proficient strains. However, mutation rates to antibiotic resistance were enhanced upon UV treatment, a phenomenon not observed in the recB-proficient counterpart (Fig. 5D). The absence of an SOS-like pathway in Neisseria (2) may account for the lack of UV-induced mutagenesis in the recB-proficient strain. The moderate increase in mutation frequencies following UV exposure in the congenic recBET-37 strain may be due to the activity of some error-prone DNA repair system unmasked by the defect of the RecBCD pathway. Future work will attempt to shed light on this repair system.
The extent of the involvement of the RecBCD pathway in the genetic transformation of Neisseria is controversial. Knockout of the three distinct recB, recC, and recD genes in gonococci led to an important transformation defect that was comparable in all mutants. Transformation frequencies were more than 40-fold lower than in the parental RecBCD-proficient strain FA1090 (18). Intriguingly, at variance with E. coli recD mutants, which are wild type for most phenotypes of recB or recC mutants, the gonococcal recD mutant resembled a recB or recC mutant in tested phenotypes. In another report, inactivation of the gonococcal recD gene of strain MS11 decreased the frequency of DNA transformation only approximately threefold (5). We have found about a fivefold decrease in the frequency of natural transformation in meningococci harboring the recBET-37 allele compared to that of the RecB-proficient parental strain, when the transforming DNA was derived from the rifampin-resistant variant of the parental strain (Table 3). The discrepancy between the available data might be, at least in part, due to (i) a different genetic background or (ii) residual activity of the RecBET-37 protein. However, consistent with the molecular defect of the mutated allele that disrupts a critical region of the protein, the RecBET-37 protein was not able at all to complement the recombination defect of the E. coli recB21 mutation, suggesting an absolute lack of activity in the mutated protein (Table 2).
The recBET-37 allele-associated transformation defect, although moderate, may contribute to the genetic isolation of meningococci of the ET-37 complex, which has been attributed to specific restriction modification systems (6). Our finding supports the view that this group of bacteria, although easily transformable in the laboratory, incorporates foreign DNA less frequently in nature than serogroup A meningococci do (6, 15). The transformation defect was mostly evident when sources of transforming DNA were several nonpathogenic Neisseria strains and transformation was very inefficient also in the RecB-proficient strain, possibly because of many heterologies in the DNA sequence (Table 3). By contrast, in Enterobacteriaceae the RecBCD nuclease, together with the MutS and MutL proteins and the UvrD helicase, contributes to the supervision of the legitimacy of RecA-promoted recombination between homologous sequences (34, 42). This comparison emphasizes a biological difference between these microorganisms.
Previous studies have demonstrated that the RecBCD pathway is not essential for gonococcal pilin antigenic variation. Using a colony-based PCR assay to determine the ability to generate recombinant pilin genes, knockout of recB, recC, or recD did not give detectable results. By contrast, recombinant pilin genes could not be detected in gonococci harboring a nonfunctional recA, recQ, or recO gene (18). However, in a different assay, disruption of recD in MS11 resulted in a 12-fold increase in the frequency at which nonparental pilin phenotypes were spawned (5). By using a PCR assay to estimate the frequency of pilin gene recombination, we demonstrated that the presence of the defective recBET-37 allele enhanced by about 2 orders of magnitude the recombination frequency at the pilin loci (Fig. 6). It is conceivable that the recBET-37 allele might affect the frequency of antigenic variation, which occurs via recombination mechanisms, of other surface molecules, such as Opa proteins that are exceedingly variable in the ET-37 complex (39).
At least two different mechanisms might account for the observed increase in pilin gene recombination. One possible explanation is a compensatory enhancement of the RecF-like pathway of recombination in the RecBCD-deficient strain, which might be in turn responsible for increased rates of pilin antigenic variation. A similar enhancement in the activity of the normally cryptic RecF pathway is observed in E. coli Exo V-deficient mutants in the presence of mutations in sbcB and -C genes, which inactivate two exonucleases (7, 16, 24, 40). As no sbcB and -C homologues exist in the neisserial database, it is conceivable that the RecF-like pathway is up regulated in RecBCD-deficient strains.
An alternative hypothesis involves possible effects of the recBET-37 allele on chromosomal recombination. In E. coli the Exo V nuclease plays an important role in chromosomal replication, being involved in the repair of disintegrated replication forks. If one of the two replication forks in a replication bubble has disintegrated while the other continues to function, chromosome overreplication occurs due to the switch of the chromosomal replication from theta to sigma mode. The phenomenon of the “sigma replication trap” upon collapse of one of the replication forks accounts for the reduced viability of the recB or recC mutants, more than their inability to repair double-strand breaks (12). Conceptually, although it must be verified to occur in meningococci, a defective RecB protein may lead to multiple chromosomes within a single cell, a condition that is predicted to facilitate unequal levels of crossing over between a silent pilin gene copy and the expression locus (31).
Reports from recent studies suggest that genetic lesions in DNA repair pathways may occur in natural bacterial populations (3, 13, 14, 22, 26, 35). Our finding supports the recent view that the occurrence of defective alleles of DNA repair and recombination genes is the rule, not an exception, in natural bacterial populations. These alleles, or possibly their combination, play an important role in the long-term evolution of pathogenic traits. They affect the general mutability and/or the frequency of genetic exchanges. They also influence by more specialized mechanisms of genetic (phase and antigenic) variation short-term adaptation, even within a single infection cycle, of pathogenic microorganisms to their specific hosts. The cost of a reduced individual fitness, which is invariably associated to genetic lesions in DNA repair and recombination systems, is balanced by these extra benefits.
Acknowledgments
We thank P. Di Nocera for useful suggestions and critical reading of the manuscript. We thank M. Frosch, J. C. Chapalain, J. M. Alonzo, P. Nicolas, and P. Mastrantuono for providing us with meningococcal strains.
This work was partially supported by grants from the MURST-PRIN program (Decreto Ministeriale no. 503 DAE-UFFIII, 18 October 1999), from the MURST-CNR biotechnology program (Legge 95/95), and from a project funded by the Regione Campania (Legge Regionale 41, 31.02.1994).
Editor: J. T. Barbieri
REFERENCES
- 1.Achtman, M. 1995. Epidemic spread and antigenic variability of Neisseria meningitidis. Trends Microbiol. 3:186-192. [DOI] [PubMed] [Google Scholar]
- 2.Black, C. G., J. A. Fyfe, and J. K. Davies. 1998. Absence of an SOS-like system in Neisseria gonorrhoeae. Gene 208:61-66. [DOI] [PubMed] [Google Scholar]
- 3.Bucci, C., A. Lavitola, P. Salvatore, L. Del Giudice, D. R. Massardo, C. B. Bruni, and P. Alifano. 1999. Hypermutation in pathogenic bacteria: frequent phase variation in meningococci is a phenotypic trait of a specialized mutator biotype. Mol. Cell 3:435-445. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhury, A. M., and G. R. Smith. 1984. Escherichia coli recBC deletion mutants. J. Bacteriol. 160:788-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaussee, M. S., J. Wilson, and S. A. Hill. 1999. Characterization of the recD gene of Neisseria gonorrhoeae MS11 and the effect of recD inactivation on pilin variation and DNA transformation. Microbiology 145:389-400. [DOI] [PubMed] [Google Scholar]
- 6.Claus, H., J. Stoevesandt, M. Frosch, and U. Vogel. 2001. Genetic isolation of meningococci of the electrophoretic type 37 complex. J. Bacteriol. 183:2570-2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connelly, J. C., E. S. de Leau, E. A. Okely, and D. R. Leach. 1997. Overexpression, purification, and characterization of the SbcCD protein from Escherichia coli. J. Biol. Chem. 272:19819-19826. [DOI] [PubMed] [Google Scholar]
- 8.Frosch, M., E. Schultz, E. Glenn-Calvo, and T. F. Meyer. 1990. Generation of capsule-deficient Neisseria meningitidis strains by homologous recombination. Mol. Microbiol. 4:1215-1218. [DOI] [PubMed] [Google Scholar]
- 9.Heelis, P. F., S. T. Kim, T. Okamura, and A. Sancar. The photo repair of pyrimidine dimers by DNA photolyase and model systems. J. Photochem. Photobiol. 17:219-228. [DOI] [PubMed]
- 10.Koomey, M., E. C. Gotschlich, K. Robbins, S. Bergstrom, and J. Swanson. 1987. Effects of recA mutations on pilus antigenic variation and phase transitions in Neisseria gonorrhoeae. Genetics 117:391-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kushner, S. R., H. Nagaishi, A. Templin, and A. J. Clark. 1971. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc. Natl. Acad. Sci. USA 68:824-827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuzminov, A. 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage lambda. Microbiol. Mol. Biol. Rev. 63:751-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeClerc, J. E., and T. A. Cebula. 2000. Pseudomonas survival strategies in cystic fibrosis. Science 289:391-392. [DOI] [PubMed] [Google Scholar]
- 14.LeClerc, J. E., B. Li, W. L. Payne, and T. A. Cebula. 1996. High mutation frequencies among Escherichia coli and Salmonella pathogens. Science 274:1208-1211. [DOI] [PubMed] [Google Scholar]
- 15.Linz, B., M. Schenker, P. Zhu, and M. Achtman. 2000. Frequent interspecific genetic exchange between commensal Neisseriae and Neisseria meningitidis. Mol. Microbiol. 36:1049-1058. [DOI] [PubMed] [Google Scholar]
- 16.Lloyd, R. G., and C. Buckman. 1985. Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol. 164:836-844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazzone, M., E. De Gregorio, A. Lavitola, C. Pagliarulo, P. Alifano, and P. P. Di Nocera. 2001. Whole-genome organization and functional properties of miniature DNA insertion sequences conserved in pathogenic Neisseriae. Gene 278:211-222. [DOI] [PubMed] [Google Scholar]
- 18.Mehr, I. J., and H. S. Seifert. 1998. Differential roles of homologous recombination pathways in Neisseria gonorrhoeae pilin antigenic variation, DNA transformation and DNA repair. Mol. Microbiol. 30:697-710. [DOI] [PubMed] [Google Scholar]
- 19.Meyer, T. F., J. Pohlner, and J. P. van Putten. 1994. Biology of the pathogenic Neisseriae. Curr. Top. Microbiol. Immunol. 192:283-317. [DOI] [PubMed] [Google Scholar]
- 20.Moxon, E. R., P. B. Rainey, M. A. Nowak, and R. E. Lenski. 1994. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr. Biol. 4:24-33. [DOI] [PubMed] [Google Scholar]
- 21.Myers, R. S., and F. W. Stahl. 1994. Chi and the RecBCD enzyme of Escherichia coli. Annu. Rev. Genet. 28:49-70. [DOI] [PubMed] [Google Scholar]
- 22.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 23.Parkhill, J., M. Achtman, K. D. James, S. D. Bentley, C. Churcher, S. R. Klee, G. Morelli, D. Basham, D. Brown, T. Chillingworth, R. M. Davies, P. Davis, K. Devlin, T. Feltwell, N. Hamlin, S. Holroyd, K. Jagels, S. Leather, S. Moule, K. Mungall, M. A. Quail, M. A. Rajandream, K. M. Rutherford, M. Simmonds, J. Skelton, S. Whitehead, B. G. Spratt, and B. G. Barrell. 2000. Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 404:502-506. [DOI] [PubMed] [Google Scholar]
- 24.Phillips, G. J., D. C. Prasher, and S. R. Kushner. 1988. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J. Bacteriol. 170:2089-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Potts, W. J., and J. R. Saunders. 1988. Nucleotide sequence of the structural gene for class I pilin from Neisseria meningitidis: homologies with the pilE locus of Neisseria gonorrhoeae. Mol. Microbiol. 2:647-653. [DOI] [PubMed] [Google Scholar]
- 26.Richardson, A. R., and I. Stojiljkovic. 2001. Mismatch repair and the regulation of phase variation in Neisseria meningitidis. Mol. Microbiol. 40:645-655. [DOI] [PubMed] [Google Scholar]
- 27.Robertson, B. D., and T. F. Meyer. 1992. Genetic variation in pathogenic bacteria. Trends Genet. 8:422-427. [DOI] [PubMed] [Google Scholar]
- 28.Salvatore, P., C. Pagliarulo, R. Colicchio, P. Zecca, G. Cantalupo, M. Tredici, A. Lavitola, C. Bucci, C. B. Bruni, and P. Alifano. 2001. Identification, characterization, and variable expression of a naturally occurring inhibitor protein of IS1106 transposase in clinical isolates of Neisseria meningitidis. Infect. Immun. 69:7425-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 30.Saunders, N. J., A. C. Jeffries, J. F. Peden, D. Hood, H. Tettelin, R. Rappuoli, and E. R. Moxon. 2000. Repeat-associated phase variable genes in the complete genome sequence of Neisseria meningitidis strain MC58. Mol. Microbiol. 37:207-215. [DOI] [PubMed] [Google Scholar]
- 31.Seifert, H. S. 1996. Questions about gonococcal pilus phase- and antigenic variation. Mol. Microbiol. 21:433-440. [DOI] [PubMed] [Google Scholar]
- 32.Serkin, C. D., and H. S. Seifert. 1998. Frequency of pilin antigenic variation in Neisseria gonorrhoeae. J. Bacteriol. 180:1955-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, K. C., T. V. Wang, and R. C. Sharma. 1987. recA-dependent DNA repair in UV-irradiated Escherichia coli. J. Photochem. Photobiol. 1:1-11. [DOI] [PubMed] [Google Scholar]
- 34.Stambuk, S., and M. Radman. 1998. Mechanism and control of interspecies recombination in Escherichia coli. I. Mismatch repair, methylation, recombination and replication functions. Genetics 150:533-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taddei, F., I. Matic, B. Godelle, and M. Radman. 1997. To be a mutator, or how pathogenic and commensal bacteria can evolve rapidly. Trends Microbiol. 5:427-428. [DOI] [PubMed] [Google Scholar]
- 36.Tettelin, H., N. J. Saunders, J. Heidelberg, A. C. Jeffries, K. E. Nelson, J. A. Eisen, K. A. Ketchum, D. W. Hood, J. F. Peden, R. J. Dodson, W. C. Nelson, M. L. Gwinn, R. DeBoy, J. D. Peterson, E. K. Hickey, D. H. Haft, S. L. Salzberg, O. White, R. D. Fleischmann, B. A. Dougherty, T. Mason, A. Ciecko, D. S. Parksey, E. Blair, H. Cittone, E. B. Clark, M. D. Cotton, T. R. Utterback, H. Khouri, H. Qin, J. Vamathevan, J. Gill, V. Scarlato, V. Masignani, M. Pizza, G. Grandi, L. Sun, H. O. Smith, C. M. Fraser, E. R. Moxon, R. Rappuoli, and J. C. Venter. 2000. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 287:1809-1815. [DOI] [PubMed] [Google Scholar]
- 37.Wainwright, L. A., K. H. Pritchard, and H. S. Seifert. 1994. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol. Microbiol. 13:75-87. [DOI] [PubMed] [Google Scholar]
- 38.Wainwright, L. A., J. V. Frangipane, and H. S. Seifert. 1997. Analysis of protein binding to the Sma/Cla DNA repeat in pathogenic Neisseriae. Nucleic Acids Res. 25:1362-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, J. F., D. A. Caugant, G. Morelli, B. Koumare, and M. Achtman. 1993. Antigenic and epidemiologic properties of the ET-37 complex of Neisseria meningitidis. J. Infect. Dis. 167: 1320-1329. [DOI] [PubMed] [Google Scholar]
- 40.Wang, T. C., and K. C. Smith. 1985. Mechanism of sbcB-suppression of the recBC-deficiency in postreplication repair in UV-irradiated Escherichia coli K-12. Mol. Gen. Genet. 201:186-191. [DOI] [PubMed] [Google Scholar]
- 41.Wong, I., M. Amaratunga, and T. M. Lohman. 1993. Heterodimer formation between Escherichia coli Rep and UvrD proteins. J. Biol. Chem. 268:20386-20391. [PubMed] [Google Scholar]
- 42.Zahrt, T. C., and S. Maloy. 1997. Barriers to recombination between closely related bacteria: MutS and RecBCD inhibit recombination between Salmonella typhimurium and Salmonella typhi. Proc. Natl. Acad. Sci. USA 94:9786-9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zieg, J., and S. R. Kushner. 1977. Analysis of genetic recombination between two partially deleted lactose operons of Escherichia coli K12. J. Bacteriol. 131:123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]