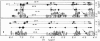Abstract
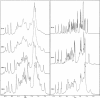
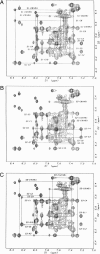
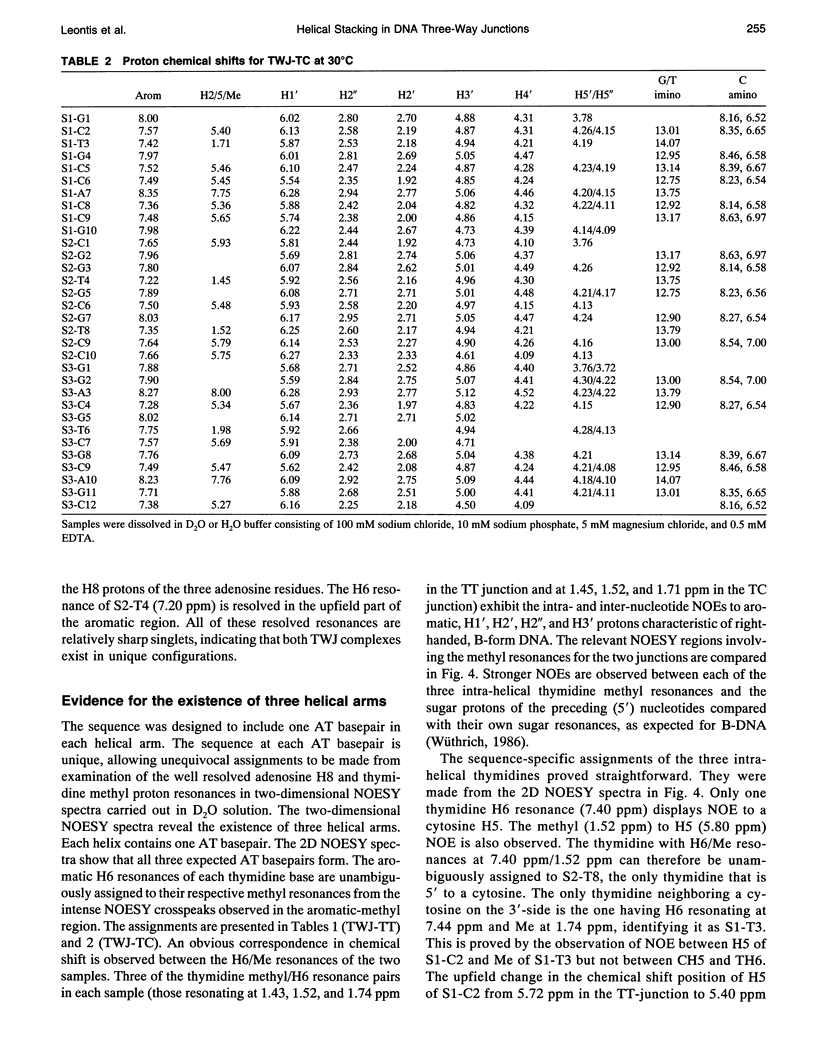
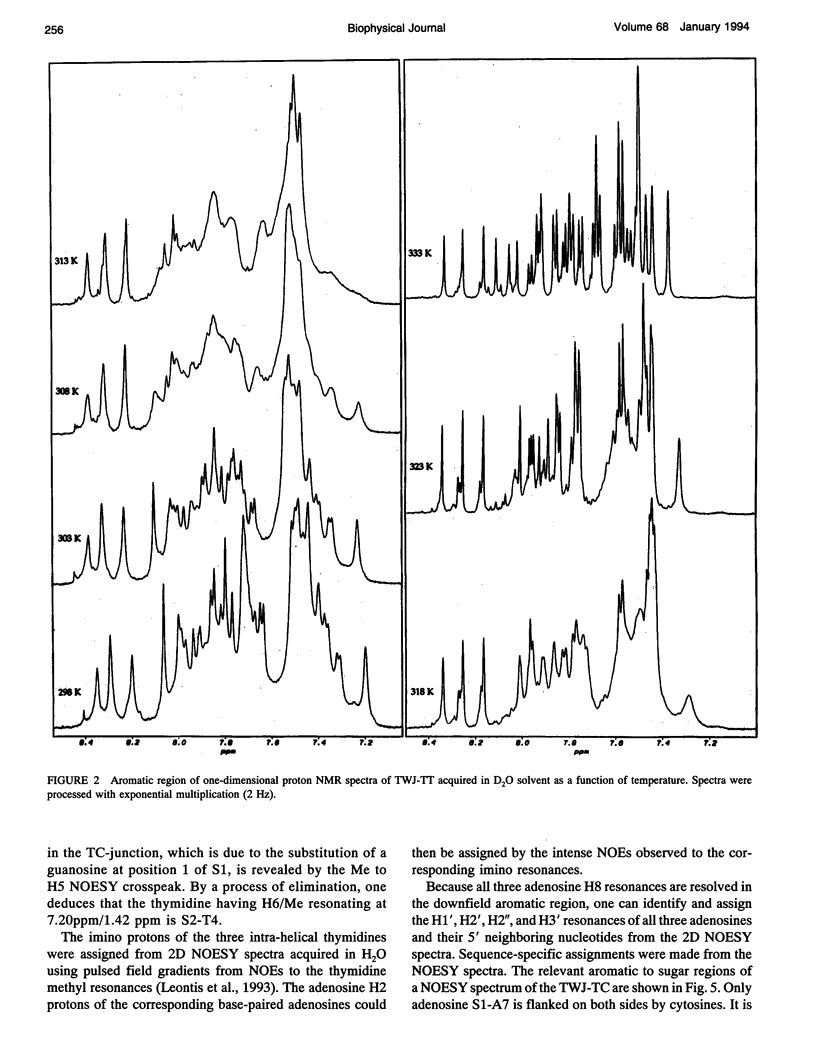
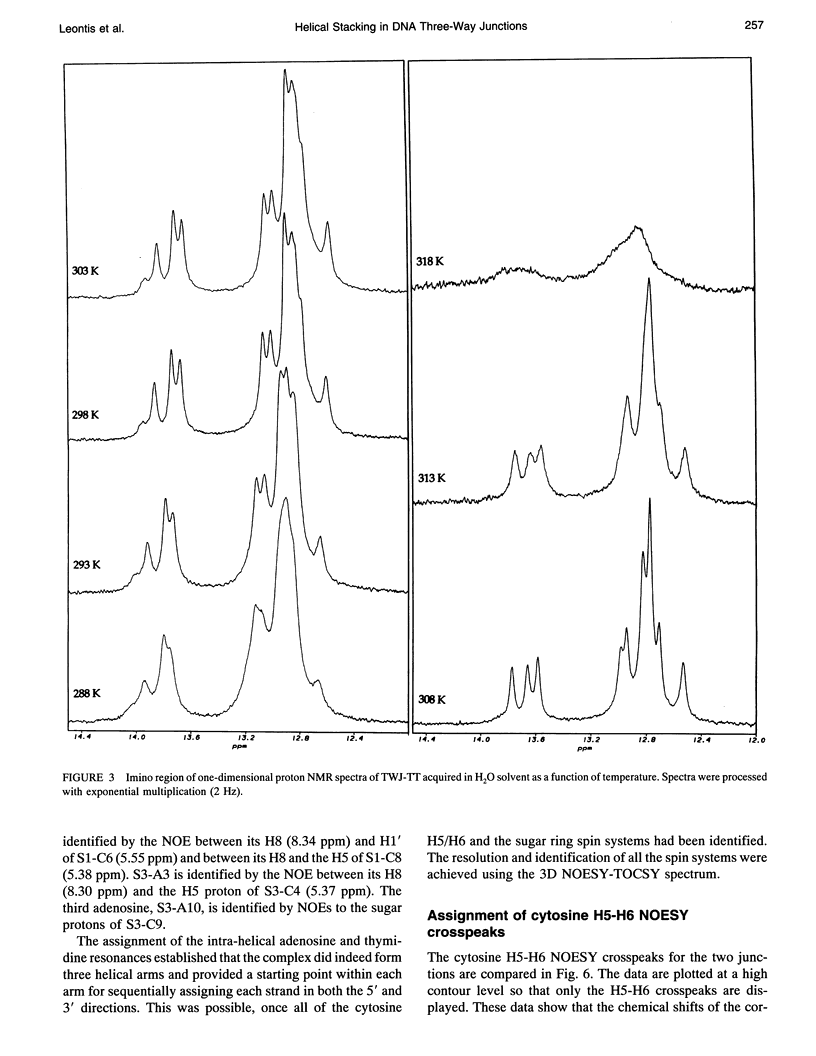
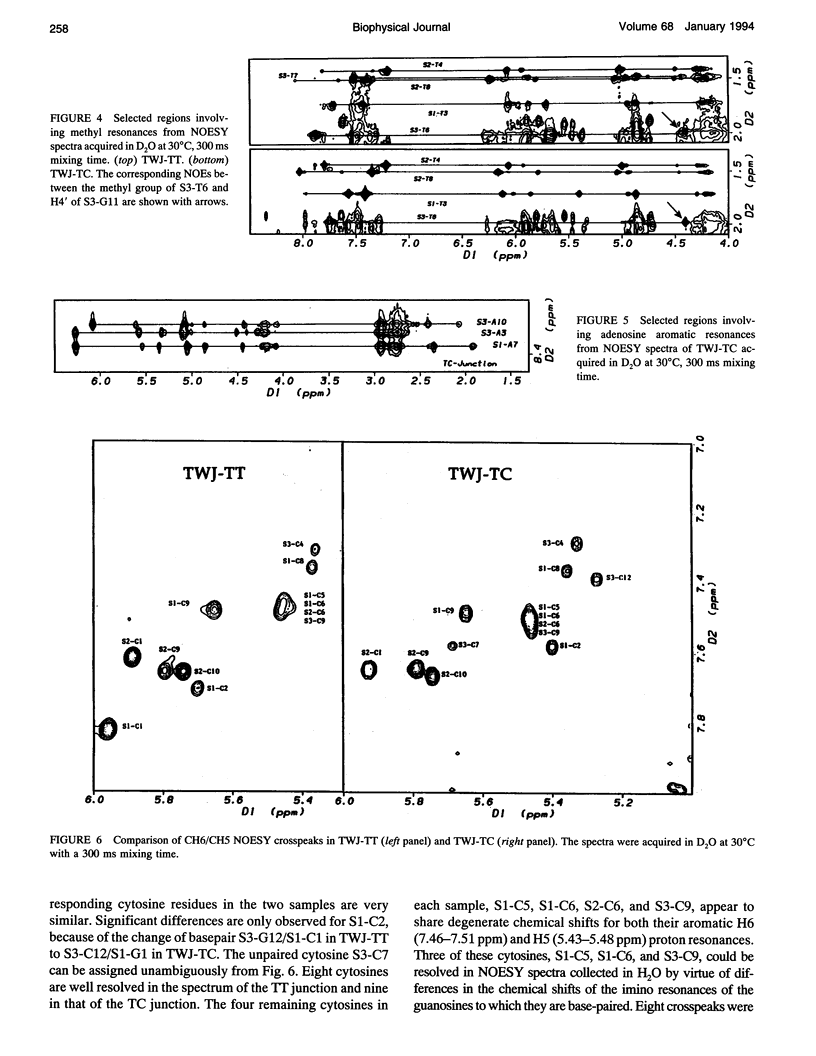
The proton NMR spectra of DNA three-way junction complexes (TWJ) having unpaired pyrimidines, 5'-TT- and 5'-TC- on one strand at the junction site were assigned from 2D NOESY spectra acquired in H2O and D2O solvents and homonuclear 3D NOESY-TOCSY and 3D NOESY-NOESY in D2O solvent. TWJ are the simplest branched structures found in biologically active nucleic acids. Unpaired nucleotides are common features of such structures and have been shown to stabilize junction formation. The NMR data confirm that the component oligonucleotides assemble to form conformationally homogeneous TWJ complexes having three double-helical, B-form arms. Two of the helical arms stack upon each other. The unpaired pyrimidine bases lie in the minor groove of one of the helices and are partly exposed to solvent. The coaxial stacking arrangement deduced is different from that determined by Rosen and Patel (Rosen, M.A., and D.J. Patel. 1993. Biochemistry. 32:6576-6587) for a DNA three-way junction having two unpaired cytosines, but identical to that suggested by Welch et al. (Welch, J. B., D. R. Duckett, D. M. J. Lilley. 1993. Nucleic Acids Res. 21:4548-4555) on the basis of gel electrophoretic studies of DNA three-way junctions containing unpaired adenosines and thymidines.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baudin F., Romaniuk P. J., Romby P., Brunel C., Westhof E., Ehresmann B., Ehresmann C. Involvement of "hinge" nucleotides of Xenopus laevis 5 S rRNA in the RNA structural organization and in the binding of transcription factor TFIIIA. J Mol Biol. 1991 Mar 5;218(1):69–81. doi: 10.1016/0022-2836(91)90874-6. [DOI] [PubMed] [Google Scholar]
- Brunel C., Romby P., Westhof E., Ehresmann C., Ehresmann B. Three-dimensional model of Escherichia coli ribosomal 5 S RNA as deduced from structure probing in solution and computer modeling. J Mol Biol. 1991 Sep 5;221(1):293–308. doi: 10.1016/0022-2836(91)80220-o. [DOI] [PubMed] [Google Scholar]
- Chen S. M., Chazin W. J. Two-dimensional 1H NMR studies of immobile Holliday junctions: nonlabile proton assignments and identification of crossover isomers. Biochemistry. 1994 Sep 27;33(38):11453–11459. doi: 10.1021/bi00204a007. [DOI] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J Mol Biol. 1987 Dec 20;198(4):711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- Egebjerg J., Christiansen J., Brown R. S., Larsen N., Garrett R. A. Protein L18 binds primarily at the junctions of helix II and internal loops A and B in Escherichia coli 5 S RNA. Implications for 5 S RNA structure. J Mol Biol. 1989 Apr 20;206(4):651–668. doi: 10.1016/0022-2836(89)90573-1. [DOI] [PubMed] [Google Scholar]
- Flemming M., Deumling B., Kemper B. Function of gene 49 of bacteriophage T4 III. Isolation of Holliday structures from very fast-sedimenting DNA. Virology. 1993 Oct;196(2):910–913. doi: 10.1006/viro.1993.1557. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of plus and minus RNAs of a virusoid and a structural model for the active sites. Cell. 1987 Apr 24;49(2):211–220. doi: 10.1016/0092-8674(87)90562-9. [DOI] [PubMed] [Google Scholar]
- Guthrie C., Patterson B. Spliceosomal snRNAs. Annu Rev Genet. 1988;22:387–419. doi: 10.1146/annurev.ge.22.120188.002131. [DOI] [PubMed] [Google Scholar]
- Jensch F., Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986 Jan;5(1):181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis N. B., Hills M. T., Piotto M., Malhotra A., Nussbaum J., Gorenstein D. G. A model for the solution structure of a branched, three-strand DNA complex. J Biomol Struct Dyn. 1993 Oct;11(2):215–223. doi: 10.1080/07391102.1993.10508722. [DOI] [PubMed] [Google Scholar]
- Leontis N. B., Kwok W., Newman J. S. Stability and structure of three-way DNA junctions containing unpaired nucleotides. Nucleic Acids Res. 1991 Feb 25;19(4):759–766. doi: 10.1093/nar/19.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of branched DNA species. Q Rev Biophys. 1993 May;26(2):131–175. doi: 10.1017/s0033583500004054. [DOI] [PubMed] [Google Scholar]
- Lilley D. M., Clegg R. M. The structure of the four-way junction in DNA. Annu Rev Biophys Biomol Struct. 1993;22:299–328. doi: 10.1146/annurev.bb.22.060193.001503. [DOI] [PubMed] [Google Scholar]
- Lu M., Guo Q., Kallenbach N. R. Interaction of drugs with branched DNA structures. Crit Rev Biochem Mol Biol. 1992;27(3):157–190. doi: 10.3109/10409239209082562. [DOI] [PubMed] [Google Scholar]
- Luehrsen K. R., Fox G. E. Secondary structure of eukaryotic cytoplasmic 5S ribosomal RNA. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2150–2154. doi: 10.1073/pnas.78.4.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa T., Murakami A., Ryo Y., Yamagishi H. Structural features of very fast sedimenting DNA formed by gene 49 defective T4. Virology. 1983 Apr 15;126(1):183–193. doi: 10.1016/0042-6822(83)90470-1. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Ribosomal RNA and translation. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- Noller H. F. Structure of ribosomal RNA. Annu Rev Biochem. 1984;53:119–162. doi: 10.1146/annurev.bi.53.070184.001003. [DOI] [PubMed] [Google Scholar]
- Nussbaum J. M., Newport M. E., Mackie M., Leontis N. B. Structure-specific binding and photosensitized cleavage of branched DNA three-way junction complexes by cationic porphyrins. Photochem Photobiol. 1994 May;59(5):515–528. doi: 10.1111/j.1751-1097.1994.tb02977.x. [DOI] [PubMed] [Google Scholar]
- Puglisi J. D., Tinoco I., Jr Absorbance melting curves of RNA. Methods Enzymol. 1989;180:304–325. doi: 10.1016/0076-6879(89)80108-9. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Patel D. J., Gao X. Three-dimensional homonuclear NOESY-TOCSY of an intramolecular pyrimidine.purine.pyrimidine DNA triplex containing a central G.TA triple: nonexchangeable proton assignments and structural implications. Biochemistry. 1992 Mar 10;31(9):2514–2523. doi: 10.1021/bi00124a011. [DOI] [PubMed] [Google Scholar]
- Rosen M. A., Patel D. J. Conformational differences between bulged pyrimidines (C-C) and purines (A-A, I-I) at the branch point of three-stranded DNA junctions. Biochemistry. 1993 Jul 6;32(26):6563–6575. doi: 10.1021/bi00077a010. [DOI] [PubMed] [Google Scholar]
- Rosen M. A., Patel D. J. Structural features of a three-stranded DNA junction containing a C-C junctional bulge. Biochemistry. 1993 Jul 6;32(26):6576–6587. doi: 10.1021/bi00077a011. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Kallenbach N. R. DNA branched junctions. Annu Rev Biophys Biomol Struct. 1994;23:53–86. doi: 10.1146/annurev.bb.23.060194.000413. [DOI] [PubMed] [Google Scholar]
- Seeman N. C., Kallenbach N. R. Design of immobile nucleic acid junctions. Biophys J. 1983 Nov;44(2):201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z., Hagerman P. J. Conformation of the central, three-helix junction of the 5 S ribosomal RNA of Sulfolobus acidocaldarius. J Mol Biol. 1994 Aug 19;241(3):415–430. doi: 10.1006/jmbi.1994.1517. [DOI] [PubMed] [Google Scholar]
- Sigal N., Alberts B. Genetic recombination: the nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol. 1972 Nov 28;71(3):789–793. doi: 10.1016/s0022-2836(72)80039-1. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Luehrsen K. R., Woese C. R., Pace N. R. An unusual 5S rRNA, from Sulfolobus acidocaldarius, and its implications for a general 5S rRNA structure. Nucleic Acids Res. 1981 Nov 25;9(22):6129–6137. doi: 10.1093/nar/9.22.6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Welch J. B., Duckett D. R., Lilley D. M. Structures of bulged three-way DNA junctions. Nucleic Acids Res. 1993 Sep 25;21(19):4548–4555. doi: 10.1093/nar/21.19.4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhof E., Romby P., Romaniuk P. J., Ebel J. P., Ehresmann C., Ehresmann B. Computer modeling from solution data of spinach chloroplast and of Xenopus laevis somatic and oocyte 5 S rRNAs. J Mol Biol. 1989 May 20;207(2):417–431. doi: 10.1016/0022-2836(89)90264-7. [DOI] [PubMed] [Google Scholar]
- Yang J. H., Usman N., Chartrand P., Cedergren R. Minimum ribonucleotide requirement for catalysis by the RNA hammerhead domain. Biochemistry. 1992 Jun 2;31(21):5005–5009. doi: 10.1021/bi00136a013. [DOI] [PubMed] [Google Scholar]
- von Kitzing E., Lilley D. M., Diekmann S. The stereochemistry of a four-way DNA junction: a theoretical study. Nucleic Acids Res. 1990 May 11;18(9):2671–2683. doi: 10.1093/nar/18.9.2671. [DOI] [PMC free article] [PubMed] [Google Scholar]