Abstract
Neonatal Escherichia coli meningitis continues to be a diagnostic and treatment challenge despite the availability of active antibiotics. Our earlier studies have shown that outer membrane protein A (OmpA) is one of the major factors responsible for Escherichia coli traversal across the blood-brain barrier that constitutes a lining of brain microvascular endothelial cells (BMEC). In this study we showed that OmpA binds to a 95-kDa human BMEC (HBMEC) glycoprotein (Ecgp) for E. coli invasion. Ecgp was partially purified by wheat germ agglutinin and Maackia amurensis lectin (MAL) affinity chromatography. The MAL affinity-purified HBMEC proteins bound to OmpA+ E. coli but not to OmpA− E. coli. In addition, the deglycosylated MAL-bound proteins still interact with OmpA+ E. coli, indicating the role of protein backbone in mediating the OmpA binding to HBMEC. Interestingly, the MAL affinity-bound fraction showed one more protein, a 65-kDa protein that bound to OmpA+ E. coli in addition to Ecgp. Further, the 65-kDa protein was shown to be a cleavage product of Ecgp. Immunocytochemistry of HBMEC infected with OmpA+ E. coli by using anti-Ecgp antibody suggests that Ecgp clusters at the E. coli entry site. Anti-Ecgp antibody also reacted to microvascular endothelium on human brain tissue sections, indicating the biological relevance of Ecgp in E. coli meningitis. Partial N-terminal amino acid sequence of Ecgp suggested that it has 87% sequence homology to gp96, an endoplasmic reticulum-resident molecular chaperone that is often expressed on the cell surface. In contrast, the 65-kDa protein, which could be the internal portion of Ecgp, showed 70% sequence homology to an S-fimbria-binding sialoglycoprotein reported earlier. These results suggest that OmpA interacts with Ecgp via the carbohydrate epitope, as well as with the protein portion for invading HBMEC.
Escherichia coli K1 meningitis is the most common infection of the central nervous system in neonates. The mortality and morbidity associated with this disease have remained unchanged despite advances in antimicrobial chemotherapy (5, 10, 11, 13, 26). The reasons for the poor outcome has been attributed to limited knowledge of pathogenesis and pathophysiology of the disease. Although most cases of E. coli meningitis occur via hematogenous spread, it is not clear what microbial and host factors are responsible for the ability of neurotropic strains of E. coli to cross the blood-brain barrier, which is formed by a single layer of brain microvascular endothelial cells (BMEC). The interaction of specific E. coli determinants with their corresponding BMEC receptors may dictate the tissue tropism in neonatal E. coli meningitis. Non-brain endothelial cells, which have generally been used to study the interaction of E. coli that causes meningitis, are not an ideal target cell culture model because they differ considerably from BMEC (1, 12, 28, 34). Thus, we developed an in vitro model of the blood-brain barrier using BMEC derived from humans, cows, and rats (24, 25).
Several investigators have made use of cultured mammalian cells to identify the mechanisms of bacterial entry into these cells (2, 3, 9, 19, 20). Many microorganisms utilize integrins on host cells as the receptor molecule for binding to and invasion of eukaryotic cells, e.g., Yersinia, Shigella, Bordetella, and Neisseria spp. and enteropathogenic E. coli (2, 3, 27, 29, 31, 32). We have shown that S fimbriae are required for E. coli binding to NeuAcα2,3-galactose-containing glycoproteins and sulfated glycolipids of BMEC (16, 25). We further showed that S fimbria interacts with a 65-kDa BMEC glycoprotein specifically expressed on brain endothelial cells but not on systemic endothelial cells (15). However, The binding via S fimbriae was not accompanied by invasion in vitro, suggesting that S fimbriae might mediate adherence of E. coli to BMEC in vivo.
After initial adherence mediated by S fimbriae, additional cell surface molecules are thought to contribute to the invasion of bacteria into host cells. Several nonfimbriated E. coli determinants have been subsequently identified that contribute to the E. coli invasion of BMEC, e.g., OmpA, IbeA, IbeB, and Yijp (7, 8, 17, 30). Except for OmpA, neither the surface localization nor mode of action of other determinants is known. OmpA, a 35-kDa cell surface transmembrane protein with four extracellular loops, is highly conserved on many gram-negative bacteria. We showed that OmpA expression enhances E. coli invasion of BMEC by 25- to 50-fold compared E. coli strains without OmpA (17). This OmpA-mediated E. coli invasion occurs via the interaction of N-terminal loops of OmpA with GlcNAcβ1,4-GlcNAc epitopes of BMEC surface glycoproteins (18). Receptor analogues, the chitooligomers (GlcNAcβ1,4-GlcNAc polymers), although at high concentrations, blocked E. coli invasion of BMEC both in vitro and in the newborn rat model of hematogenous meningitis, suggesting that the glycoproteins bearing these epitopes are indeed involved in E. coli entry into the central nervous system. Furthermore, molecular modeling of GlcNAcβ1,4-GlcNAc sugar interaction with the canyon formed by the loops 1 and 2 of OmpA showed favorable energy levels and conformations compared to any other area. (D. Datta, N. Vaidehi, W. B. Florino, N. V. Prasadarao, and W. A. Goddard III, unpublished results).
One of the salient findings of our studies so far has been that S-fimbriae-mediated binding and OmpA-contributed invasion of E. coli are specific to BMEC but not to systemic endothelial cells of non-brain origin, e.g., human umbilical vein endothelial cells (HUVEC), human arterial aortic endothelial cells, and human ileac vein endothelial cells (18). Therefore, we speculated that selective invasion of E. coli into BMEC compared to systemic endothelial cells might be a result of specific interaction of E. coli determinants with corresponding ligands specifically expressed on BMEC.
I report here the identification of a 95-kDa human BMEC (HBMEC) surface protein (Ecgp) that binds to OmpA on E. coli that was partially purified by lectin affinity chromatography. Partial N-terminal amino acid sequence of Ecgp suggests that it is a gp96-like molecule and its homologue Hsp-90. Inhibition of E. coli invasion into HBMEC by affinity-purified Ecgp and the presence of Ecgp on microvessels in human brain sections strongly suggest its role in the development of E. coli meningitis.
MATERIALS AND METHODS
Bacteria.
E44 is a rifampin-resistant mutant of E. coli K1 strain RS 218 (serotype O18:K1:H7), which was isolated from the cerebrospinal fluid of a neonate with meningitis and invades HBMEC in a cell culture model (17). E91 is a noninvasive derivative of E44 that expresses no OmpA, since the ompA gene is disrupted (17). HB101 (K-12 capsular polysaccharide), a laboratory strain, is noninvasive in HBMEC. All bacteria were grown in brain heart infusion broth with appropriate antibiotics as necessary. All bacterial media were purchased from Difco Laboratories (Detroit, Mich.).
E. coli invasion assays.
Confluent HBMEC in 24-well plates were incubated with 107 E. coli in experimental medium (1:1 mixture of Ham F-12 and M-199 containing 5% heat-inactivated fetal bovine serum) for 90 min at 37°C. Monolayers were washed three times with RPMI 1640 and incubated in experimental medium containing gentamicin (100 μg/ml) for 1 h to kill extracellular bacteria. The monolayers were washed again and lysed with 0.5% Triton X-100. The intracellular bacteria were enumerated by plating on sheep blood agar plates. In some experiments, either the bacteria or the HBMEC were pretreated with various protein fractions for 30 min prior to performing the invasion assays. The effects of these reagents on HBMEC were assessed by the trypan blue dye exclusion method, and the effects on bacterial viability were tested by colony counting.
Binding of E. coli to biotinylated HBMEC surface proteins.
Confluent HBMEC monolayers in T75 flasks were washed with RPMI 1640 three times and biotinylated with NHS-LC-biotin (Pierce, Rockford, Ill.) as per the instructions for 45 min on ice. The monolayers were washed again with RPMI 1640 containing 5% heat-inactivated serum, followed treatment with RPMI 1640 alone. The monolayers were lysed in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 (PBS-Tx), sonicated briefly, and centrifuged at 5, 000 × g for 10 min at 4°C to remove the debris. The supernatant was collected and centrifuged at 100,000 × g for 1 h at 4°C. The membrane pellet was dissolved in PBS-Tx and sonicated very briefly. Approximately 0.5 mg of biotinylated HBMEC membrane proteins in 1 ml of PBS-Tx were incubated with either OmpA+ or OmpA− E. coli strains for 1 h at 4°C, followed by centrifugation and washing with PBS-Tx three times. The proteins that bound E. coli were released by adding 50 μl of Laemmli buffer and vortexing them for a few minutes. The bacteria were removed by centrifugation, after which the buffer containing the proteins was collected and loaded onto a sodium dodecyl sulfate (SDS)-10% polyacrylamide gel. After electrophoresis, the proteins were transferred to nitrocellulose and probed with streptavidin-peroxidase. The bands were visualized by chemiluminescence.
Preparation of proteoliposomes for inhibition studies.
E. coli outer membrane proteins were isolated, and proteoliposomes containing the outer membrane proteins were prepared as described previously (17). Briefly, a mixture of 2.5 μl of acetone washed egg phosphatidylcholine (type XVI-E; Sigma), and 0.2 μl of diacetylphosphate was dried under nitrogen. The lipids were dissolved in anhydrous benzene, dried to remove traces of water, dissolved in ethyl ether, and then dried to produce an even film. The outer membrane proteins from either OmpA+ E. coli or OmpA− E. coli dissolved in buffer containing detergent were passed through Extractigel-D to remove most of the detergent and resuspended in water. A portion of water (0.2 ml) containing the outer membrane proteins was added to the lipid film, sonicated briefly, and then dried in a desiccator under vacuum. The dried protein-lipid film was resuspended in 5 mM Tris-HCl buffer (pH 8.0) containing 15% dextran 40. Proteoliposomes were used after having been left at room temperature for 1 h. Liposomes containing 10 to 20 μg of outer membrane proteins were incubated with biotinylated HBMEC membranes before incubation with the bacteria.
Lectin affinity chromatography.
HBMEC membrane proteins were prepared as described above except that the cells were not biotinylated. A total of 5 mg of the membrane proteins was incubated with wheat germ agglutinin (WGA; specific to GlcNAc1,4GlcNAc)-Sepharose for 1 h at 4°C in a column and then washed with 10 bed volumes of Tris buffer (pH 7.4) containing 0.01% Triton X-100 (Tris-Tx). The bound proteins were eluted with 200 mM N-acetylglucosamine in Tris-Tx, and the eluted proteins were dialyzed against PBS extensively and concentrated. For Maackia amurensis lectin (MAL; specific to NeuAc2,3-galactose) chromatography, 1 mg of WGA-Sepharose-bound HBMEC proteins was applied to the prewashed column in 0.5 ml of PBS containing 0.03% Triton X-100, along with a cocktail of protease inhibitor mixture. The column was washed with loading buffer several times, and the proteins were eluted with glycine-HCl buffer (pH 2.5). The eluted fractions were collected into a tube containing a 1/20 volume of 1 M phosphate buffer (pH 7.9) containing 0.01% Triton X-100. The eluant was dialyzed against PBS for 48 h with several changes of buffer at 4°C and then concentrated by using Centricon tubes. In some experiments the biotinylated HBMEC membrane proteins were subjected to lectin affinity chromatography as described above. The N-linked oligosaccharides of Ecgp were cleaved with 5 U of peptide N-glycosidase F in 50 mM sodium phosphate buffer (pH 7.8) at 37°C overnight (15). The cleavage efficiency was monitored by WGA blotting.
Anti-Ecgp antibody affinity chromatography.
Polyclonal antibodies (Ecgp-Ab) raised against WGA-Sepharose-bound HBMEC membrane proteins, which reacted to both 95- and 65-kDa proteins (15), were coupled to CNBr-activated Sepharose by a method described earlier (15). HBMEC membrane proteins (5 mg) were initially passed through Sepharose columns to remove nonspecific binding proteins, and the proteins were subjected to anti-Ecgp affinity-Sepharose chromatography. The bound proteins were released with glycine-HCl (pH 2.5), dialyzed immediately against PBS, and concentrated with Centricon tubes, and the protein content was estimated by using Bio-Rad reagent.
Immunocytochemistry.
HBMEC were grown in eight-well chamber slides and infected as described above for different time periods. The infection was stopped by three washes with RPMI 1640, and then the cells were fixed in 2% paraformaldehyde in PBS for at least 20 min at room temperature. Fixed cells were incubated with 3% bovine serum albumin (BSA) in PBS for 30 min at room temperature to block nonspecific binding sites. The cells were then incubated with anti-Ecgp antibody (1:500 dilution in BSA-PBS) for 1 h, washed, and further incubated with Cy3-conjugated anti-rabbit immunoglobulin G (IgG) for 45 min at room temperature. Stained slides were mounted in solution containing Vectashield (Vector Labs) antifade solution containing DAPI (4′,6′-diamidino-2-phenylindole). Cells were viewed by a Leica (Wetzlar, Germany) DMRA microscope with Plan-Apochromat 40×/1.25 NA and 63×/1.40 NA oil immersion objective lenses. Image acquisition was with a SkyVision-2/VDS digital charge-coupled device (12-bit 1,280 by 1,024 pixel) camera in unbinned or 2-by-2 binned models into EasyFISH software, saved as 16-bit monochrome, and merged as 24-bit RGB TIFF images (Applied Spectral Imaging Inc., Carlsbad, Calif.). Human brain tissue was obtained from the leftover tissues devoid of any pathological condition from children who underwent surgery for seizure disorders at Childrens Hospital at Los Angeles. The tissue was fixed in formalin overnight and embedded in paraffin. Dewaxed sections were placed on a glass slide and incubated with 2% normal goat serum in PBS (NGS-PBS) for 1 h, followed by incubation with anti-Ecgp antibody in NGS-PBS for 2 h at room temperature. The sections were washed with PBS, and primary antibodies were detected by incubation with horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad), followed by a 3,3′-diaminobenzidine reaction in PBS. Control sections were incubated with unrelated polyclonal antibody. In some experiments, anti-Ecgp antibody was preincubated with MAL-bound HBMEC proteins prior to adding them to the brain sections. Photographs of sections were taken on an Olympus upright microscope. Photographs were scanned, and the figures were assembled and labeled by using Adobe Photoshop 6.0.
Electroelution and N-terminal amino acid sequencing.
Ecgp was eluted from MAL-bound fraction for examination of the cleavage pattern of Ecgp with Hoefer Sixpac Gel Eluter GE 200. The MAL-bound proteins were separated on a SDS-10% polyacrylamide gel and briefly stained with freshly prepared Coomassie brilliant blue solution. The band was excised and cut into small pieces. The gel slices were placed into the inner tubes containing 300 μl of Laemmli buffer. The inner tube was place in the outer tube containing 100 μl of 4× Laemmli buffer, and the electroelution was carried out according to the instruction manual. After elution, the protein was stored at −20°C overnight in a solution containing a cocktail mixture of protease inhibitors, followed by thawing, and then subjected to SDS-polyacrylamide gel electrophoresis (PAGE). For microsequencing, the proteins in a duplicate gel were transferred to a nitrocellulose sheet and briefly stained with Coomassie brilliant blue solution. The bands corresponding to Ecgp and 65-kDa proteins were excised and subjected to partial N-terminal amino acid sequencing.
RESULTS
(i) OmpA+ E. coli binds to a ∼95-kDa HBMEC surface protein.
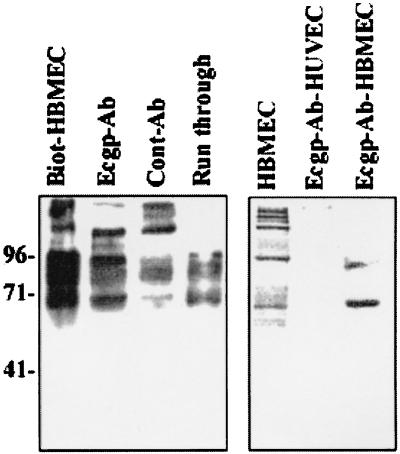
Our studies with isogenic OmpA+ and OmpA− E. coli strains suggested that the expression of OmpA enhances the E. coli invasion of HBMEC by 25- to 50-fold compared to strains lacking OmpA (17). Thus, it is possible that OmpA might be interacting with HBMEC surface structures for E. coli invasion. To identify the OmpA-binding molecules on HBMEC, the cell surface proteins were biotinylated, and the membrane proteins were prepared. OmpA+ and OmpA− E. coli strains were then incubated with the biotinylated HBMEC membrane proteins for 1 h at 4°C. The E. coli bound proteins were released with glycine-HCl (pH 2.5), dissolved in SDS buffer, and analyzed by immunoblotting with streptavidin-peroxidase, followed by chemiluminescence. As shown in Fig. 1, OmpA+ E. coli (E44) bound to a ∼95-kDa protein, whereas OmpA− E. coli (E91) showed no proteins on the blot. Biotinylated BSA was used as a negative control and did not show any association with either of these bacteria. In addition, HBMEC membrane proteins preincubated with liposomes reconstituted with outer membrane proteins from OmpA+ E. coli inhibited the binding of E44 to the 95-kDa protein. In contrast, outer membrane proteins of E91 reconstituted in liposomes did not show such blocking activity. These results suggest that OmpA is specifically interacting with a 95-kDa HBMEC surface protein.
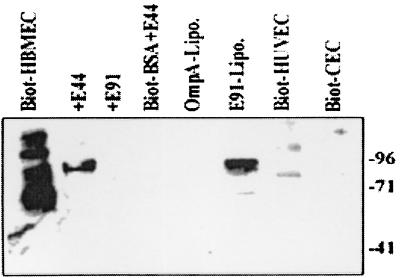
FIG. 1.
Binding of HBMEC proteins to OmpA+ E. coli. Confluent monolayers of HBMEC were biotinylated, and membrane proteins were prepared and incubated with OmpA+ and OmpA− E. coli strains as described in Materials and Methods. Biotinylated BSA was used as a control. In some experiments, the HBMEC membrane proteins were preincubated with either outer membrane proteins of E44 (OmpA-Lipo.) or E91 (E91-Lipo.) reconstituted in liposomes for 30 min on ice, followed by incubation with E44. Similarly biotinylated HUVEC and CEC membranes were also analyzed by immunoblotting with streptavidin-peroxidase for their binding capacity to E44. The bound proteins were identified by diaminobenzidene and hydrogen peroxide. The molecular weight markers are indicated on the right.
We have previously shown that OmpA+ E. coli invasion was specific to HBMEC, but not to cells of non-brain origin (18). To determine whether this phenomenon could be due to lack of OmpA binding molecules and/or receptors on non-brain cells, we biotinylated the surface proteins of HUVEC (systemic endothelial cells) and conjunctival epithelial cells (CEC; non-brain cells). The membrane proteins were incubated and analyzed as described above. The OmpA+ E. coli strain bound to some proteins from both HUVEC or CEC but in very small quantities and of different molecular masses than that of OmpA-binding proteins, suggesting that the 95-kDa OmpA-binding protein may be specifically present or significantly expressed on HBMEC. We refer to the 95-kDa protein below as Ecgp.
(ii) OmpA-binding Ecgp is a glycoprotein.
Since we have previously shown that OmpA interacts with GlcNAc1,4GlcNAc epitopes of HBMEC glycoproteins (19), the Ecgp could be a glycoprotein. Thus, to isolate HBMEC glycoproteins, WGA-affinity chromatography was carried out. The WGA-bound HBMEC membrane fraction showed several proteins, but 95- and 65-kDa proteins appeared to be the major proteins determined by SDS-PAGE, followed by Coomassie staining (Fig. 2A, second lane). A portion of the WGA-bound HBMEC fraction was biotinylated, incubated with OmpA+ E. coli, and analyzed as described above. Similar to previous results E44 bound to Ecgp; however, an additional band of 65-kDa protein was also observed, along with two other faint bands (Fig. 2B). To examine whether the 65-kDa protein was a degradation product of Ecgp, the 95-kDa protein was electoeluted from the SDS gel of WGA-bound fraction, frozen at −20°C overnight, thawed on ice, and subjected to SDS-PAGE. After being stained with Coomassie brilliant blue, the gel showed three proteins: a 95-kDa protein (faint band), a 65-kDa protein, and a 30-kDa protein (Fig. 2A). These results suggest that 95-kDa Ecgp degrades into two small-molecular-size proteins very easily despite the presence of various protease inhibitors. The absence of 30-kDa protein in similar quantities to that of 65-kDa in WGA-bound or E44-bound fraction could be due to the lack of glycosylation sites on the 30-kDa fragment that are necessary for interaction.
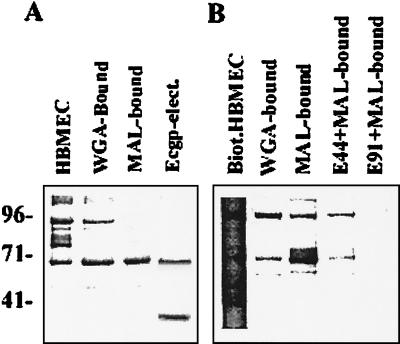
FIG. 2.
Identification of OmpA-bound proteins from MAL affinity chromatography-purified proteins. HBMEC membrane proteins either unbiotinylated or biotinylated were initially subjected to WGA affinity chromatography, and the WGA-bound proteins were further subjected to MAL chromatography. The lectin-bound protein fractions were separated by SDS-10%PAGE. The unbiotinylated proteins were stained with Coomassie brilliant blue (A), and the biotinylated proteins were transferred to nitrocellulose, followed by probing with streptavidin-peroxidase (B). In addition, the 95-kDa protein from the unbiotinylated MAL-bound fraction was electroeluted, frozen at −20°C overnight, thawed, and subjected to SDS-PAGE (Ecgp-elect.).
Our previous studies also showed that S fimbriae bind to NeuAc2,3-galactose residues of HBMEC surface glycoproteins (25). Interestingly, the molecular size of the sialoglycoprotein that interacts with S fimbriae is also 65 kDa. Thus, we speculated that the 65-kDa protein bound to OmpA and the 65-kDa protein bound to S fimbriae might be the same molecule. To examine this, the WGA-bound HBMEC proteins were further purified by MAL chromatography as described earlier (18). The MAL-bound HBMEC proteins showed a major 65-kDa protein and a faint 95-kDa protein (Fig. 2A, third lane). A portion of MAL-bound fraction was biotinylated and incubated with either E44 or E91, released the bound proteins, and analyzed as described above. Two proteins (95 and 65 kDa) specifically bound E44 in significant quantities compared to E91 (Fig. 2B), suggesting that OmpA-binding Ecgp is also a sialoglycoprotein. Some other high-molecular-weight proteins were also observed in this fraction; however, they appear to be nonspecifically interacting with affinity medium or bacteria. In our previous studies, a polyclonal antibody (anti-S-fimbria-binding protein [SBP] antibody) raised against WGA-bound fraction recognizes a 130-kDa protein, a 95- to 97-kDa protein (minor), and a 65-kDa protein (15). In the present study the question of whether the anti-SBP antibody also reacts to OmpA-binding proteins by incubating the MAL-bound HBMEC membrane proteins with either OmpA+ or OmpA− E. coli, followed by the release of bound proteins and Western analysis, was investigated. OmpA bound to both 95-kDa (Ecgp) and 65-kDa proteins from the MAL-bound fraction and reacted to anti-SBP antibody (referred as Ecgp-Ab hereafter) (Fig. 3). E91 showed some proteins in small quantities but not of the same molecular sizes. These results suggest that the 95-kDa protein (Ecgp) might be a binding molecule for both S fimbriae and OmpA for E. coli invasion via NeuAc2,3-galactose and GlcNAc1,4GlcNAc epitopes, respectively. In contrast, MAL affinity chromatography of HUVEC and CEC proteins showed several proteins by SDS-PAGE (gel not shown) but did not show any binding to OmpA+E. coli. Thus, Ecgp is specifically present or differentially expressed on HBMEC but not on systemic endothelial cells.
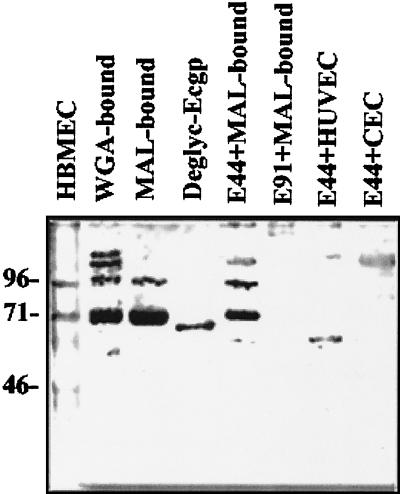
FIG. 3.
Western analysis of various HBMEC fractions with anti-Ecgp antibody. HBMEC membrane proteins were subjected to WGA and MAL affinity chromatography as described in Materials and Methods. The MAL-bound fraction (0.5 mg) was incubated with E44 (E44+MAL-bound) or E91 (E91+MAL-bound), the bound proteins were released and separated by SDS-PAGE. After the proteins were transferred to nitrocellulose, the blot was subjected to immunoblotting with anti-Ecgp antibody. Similarly, deglycosylated Ecgp (Deglyc-Ecgp), MAL-bound either HUVEC (E44+HUVEC), or CEC (E44+CEC) membrane proteins were also analyzed as described. WGA-bound and MAL-bound fractions were also loaded onto the gel to examine the protein pattern. The molecular weight markers are indicated on the left.
(iii) MAL-bound HBMEC proteins block E. coli invasion of HBMEC.
Our previous studies showed that the MAL-bound HBMEC membrane proteins blocked S-fimbria-mediated binding of E. coli to HBMEC (15). Here, the effect of Ecgp (MAL-bound fraction) on E. coli invasion was tested by incubating different concentrations of Ecgp with either OmpA+ or OmpA− E. coli strains before adding them to the HBMEC monolayers. As shown in Fig. 4, Ecgp blocked the E. coli invasion in a dose-dependent manner with a 50% inhibition at 5 μg/well and a 75% inhibition at 10 μg/well (10,750 ± 1,250 CFU/well for control versus 2,175 ± 895 CFU/well for Ecgp; P < 0.01). In contrast, the MAL-bound fraction did not show any effect on E91 invasion (2,560 ± 450 CFU/well [background invasion]). Similarly, MAL-bound HUVEC proteins also did not show significant effects on the invasion. We further verified whether Ecgp exhibited any antibacterial effect by incubating the E. coli with Ecgp, followed by colony plating. The results showed no killing of bacteria by Ecgp (data not shown). In addition, fetuin that contains significant quantities of oligosaccharides with NeuAc2,3-galactose and GlcNAc1,4GlcNAc sequences was also used in inhibition studies to assess the role of carbohydrate moieties in the invasion. Fetuin reduced the invasion by only 30% at a 100-μg concentration (9,890 ± 895 CFU/well for control with no protein versus 7,500 ± 700 CFU/well with fetuin; P < 0.05). These results suggest that the carbohydrate epitopes of fetuin are in a different conformation from that of Ecgp so that they do not interact with OmpA. Alternatively, the protein portion of Ecgp may contribute to the specificity of the interaction.
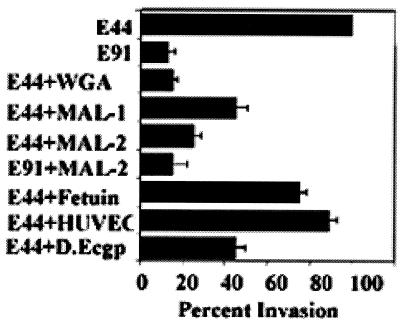
FIG. 4.
Inhibition of E. coli with WGA- and MAL-bound fractions. OmpA+ E. coli (107 CFU in 100 μl) samples were incubated with either 5 μg (MAL-1) or 10 μg (MAL-2) of MAL-bound HBMEC fractions or deglycosylated MAL-bound fraction (D.Ecgp) for 1 h on ice and then added to HBMEC monolayers. In some experiments, WGA-bound either HBMEC (E44+WGA) or HUVEC (E44+HUVEC) fractions were incubated with OmpA+ E. coli strain. The invasion assays were carried out as described in Materials and Methods. Fetuin, a control protein, was used at a 100-μg concentration. The experiments were carried out at least three times in triplicate and expressed as a percentage of control with no protein (10,750 ± 1250 CFU/well). The error bars represent the standard deviation. The inhibition of E. coli invasion by WGA- and MAL-bound fractions and deglycosylated Ecgp was statistically significant compared to the control (P < 0.05 by two-tailed, unpaired t test).
Thus, to examine the role of the protein backbone of Ecgp, the MAL-bound protein fraction was deglycosylated by using PNGase F to cleave the Asn-linked oligosaccharide chains, and the protein was used in E. coli invasion assays. The efficiency of deglycosylation was confirmed by lectin blotting with WGA. The electrophoretic mobility of deglycosylated Ecgp is slightly faster than that of glycosylated protein and reacted to anti-Ecgp antibody (Fig. 3, fourth lane). The deglycosylated Ecgp also showed a significant inhibitory effect on E. coli invasion, although 30% less than that of native Ecgp (9,870 ± 1520 CFU/well for control cells versus 4,150 ± 590 CFU/well for deglycosylated Ecgp; P < 0.01). Taken together, these results suggest that Ecgp interacts with OmpA via both chitobiose epitopes and the protein backbone of Ecgp.
(iv) Ecgp antibody and Ecgp antibody-affinity-purified HBMEC proteins block the E. coli invasion.
First, to further confirm that the proteins that interact with Ecgp-Ab are surface proteins, biotinylated HBMEC proteins were immunoprecipitated with Ecgp-Ab. The immunocomplexes were separated on SDS-PAGE and probed with streptavidin-peroxidase. Ecgp-Ab pulled down approximately 95 and 65-kDa proteins (Fig. 5A). However, a high-molecular-weight protein was also immunoprecipitated with the antibody. This protein appears to be nonspecifically interacting with the antibody as the control antibody also showed similar band. This suggests that Ecgp-Ab recognizes a surface protein on HBMEC.
FIG. 5.
Anti-Ecgp antibody recognizes HBMEC surface proteins. (A) Biotinylated HBMEC proteins (0.2 mg) were preincubated with 1 ml of agarose to preclear the nonspecific binding proteins. The supernatant was incubated with 1 μg of anti-Ecgp antibody overnight at 4°C, followed by incubation with protein A-agarose. The immunocomplexes were released by Laemmli buffer, separated by SDS-PAGE, and probed with streptavidin-peroxidase. An unrelated antibody was used as a control (Cont-Ab). (B) HBMEC and HUVEC membrane proteins were subjected to Ecgp-Ab affinity chromatography as described in Materials and Methods. The bound proteins were eluted and subjected to immunoblotting with Ecgp-Ab.
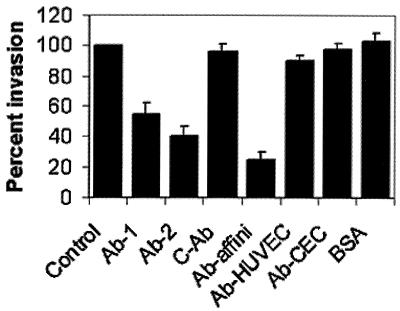
Next, I examined whether the Ecgp-Ab blocks the E. coli invasion by incubating different concentrations of Ecgp-Ab with HBMEC monolayers before the addition of the OmpA+ and OmpA− E. coli. Interestingly, the Ecgp-Ab showed only 45 to 50% inhibition of the invasion, whereas the control antibody showed no inhibition (Fig. 6). Partial inhibition of E. coli invasion by Ecgp-Ab suggests that the epitope recognized by Ecgp-Ab is slightly different from that of the OmpA-binding site. To verify this possibility, Ecgp-Ab affinity chromatography of HBMEC membrane proteins was performed, and the bound HBMEC proteins were used in E. coli invasion assays. The Ecgp-bound fraction showed two proteins with molecular masses of 95 and 65 kDa, which reacted to Ecgp antibody (Fig. 5B). The Ecgp proteins obtained from the Ecgp-Ab affinity column significantly blocked the E. coli invasion of HBMEC (>70% inhibition with a 16-μg/well concentration; Fig. 6). In contrast, the Ecgp-Ab column-bound HUVEC proteins showed no significant proteins that reacted to Ecgp-Ab and did not show any significant effect on E. coli invasion. Similar results were obtained with Ecgp-Ab-bound CEC membrane proteins. BSA was used as a negative control and showed no effect on the invasion. These results suggest that Ecgp is present specifically on BMEC but not on other cells. In addition, the data indicate that the epitopes recognized by Ecgp-Ab and OmpA might be slightly away from each other; thus, the difference in the efficiency of blocking E. coli invasion by Ecgp-Ab versus Ecgp-Ab affinity-purified proteins. The Ecgp-Ab-bound HBMEC membrane proteins also showed significant inhibition of E. coli binding to HBMEC in a previous study (15). Taken together, these studies suggest that both S fimbriae (NeuAc2,3-Galactose) and OmpA (GlcNAc1,4GlcNAc) may interact with the 95-kDa HBMEC sialoglycoprotein, Ecgp.
FIG. 6.
Inhibition of E. coli invasion of HBMEC by Ecgp-Ab and Ecgp-Ab affinity-purified proteins. Confluent monolayers of HBMEC were incubated with either 10 μg (Ab-1) or 20 μg (Ab-2) of Ecgp-Ab or control antibody (C-Ab) for 1 h at 37°C prior to infection with the bacteria. In some experiments, Ecgp-Ab affinity-purified proteins from HBMEC (10 μg of Ab-affini-2), HUVEC (10 μg of Ab-HUVEC), CEC (10 μg of Ab-CEC), or BSA (10 μg) were incubated with E44 for 1 h on ice before adding to the HBMEC monolayers. Invasion assays were carried out as described in Materials and Methods. The experiments were carried out at least two times in triplicate and are expressed as a percentage of control with no protein (10,400 ± 2,000 CFU/well). The error bars represent the standard deviation. The inhibition of E. coli invasion by Ecgp-Ab and Ecgp-Ab affinity-purified proteins was statistically significant compared to the control (P < 0.05 by two-tailed, unpaired t test).
(v) Ecgp is a surface-exposed protein interacting with OmpA.
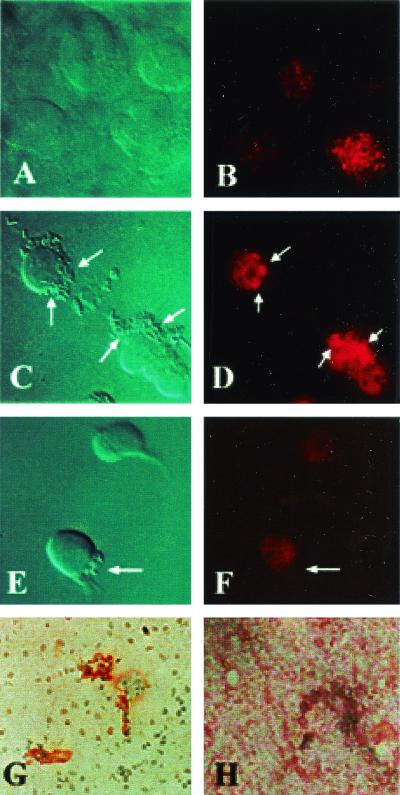
We have shown that Ecgp is on the surface of HBMEC by incubating the biotinylated HBMEC proteins with OmpA+ E. coli. In concurrence with these results, immunocytochemistry of HBMEC monolayers with Ecgp-Ab showed the reactivity on the surface of the cells in a previous study (15). To further examine whether Ecgp is associated with the bacterial entry, HBMEC were incubated with Ecgp-Ab initially for 1 h, followed by incubation with OmpA+ E. coli for 30 min. The monolayers were then washed, fixed, and stained with Cy3-conjugated anti-rabbit IgG. Uninfected HBMEC showed that Ecgp distributed throughout the cell as dense spots (Fig. 7B). Moreover, the staining of Ecgp was observed only on particular populations of cells. The monolayers infected with E44 revealed that Ecgp is colocalized directly beneath a group of bacteria (Fig. 7D). It is interesting that the Ecgp is clustered at the bacterial entry site. Very few bacteria associated with clustered Ecgp, despite the presence of several groups of bacteria on HBMEC. This could be due to the blocking of OmpA binding sites by Ecgp-Ab (functional), whereas the clusterization was due to the reactivity of nonblocking antibodies of Ecgp (nonfunctional) present in the same antiserum. OmpA− E. coli showed no such colocalization of Ecgp beneath the groups of bacteria (Fig. 7E and F). Similarly, human brain tissue sections showed that Ecgp-Ab reactivity was dense in some areas of blood vessel compared to other areas (Fig. 7G). In contrast, the control antibody did not show any staining of the blood vessels (Fig. 7H). These results suggest that Ecgp is expressing on a particular subpopulation of cells both on cultured HBMEC and on human brain microvessels.
FIG. 7.
Immunocytochemistry of HBMEC and human brain sections. Confluent untreated HBMEC monolayers (A and B) were treated with Ecgp-Ab (1:500), followed by infection with either OmpA+ E. coli (C and D) or OmpA− E. coli (E and F). The monolayers were washed and fixed with 2% paraformaldehyde. The bound Ecgp-Ab was identified by Cy3-conjugated secondary antibody, followed by visualization with laser confocal microscope. The sections of brain embedded in paraffin were stained with either Ecgp-Ab (G) or control rabbit IgG (H), followed by the addition of horseradish peroxidase-conjugated secondary antibody. The brown color was developed with diaminobenzidene and hydrogen peroxide. The pictures were edited and labeled using Adobe Photoshop 6.0. The arrows indicate either the location of invading bacteria or the clusterization of Ecgp.
(vi) Ecgp represents a gp96-like glycoprotein.
The MAL-purified 95- and 65-kDa proteins were separated by PAGE and transferred to a nitrocellulose sheet. The bands were excised and subjected to partial N-terminal amino acid sequencing. The sequences of these proteins were as follows (X is an unidentified amino acid): Ecgp, XDEVDVDGTVXXDLGFSREGSRTDDEVVXREE; gp65, XLMSVAIVDVFAAVFDLXE. A search of protein databases (PIR and Swiss-Prot) revealed that the N-terminal portion of Ecgp has 87% sequence homology to the tumor rejection antigen, gp96, and its homologue heat shock protein 90 (Hsp90) (4, 6). However, the N-terminal amino acid sequence of 65-kDa protein showed no sequence homology to any known proteins in the database but did show ∼70% sequence homology to the SBP when they are aligned together. These results suggest that OmpA binds to a gp96-like protein present on the surface of HBMEC and that the gp65 protein might be the SBP protein identified in previous studies. Taken together with other data, gp65 could be a separate protein that also binds OmpA, or it could be a cleavage product of gp96.
DISCUSSION
The molecular mechanisms underlying E. coli invasion of HBMEC to cause meningitis are not well understood. Specific surface structures on both bacteria and host cells are likely to contribute to the tissue tropism of this disease. We have previously shown that S fimbriae, the hairlike appendages on the surface of E. coli, enhance the binding to HBMEC via NeuAc2,3-galactose residues present on glycoproteins in vitro, although they are not absolute necessary in vivo (15). We further showed that OmpA is one of the E. coli factors that enhances the invasion of HBMEC (17). In addition, OmpA interacts with GlcNAc1,4-GlcNAc epitopes present on HBMEC glycoproteins for invasion of E. coli (18). Of interest, E. coli does not invade HUVEC despite the presence of glycoproteins containing GlcNAc1,4-GlcNAc epitopes, suggesting that either the carbohydrate structures are modified by the presence of additional sugars or the interaction of the protein backbone of the glycoprotein with OmpA plays a significant role in dictating the tissue tropism. However, the OmpA-binding molecules on HBMEC are not known.
In the present study, I identified an approximately 95-kDa protein (Ecgp) on the surface of HBMEC that interact with OmpA. Partial N-terminal amino acid sequencing of Ecgp indicated that it has sequence homology to the tumor rejection antigen, gp96, and its homologue, Hsp90. These studies suggested that the Ecgp is cleaved into 65- and 30-kDa fragments. Several groups have reported that gp96 also cleaves into a 65-kDa protein very easily; thus, the 65-kDa protein identified as SBP in our previous studies could be a degradation product of Ecgp. In agreement with this observation, anti-SBP antibody also reacted with Ecgp. Additional evidence comes from the partial N-terminal sequence data of 65-kDa protein observed in OmpA-binding fraction of HBMEC membrane proteins, which showed >70% sequence homology to SBP (15). Inhibition of both binding and invasion of E. coli into HBMEC by Ecgp further suggest that Ecgp might be the binding molecule for both S fimbriae and OmpA, which interact via different carbohydrate epitopes of N-linked oligosaccharides. Alternatively, it is possible that the SBP and the 65-kDa fragment of Ecgp might be two different molecules but share significant similarities. Since neither fetuin nor HUVEC membrane proteins, which contain significant quantities of N-linked oligosaccharide portion, block the invasion, the binding of OmpA to Ecgp peptide region might be crucial for the specificity of the interaction. Our previous studies showed that OmpA+ E. coli invasion of HBMEC induces actin condensation beneath the bacterial entry site by triggering the phosphorylation of focal adhesion kinase but not in HUVEC (19, 20). Inhibiting the interaction of OmpA with HBMEC by synthetic peptides that represent N-terminal portion of OmpA and excess amounts of GlcNAc1,4-GlcNAc polymers can block these events. Interestingly, the OmpA-interacting Ecgp could not be found in significant quantities on non-brain endothelial cells such as HUVEC as analyzed by WGA and MAL chromatography, as well as by direct binding to OmpA+ E. coli. Thus, the neurotropic nature of E. coli could be due to its interaction with Ecgp that is specifically or differentially expressing on HBMEC to induce cytoskeletal changes for its own internalization.
The Ecgp-Ab raised against WGA-bound HBMEC membrane fraction showed no inhibitory effect on E. coli binding to HBMEC in our previous studies (18). However, it is interesting that the same antibody showed partial inhibition on the invasion. This inhibition could be due to the binding of Ecgp-Ab to an epitope that does not completely encompass the OmpA-binding site. OmpA might also interact with the peptide region of Ecgp as shown by the binding of the deglycosylated Ecgp to OmpA+ E. coli and by the inhibition of E. coli invasion, although the effect is less compared to that seen with native Ecgp. It is possible that the presence of carbohydrate epitopes might enhance the affinity of Ecgp-OmpA interaction. Higher association constants have been demonstrated for several receptors for interaction with glycoproteins than with deglycosylated proteins (22, 23).
The ability of Ecgp-Ab to react with cultured HBMEC in previous studies suggests that Ecgp is a surface-exposed molecule (15). Furthermore, Ecgp-Ab reactivity with HBMEC even after treatment with bacteria suggests that some of the epitopes recognized by this antibody are away from the OmpA-binding site. A compelling aspect of Ecgp-Ab reactivity beneath the bacterial entry site is the clustering of Ecgp. Receptor clustering has been reported for several lectin-like molecules. OmpA acts as a lectin by interacting with carbohydrate epitopes of HBMEC glycoproteins. Thus, upon binding to HBMEC, receptors and intracellular proteins probably translocate to the bacterial entry site and form a spatially segregated supramolecular activation clusters. Formation of these clusters has been reported for several ligand-receptor interactions (14). In addition, the presence of Ecgp-Ab reactive molecules on human brain tissue sections suggests the biological relevance of this receptor in E. coli meningitis.
It is interesting to note the sequence similarity of Ecgp with both gp96 and hsp90, which are mostly retained in the endoplasmic reticulum (ER). These proteins contain ER retention signals such as KDEL; however, several studies have indicated that these proteins also express on the cell surface of several normal cells (21). Similarly, an ER-resident protein, calnexin has been shown to escape from the ER of immature thymocytes to be expressed on the plasma membrane (33). We have identified the gene sequence encoding the Ecgp by screening the cDNA library of HBMEC with Ecgp-Ab. Sequencing of this gene suggested that Ecgp does also contain the KDEL sequence (unpublished data). Overexpression of this gene in CHO cells (normally noninvasive for E. coli) make these cells susceptible to E. coli invasion, suggesting that Ecgp indeed is responsible for E. coli invasion of HBMEC. Despite the accumulation of significant data suggesting that OmpA on E. coli K1 is the crucial factor for invasion, other molecules, such as IbeA, IbeB, and CNF, also play a role. OmpA interaction with HBMEC might induce the virulence factors described above, which may further interact with HBMEC for efficient invasion. Alternatively, there might be more than one mechanism involved in E. coli invasion. Our recent studies suggest that two parallel signaling pathways operate in E. coli invasion whose convergence is needed for the efficient uptake of the bacteria; however, OmpA expression is necessary for inducing these pathways (unpublished results).
In conclusion, our data demonstrate that OmpA binds to Ecgp via a carbohydrate epitope and the peptide regions of Ecgp for further signaling events that are necessary for internalization of bacteria into HBMEC. The Ecgp-Ab reactivity on human brain tissues, especially on vascular endothelium, highlights the importance of this molecule in E. coli traversal across the blood-brain barrier. Use of soluble Ecgp as a blocking agent in the newborn rat model of E. coli meningitis would provide insights into the possibility of this using this molecule as a therapeutic target.
Acknowledgments
I thank K. S. Kim and M. F. Stins, Division of Infectious Diseases, Johns Hopkins University School of Medicine, Baltimore, Maryland, for providing HBMEC. I also thank Carol Wass for assistance in tissue culture, M. F. Stins for immunocytochemistry, and S. Filler for critical reading of the manuscript.
This work was supported by NIH grant AI40567.
Editor: E. I. Tuomanen
REFERENCES
- 1.Bar, R. S., B. L. Dake, and R. G. Rapanheimer. 1985. Sulfated glycosaminoglycans in cultured endothelial cells from capillaries and large vessels of human and bovine origin. Atherosclerosis 56:11-26. [DOI] [PubMed] [Google Scholar]
- 2.Clark, M. A., B. H. Hirst, and M. A. Jepson. 1998. M-cell surface betal integrin expression and invasin-mediated targeting of Yersinia pseudotuberculosis to mouse Peyer's patch M cells. Infect. Immun. 66:1237-1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Everest, P., J. Li, and G. Douce. 1996. Bordetella pertussis P69/pertactin protein and the P69/pertactin RGD motif in the adherence to and invasion of mammalian cell. Microbiology 142:3261-3268. [DOI] [PubMed] [Google Scholar]
- 4.Feldweg, A. M., and P. K. Srivastava. 1995. Molecular heterogeneity of tumor rejection antigen/heat shock protein GP96. Int. J. Cancer 63:310-314. [DOI] [PubMed] [Google Scholar]
- 5.Gladstone, I. M., R. A. Ehrenkranz, S. C. Edberg, and R. S. Baltimore. 1990. A ten-year review of neonatal sepsis and comparison with previous fifty-year experience. Pediatr. Infect. Dis. J. 9:819-825. [DOI] [PubMed] [Google Scholar]
- 6.Graham, L. D. 1994. Tumor rejection antigens of the hsp90 family (gp96) closely resemble tumor-associated heparanase enzymes. Biochem. J. 301:917-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang, S. H., C. A. Wass, Q. Fu, N. V. Prasadarao, M. F. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang, S. H., Y. H. Chen, Q. Fu, M. F. Stins, Y. Wang, C. A. Wass, and K. S. Kim. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaper, J. B. 1998. EPEC delivers the goods. Trends Microbiol. 6:169-172. [DOI] [PubMed] [Google Scholar]
- 10.Koedel, U., and H. W. Pfister. 1999. Models of experimental bacterial meningitis: role and limitations. Infect. Dis. Clin. N. Am. 13:549-577. [DOI] [PubMed] [Google Scholar]
- 11.Leib, S. L., and M. G. Tauber. 1999. Pathogenesis of bacterial meningitis. Infect. Dis. Clin. N. Am. 13:527-547. [DOI] [PubMed] [Google Scholar]
- 12.McCarthy, S. A., I. Kuzu, K. C. Gatter, and R. Bicknell. 1991. Hetergeneity of the endothelial cell and its role in organ preference of tumor metastasis. Trends Pharmacol. Sci. 12:462-467. [DOI] [PubMed] [Google Scholar]
- 13.Pong, A., and J. S. Bradley. 1999. Bacterial meningitis and the newborn infant. Infect. Dis. Clin. N. Am. 13:711-733. [DOI] [PubMed] [Google Scholar]
- 14.Potter, T. A., K. Grebe, B. Freiberg, and A. Kupfer. 2001. Formation of supramolecular activation clusters on fresh ex vivo CD8+ T cells after engagement of the T-cell antigen receptor and CD8 by antigen-presenting cells. Proc. Natl. Acad. Sci. USA 98:12624-12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1997. Identification and characterization of S-fimbria-binding sialoglycoproteins on brain microvascular endothelial cells. Infect. Immun. 65:2852-2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prasadarao, N. V., C. A. Wass, J. Hacker, K. Jann, and K. S. Kim. 1993. Adhesion of S-fimbriated Escherichia coli to brain glycolipids mediated by sfaA gene-encoded protein of S-fimbriae. J. Biol. Chem. 268:10356-10363. [PubMed] [Google Scholar]
- 17.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prasadarao, N. V., C. A. Wass, and K. S. Kim. 1996. Endothelial cell GlcNAcβ1-4GlcNAc epitopes for outer membrane protein A enhance traversal of Escherichia coli across the blood-brain barrier. Infect. Immun. 64:154-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prasadarao, N. V., C. A. Wass, M. F. Stins, H. Shimada, and K. S. Kim. 1999. Outer membrane protein A-promoted actin condensation of brain microvascular endothelial cells is required for Escherichia coli invasion. Infect. Immun. 67:5775-5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy, M. A., C. A. Wass, K. S. Kim, D. D. Schlaepfer, and N. V. Prasadarao. 2000. Involvement of focal adhesion kinase in Escherichia coli invasion of brain microvascular endothelial cells. Infect. Immun. 68:6423-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robert, J. and A. Minoret. 1999. Cell surface expression of the endoplasmic reticular heat shock protein gp96 is phylogenetically conserved. J. Immunol. 163:4133-4139. [PubMed] [Google Scholar]
- 22.Roseman, D. S., and J. U. Baenziger. 2000. Molecular basis of lutropin by the Man/GalNAc-4SO4 receptor. Proc. Natl. Acad. Sci. USA 297:9949-9954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stanley, P. 1993. Use of mammalian cell mutants to study the functions of N- and O-linked glycosylation, p. 181-222. In D. D. Roberts and R. P. Meacham (ed.), Cell surface extracellular glycoconjugates: structure and function. Academic Press, Inc., New York, N.Y.
- 24.Stins, M. F., N. V. Prasadarao, C. A. Wass, and K. S. Kim. 1999. Escherichia coli binding to and invasion of brain microvascular endothelial cells derived from humans and rats of different ages. Infect. Immun. 67:5522-5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stins, M. F., N. V. Prasadarao, L. Ibric, C. A. Wass, P. Luckett, and K. S. Kim. 1994. Binding characteristics of S-fimbriated Escherichia coli to isolated brain microvascular endothelial cells. Am. J. Pathol. 145:1228-1236. [PMC free article] [PubMed] [Google Scholar]
- 26.Unhanand, M., M. M. Mustafa, G. H. McCracken, J. D. Nelsen. 1993. Gram-negative enteric bacillary meningitis: a twenty-year experience. J. Pediatr. 122:15-21. [DOI] [PubMed] [Google Scholar]
- 27.van Putten, J. P. M., and S. M. Paul. 1995. Binding of syndecan like cell surface proteoglycan receptors is required for Neisseria gonorrhoeae entry into human mucosal cells. EMBO J. 14:2144-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vercellotti, G. M., M. Dobson, A. E. Schorer, and C. F. Moldow. 1988. Endothelial cell heterogeneity: antioxidant profiles determine vulnerability to oxidant injury. Proc. Soc. Exp. Biol. Med. 187:181-189. [DOI] [PubMed] [Google Scholar]
- 29.Virji, M., K. Makepeace, and E. R. Moxon. 1994. Distinct mechanisms of interactions of Ope-expressing meningococci at apical and basolateral surfaces of human endothelial cells: the role of integrins in apical interactions. Mol. Microbiol. 14:173-184. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Y., S. H. Huang, C. A. Wass, M. F. Stins, and K. S. Kim. 1999. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 67:4751-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watarai, M., S. Funato, and C. Sasakawa. 1996. Interaction of Ipa proteins of Shigella flexneri with α5β1 integrin promotes entry of the bacteria into mammalian cells. J. Exp. Med. 183:991-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weel, J. F., C. T. P Hopman, and J. P. M. van Putten. 1991. In situ expression and localization of Neisseria gonorrhoeae opacity proteins in infected epithelial cells: apparent role of Opa proteins in cellular invasion. J. Exp. Med. 173:13905-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiest, D. L., A. Bhandoola, J. Punt, G Kreibich, D. McKean, and A. Singer. 1997. Incomplete endoplasmic reticulum (ER) retention in immature thymocytes as revealed by surface expression of “ER-resident” molecular chaperones. Proc. Natl. Acad. Sci. USA 94:1884-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zetter, B. R. 1988. Endothelial heterogeneity: influence of vessel size, organ localization, and species specificity on the properties of cultured endothelial cells, p. 63-79. In U. S. Ryan (ed.), Endothelial cell, vol. II. CRC Press, Boca Raton, Fla. [Google Scholar]