Abstract
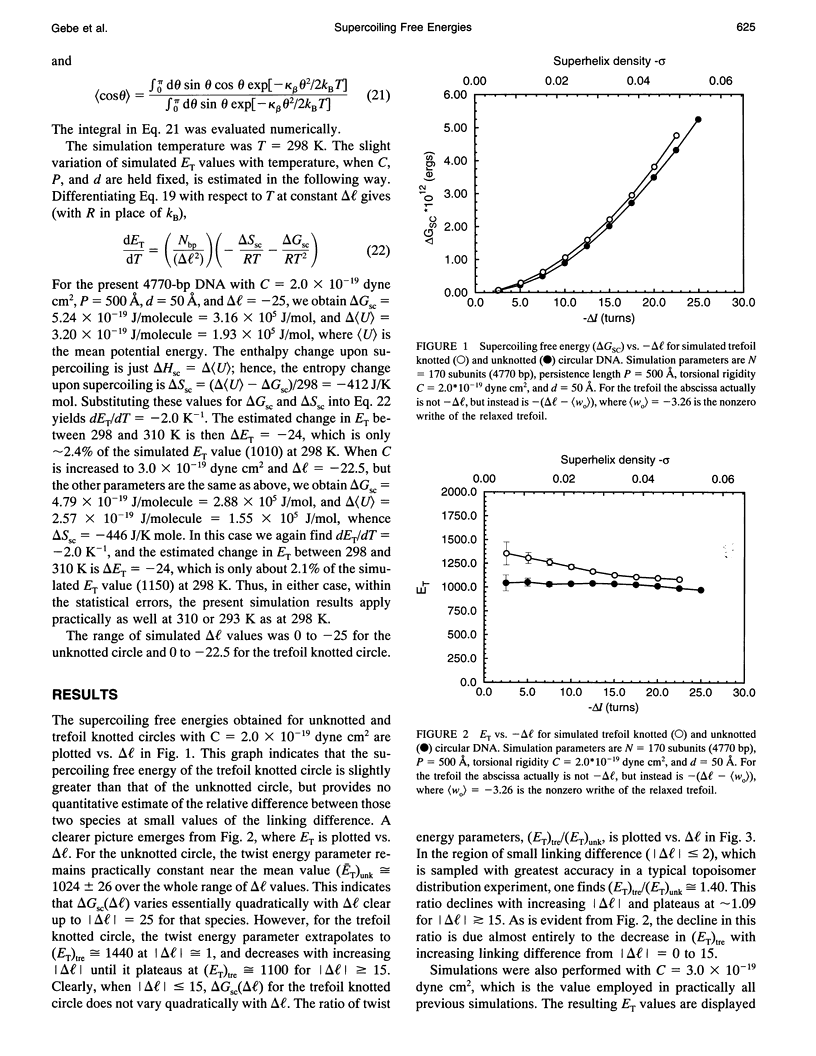
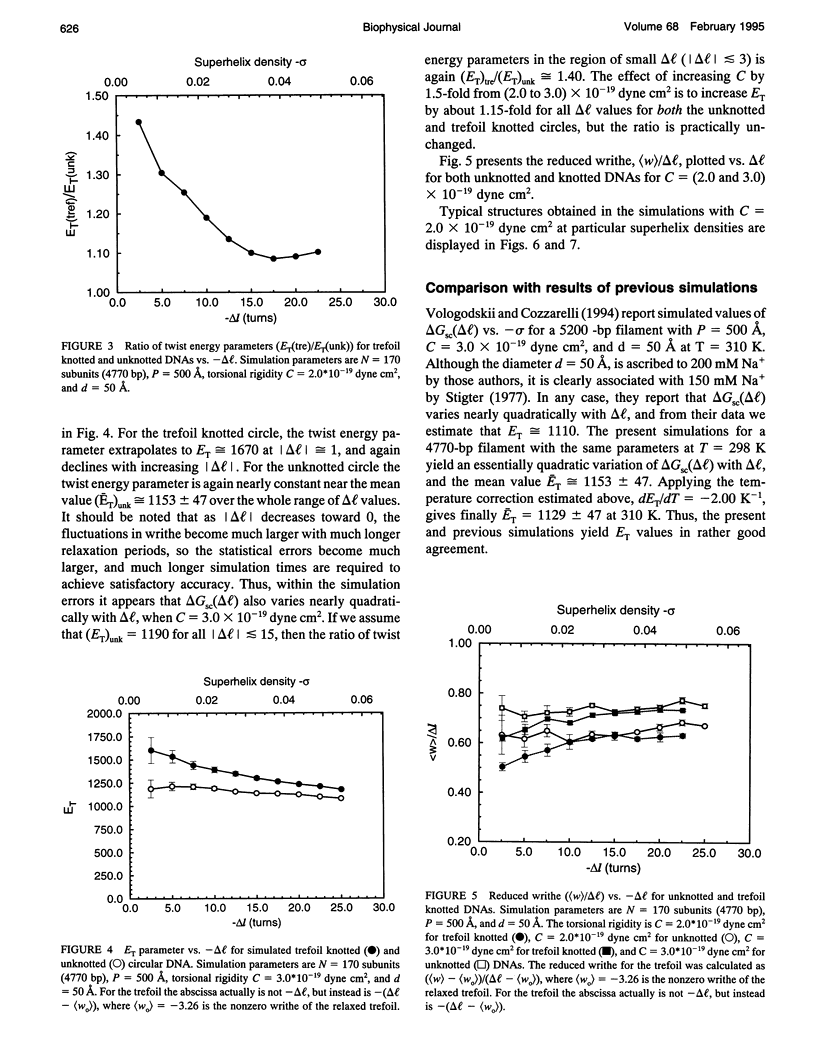
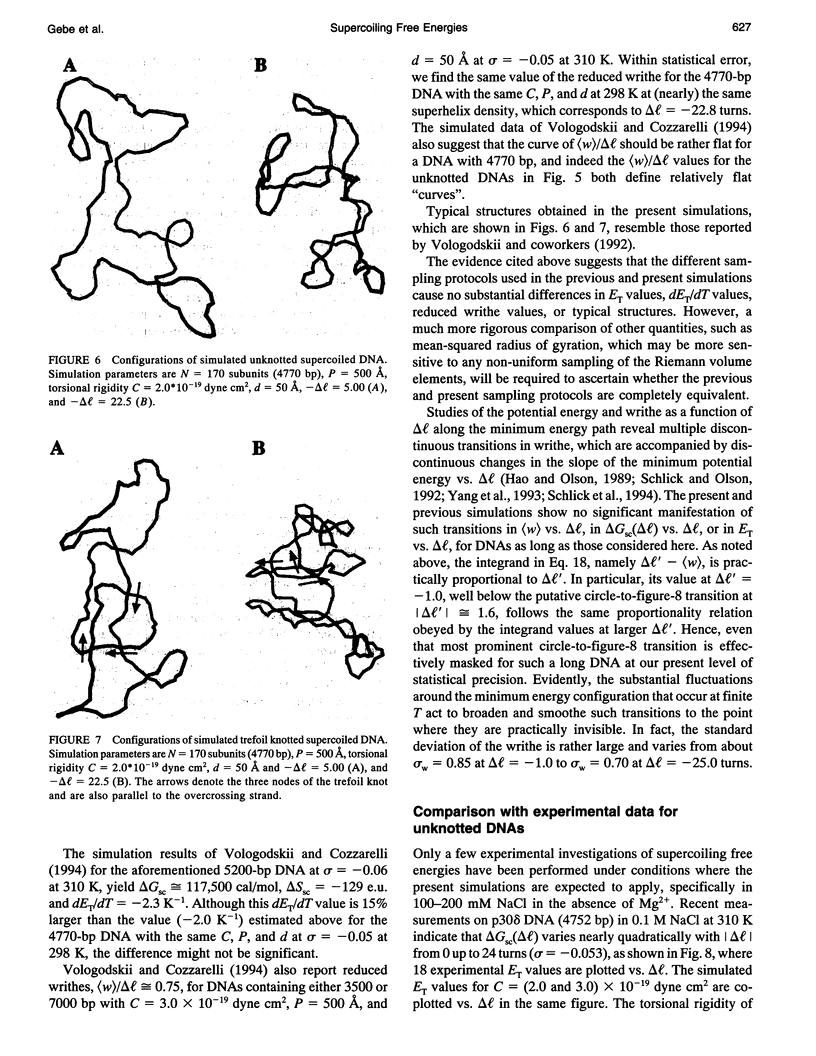
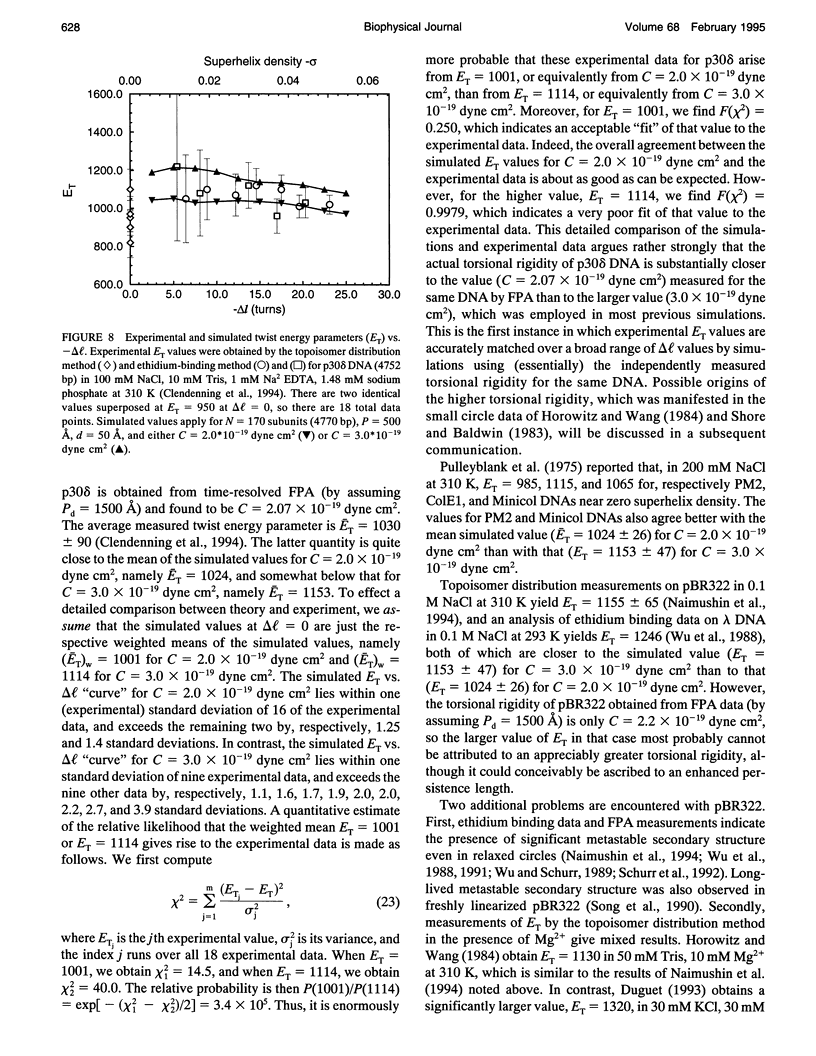
A new Monte Carlo (MC) algorithm is proposed for simulating inextensible circular chains with finite twisting and bending rigidity. This new algorithm samples the relevant Riemann volume elements in a uniform manner, when the constraining potential vanishes. Simulations are performed for filaments comprising 170 subunits, each containing approximately 28 bp, which corresponds to a DNA length of 4770 bp. The bending rigidity is chosen to yield a persistence length, P = 500 A, and the intersubunit potential is taken to be a hard-cylinder potential with diameter d = 50 A. This value of d yields the same second virial coefficient as the electrostatic potential obtained by numerical solution of the Poisson-Boltzmann equation for 150 mM salt. Simulations are performed for unknotted circles and also for trefoil knotted circles using two different values of the torsional rigidity, C = (2.0 and 3.0) x 10(-19) dyne cm2. In the case of unknotted circles, the simulated supercoiling free energy varies practically quadratically with linking difference delta l. The simulated twist energy parameter ET, its slope dET/dT, and the mean reduced writhe <w>/delta l for C = 3 x 10(-19) dyne cm2 all agree well with recent simulations for unknotted circles using the polygon-folding algorithm with identical P, d, and C. The simulated ET vs. delta l data for C = 2.0 x 10(-19) dyne cm2 agree rather well with recent experimental data for p30 delta DNA (4752 bp), for which the torsional rigidity, C = 2.07 x 10(-19) dyne cm2, was independently measured. The experimental data for p30 delta are enormously more likely to have arisen from C = 2.0 x 10(-19) than from C = 3.0 x 10(-19) dyne cm2. Serious problems with the reported experimental assessments of ET for pBR322 and their comparison with simulated data are noted. In the case of a trefoil knotted DNA, the simulated value, (ET)tre, exceeds that of the unknotted DNA, (ET)unk, by approximately equal to 1.40-fold at magnitude of delta l = 1.0, but declines to a plateau about 1.09-fold larger than (ET)unk when magnitude of delta l > or = 15. Although the predicted ratio, (ET)tre/(ET)unk approximately equal to 1.40, agrees fairly well with recent experimental measurements on a 5600-bp DNA, the individual measured ET values, like some of those reported for pBR322, are so large that they cannot be simulated using P = 500 A, d = 50 A, and any previous experimental estimate of C.
Full text
PDF










Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Clendenning J. B., Naimushin A. N., Fujimoto B. S., Stewart D. W., Schurr J. M. Effect of ethidium binding and superhelix density on the supercoiling free energy and torsion and bending constants of p30 delta DNA. Biophys Chem. 1994 Nov;52(3):191–218. doi: 10.1016/0301-4622(94)00038-l. [DOI] [PubMed] [Google Scholar]
- Clendenning J. B., Schurr J. M. Circularization of small DNAs in the presence of ethidium: a theoretical analysis. Biopolymers. 1994 Jul;34(7):849–868. doi: 10.1002/bip.360340705. [DOI] [PubMed] [Google Scholar]
- Duguet M. The helical repeat of DNA at high temperature. Nucleic Acids Res. 1993 Feb 11;21(3):463–468. doi: 10.1093/nar/21.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D., Lukashin A. V., Anshelevich V. V., Vologodskii A. V. Torsional and bending rigidity of the double helix from data on small DNA rings. J Biomol Struct Dyn. 1985 Feb;2(5):1005–1012. doi: 10.1080/07391102.1985.10507616. [DOI] [PubMed] [Google Scholar]
- Fujimoto B. S., Schurr J. M. Dependence of the torsional rigidity of DNA on base composition. Nature. 1990 Mar 8;344(6262):175–177. doi: 10.1038/344175a0. [DOI] [PubMed] [Google Scholar]
- Gray H. B., Jr, Hearst J. E. Flexibility of native DNA from the sedimentation behavior as a function of molecular weight and temperature. J Mol Biol. 1968 Jul 14;35(1):111–129. doi: 10.1016/s0022-2836(68)80041-5. [DOI] [PubMed] [Google Scholar]
- Hinton D. M., Bode V. C. Ethidium binding affinity of circular lambda deoxyribonucleic acid determined fluorometrically. J Biol Chem. 1975 Feb 10;250(3):1061–1070. [PubMed] [Google Scholar]
- Horowitz D. S., Wang J. C. Torsional rigidity of DNA and length dependence of the free energy of DNA supercoiling. J Mol Biol. 1984 Feb 15;173(1):75–91. doi: 10.1016/0022-2836(84)90404-2. [DOI] [PubMed] [Google Scholar]
- Klenin K. V., Vologodskii A. V., Anshelevich V. V., Dykhne A. M., Frank-Kamenetskii M. D. Computer simulation of DNA supercoiling. J Mol Biol. 1991 Feb 5;217(3):413–419. doi: 10.1016/0022-2836(91)90745-r. [DOI] [PubMed] [Google Scholar]
- Klenin K. V., Vologodskii A. V., Anshelevich V. V., Klishko VYu, Dykhne A. M., Frank-Kamenetskii M. D. Variance of writhe for wormlike DNA rings with excluded volume. J Biomol Struct Dyn. 1989 Feb;6(4):707–714. doi: 10.1080/07391102.1989.10507731. [DOI] [PubMed] [Google Scholar]
- Le Bret M. Monte Carlo computation of the supercoiling energy, the sedimentation constant, and the radius of gyration of unknotted and knotted circular DNA. Biopolymers. 1980 Mar;19(3):619–637. doi: 10.1002/bip.1980.360190312. [DOI] [PubMed] [Google Scholar]
- Levene S. D., Crothers D. M. Topological distributions and the torsional rigidity of DNA. A Monte Carlo study of DNA circles. J Mol Biol. 1986 May 5;189(1):73–83. doi: 10.1016/0022-2836(86)90382-7. [DOI] [PubMed] [Google Scholar]
- Naimushin A. N., Clendenning J. B., Kim U. S., Song L., Fujimoto B. S., Stewart D. W., Schurr J. M. Effect of ethidium binding and superhelix density on the apparent supercoiling free energy and torsion constant of pBR322 DNA. Biophys Chem. 1994 Nov;52(3):219–226. doi: 10.1016/0301-4622(94)00037-k. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson B. H., Lerman L. S., Beth A. H., Frisch H. L., Dalton L. R., Auer C. Analysis of double-helix motions with spin-labeled probes: binding geometry and the limit of torsional elasticity. J Mol Biol. 1980 May 5;139(1):19–44. doi: 10.1016/0022-2836(80)90113-8. [DOI] [PubMed] [Google Scholar]
- Schlick T., Olson W. K. Supercoiled DNA energetics and dynamics by computer simulation. J Mol Biol. 1992 Feb 20;223(4):1089–1119. doi: 10.1016/0022-2836(92)90263-j. [DOI] [PubMed] [Google Scholar]
- Schlick T., Olson W. K., Westcott T., Greenberg J. P. On higher buckling transitions in supercoiled DNA. Biopolymers. 1994 May;34(5):565–597. doi: 10.1002/bip.360340502. [DOI] [PubMed] [Google Scholar]
- Seidl A., Hinz H. J. The free energy of DNA supercoiling is enthalpy-determined. Proc Natl Acad Sci U S A. 1984 Mar;81(5):1312–1316. doi: 10.1073/pnas.81.5.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S. Y., Wang J. C. Knotting of a DNA chain during ring closure. Science. 1993 Apr 23;260(5107):533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- Shimada J., Yamakawa H. Moments for DNA topoisomers: the helical wormlike chain. Biopolymers. 1988 Apr;27(4):657–673. doi: 10.1002/bip.360270409. [DOI] [PubMed] [Google Scholar]
- Shimada J., Yamakawa H. Statistical mechanics of DNA topoisomers. The helical worm-like chain. J Mol Biol. 1985 Jul 20;184(2):319–329. doi: 10.1016/0022-2836(85)90383-3. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983 Nov 15;170(4):983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- Soda K., Wada A. Dynamic light-scattering studies on thermal motions of native DNAs in solution. Biophys Chem. 1984 Oct;20(3):185–200. doi: 10.1016/0301-4622(84)87023-4. [DOI] [PubMed] [Google Scholar]
- Song L., Fujimoto B. S., Wu P. G., Thomas J. C., Shibata J. H., Schurr J. M. Evidence for allosteric transitions in secondary structure induced by superhelical stress. J Mol Biol. 1990 Jul 5;214(1):307–326. doi: 10.1016/0022-2836(90)90163-g. [DOI] [PubMed] [Google Scholar]
- Stigter D. Interactions of highly charged colloidal cylinders with applications to double-stranded. Biopolymers. 1977 Jul;16(7):1435–1448. doi: 10.1002/bip.1977.360160705. [DOI] [PubMed] [Google Scholar]
- Taylor W. H., Hagerman P. J. Application of the method of phage T4 DNA ligase-catalyzed ring-closure to the study of DNA structure. II. NaCl-dependence of DNA flexibility and helical repeat. J Mol Biol. 1990 Mar 20;212(2):363–376. doi: 10.1016/0022-2836(90)90131-5. [DOI] [PubMed] [Google Scholar]
- Thomas J. C., Schurr J. M. Fluorescence depolarization and temperature dependence of the torsion elastic constant of linear phi 29 deoxyribonucleic acid. Biochemistry. 1983 Dec 20;22(26):6194–6198. doi: 10.1021/bi00295a024. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. Annu Rev Biophys Biomol Struct. 1994;23:609–643. doi: 10.1146/annurev.bb.23.060194.003141. [DOI] [PubMed] [Google Scholar]
- Vologodskii A. V., Levene S. D., Klenin K. V., Frank-Kamenetskii M., Cozzarelli N. R. Conformational and thermodynamic properties of supercoiled DNA. J Mol Biol. 1992 Oct 20;227(4):1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- Wilcoxon J., Schurr J. M. Temperature dependence of the dynamic light scattering of linear phi 29 DNA: implications for spontaneous opening of the double-helix. Biopolymers. 1983 Oct;22(10):2273–2321. doi: 10.1002/bip.360221011. [DOI] [PubMed] [Google Scholar]
- Wu P. G., Fujimoto B. S., Song L., Schurr J. M. Effect of ethidium on the torsion constants of linear and supercoiled DNAs. Biophys Chem. 1991 Dec;41(3):217–236. doi: 10.1016/0301-4622(91)85038-r. [DOI] [PubMed] [Google Scholar]
- Wu P. G., Song L., Clendenning J. B., Fujimoto B. S., Benight A. S., Schurr J. M. Interaction of chloroquine with linear and supercoiled DNAs. Effect on the torsional dynamics, rigidity, and twist energy parameter. Biochemistry. 1988 Oct 18;27(21):8128–8144. doi: 10.1021/bi00421a023. [DOI] [PubMed] [Google Scholar]
- Wu P., Schurr J. M. Effects of chloroquine on the torsional dynamics and rigidities of linear and supercoiled DNAs at low ionic strength. Biopolymers. 1989 Oct;28(10):1695–1703. doi: 10.1002/bip.360281005. [DOI] [PubMed] [Google Scholar]


