Abstract
Enteroaggregative Escherichia coli (EAEC) is recognized as an emerging cause of diarrhea in children and adults worldwide, and recent studies have implicated EAEC in persistent diarrhea in patients infected with human immunodeficiency virus (HIV). In this study, we identified aggregative adhesion fimbria type III (AAF-III) in isolate 55989, a typical EAEC strain. Analysis of the sequence of the plasmid-borne agg-3 gene cluster encoding AAF-III showed this cluster to be closely related to the agg and aaf operons and to the afa operons carried by diffusely adherent pathogenic E. coli. We investigated the adhesion properties of a collection of 25 EAEC strains isolated from HIV-infected patients presenting with persistent diarrhea. We found that a minority of strains (36%) carried sequences similar to those of the agg and aaf operons, which encode AAF-I and AAF-II, respectively. We developed PCR assays specific for the agg-3 operon. In our collection, the frequency of AAF-III strains was similar (12%) to that of AAF-I strains (16%) but higher than that of AAF-II isolates (0%). Differences between EAEC strains in terms of the virulence factors present render detection of these strains difficult with the available DNA probes. Based on comparison of the agg, aaf, and agg-3 operons, we defined an AAF probe internal to the adhesion gene clusters and demonstrated that it was efficient for the identification of EAEC strains. We investigated 32 EAEC isolates, of which only 34.4% were detected with the classical CVD432 probe (detecting pAA virulence plasmids) whereas 65.6% were detected with the AAF probe.
Escherichia coli is the predominant facultative anaerobe of the human colonic flora, and most strains are harmless. However, certain E. coli strains cause intestinal and extraintestinal diseases in humans. Diarrheagenic E. coli strains generally adhere to mucosal epithelia early in the colonization of host tissues. This pathogenic property is associated with the production of adhesins, which are specific to the particular disease. Enteroaggregative E. coli (EAEC) strains constitute an emerging pathotype responsible for enteric illness. Epidemiological studies have now implicated EAEC strains in acute and persistent diarrhea in children, in food-borne diarrhea outbreaks, and in traveler's diarrhea (36). Several recent studies have reported an association between EAEC strains and persistent diarrhea in patients infected with the human immunodeficiency virus (HIV) in developing and industrialized countries (19, 21, 28, 32, 44). Eradication of EAEC from the stools of HIV-infected patients with persistent diarrhea by antibiotic treatment, leading to the resolution or improvement of intestinal symptoms, has been reported (17, 19, 43).
EAEC displays a characteristic aggregative adhesion (AA) pattern on the surfaces of HEp-2 or HeLa cells, with the bacteria adhering to each other to form a “stacked brick wall”-like structure on epithelial cells and glass coverslips. Two types of fimbriae, AA fimbriae types I and II (AAF-I and AAF-II), have been shown to be required for expression of an AA pattern. Analysis of the distribution of the genes encoding these fimbriae among EAEC strains revealed that only a minority of isolates produced these two types of fimbriae (12, 38, 41). Thus, EAEC strains must also produce other adhesive structures. In addition to adhesins, a variety of other virulence factors have been described, including the heat-stable enterotoxin (EAST1), the plasmid-encoded toxin (Pet), and a protein involved in intestinal colonization (Pic) (36). However, none of these factors are present in all EAEC isolates. Consequently, the molecular probes developed by genetic characterization of plasmid- and chromosome-borne virulence genes cannot be used in routine clinical laboratory testing for identification of EAEC. Identification remains dependent on production of the adhesins that generate the AA pattern.
EAEC strains are heterogeneous both with respect to symptoms developed by infected hosts and with respect to virulence genes. The aim of this study was to explore this diversity and to increase our understanding of the genetic basis of virulence factors in EAEC by identifying new adhesins. This study was made possible by characterization of a collection of E. coli isolates recovered from the stools of HIV-infected patients suffering from persistent diarrhea. We carried out molecular typing for a collection of 25 EAEC isolates, using probes and PCR assays for detection of adhesion-associated genes in order to evaluate their distribution. In addition, because EAEC identification depends on production of the adhesins that generate the AA pattern, we decided to develop an EAEC-sensitive probe by comparing the two AAF-encoding operons previously described and the new operon identified in our study, in order to identify common features.
(This work was presented in part at the 100th and 101st Meetings of the American Society for Microbiology, Los Angeles, Calif., 21 to 25 May 2000, and Orlando, Fla., 20 to 24 May 2001, respectively.)
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
EAEC strains have frequently been identified as agents of persistent diarrhea (of at least 14 days' duration) in etiological studies of HIV and diarrhea in adults in Bangui, Central African Republic (19), and Dakar, Senegal (17). Yves Germani of the Pasteur Institute of Bangui and Awa Aïdara-Kane of the Pasteur Institute of Dakar kindly provided us with 32 such EAEC isolates. Twenty-five of these strains were isolated from the stools of patients with CD4 lymphocyte counts of <499 cells/mm who presented with persistent diarrhea. In each case, E. coli accounted for 90 to 100% of the bacteria isolated on the bromocresol purple agar plate onto which stool specimens were streaked. At least three of the five colonies initially selected for adhesion assays displayed an AA pattern. One of these colonies was selected and sent to our laboratory. Seven EAEC stains were isolated from nondiarrheagenic stools from immunocompromised patients.
The prototype EAEC strains used in this study were JM221 (29), which produces AAF-I, and 042 (33), which produces AAF-II. These two strains were initially isolated from the stools of children with diarrhea in Mexico and Peru, respectively. Strain A30, carrying the afa-3 operon (22), and E. coli E2348/69 (1), kindly provided by J. Kaper, were used as diffusely adherent E. coli (DAEC) and enteropathogenic E. coli (EPEC) prototypes, respectively.
E. coli strains HB101 (6) and MC1061 (27) were used as hosts for the recombinant plasmids listed in Table 1. Vectors pBR322 (5), pUC18 (46), and pACY184 (7), as well as the cosmid vector pHC79 (9), were used in cloning experiments. E. coli AAEC185 (ΔfimB-H) (4), kindly provided by D. Gally, was used as a non-type 1 pilus-producing host strain for recombinant plasmids in electron microscopy experiments and invasion assays.
TABLE 1.
Recombinant plasmids used in this study
| Plasmid | Vector | Insert | Phenotypea
|
Reference | ||
|---|---|---|---|---|---|---|
| Aggregates in EPO | Production of fimbriae | HeLa cell binding | ||||
| pILL1268 | pHC79 | 45-kb Sau3A fragment from E. coli 55989 DNA containing the agg-3 gene cluster | + | + | + | This study |
| pILL1282 | pHC79 | pILL1268 with an agg-3A gene mutated by partial deletion and insertion of the kanamycin resistance cassette | − | − | − | This study |
| pILL1279 | pBR322 | 4.7-kb NheI-SphI PCR product from pILL1268 containing the agg-3 operon | + | + | + | This study |
| pILL1277 | pUC18 | 0.5-kb EcoRI-HindIII PCR product from pILL1268 containing an agg-3C internal fragment | ND | ND | ND | This study |
| pILL1151 | pUC18 | 10.5-kb BamHI fragment carrying the afa-3 gene cluster from A30 | ND | − | + | 15 |
| pILL1218 | pACY184 | ClaI fragment carrying the AAF-I biogenesis genes with a mutation in aggA | ND | − | − | 14 |
| Cosmid D6 | pCVD301 | Sau3A fragment from pAA2 carrying the AAF-II biogenesis genes | + | + | + | 13 |
ND, not determined.
Strains were routinely cultured in Luria-Bertani (LB) broth (or on LB agar) without glucose. For autoaggregation and infection assays, strains were grown in peptone water (or on peptone agar), without shaking, at 37°C. We selected for antibiotic resistance by adding antibiotics to the medium at the following concentrations: carbenicillin, 100 mg/liter; tetracycline, 10 mg/liter; kanamycin, 20 mg/liter; and chloramphenicol, 20 mg/liter.
DNA manipulations.
We used standard recombinant DNA techniques (27). Hybridization was performed on nitrocellulose filters under stringent (65°C) conditions, unless otherwise stated, with probes labeled with [32P]dCTP by using the Megaprime DNA labeling system (Amersham). PCR was carried out as previously described (24) with Taq polymerase (Amersham) for detection assays and with the Expand High-Fidelity PCR System (Roche Molecular Biochemicals) for amplification of DNA fragments for cloning. Most of the probes and oligonucleotides used are listed in Table 2. The astA, pet, shf, aspU, and irp2 sequences were detected with primers described elsewhere (11, 45). The sequences of the primers used to amplify the pic gene were 5′-CGTAACGCCTCGCTGAACAG-3′ and 5′-ACCGGCCTGAATATGCCCGT-3′ (J. Nataro, personal communication). A genomic library, consisting of 35- to 50-kb Sau3A DNA fragments from E. coli 55989, was constructed in the pHC79 cosmid vector digested with BamHI, as previously described (23).
TABLE 2.
Primers and probes used in this study
| Virulence factor detected | Targeted region
|
PCR primers or restriction fragment | Reference | ||
|---|---|---|---|---|---|
| Genes | Description | Size | |||
| EAEC factors | |||||
| CVD432 probe | Fragment of the pAA virulence plasmida | Unknown function | 630 bp | 5′-CTGGCGAAAGACTGTATCAT-3′ | 40 |
| 5′-CAATGTATAGAAATCCGCTGTT-3′ | |||||
| AAF probe | agg-3C | AAF-III usher subunit | agg3C PCR product | This study | |
| AAF-I | aggA | AAF-I fimbrial subunit | 450 bp | 5′-TTAGTCTTCTATCTAGGG-3′ | 11 |
| 5′-AAATTAATTCCGGCATGG-3′ | |||||
| AAF-II | aafA | AAF-II fimbrial subunit | 332 bp | HindIII-EcoRI fragment from pJC2 | 10 |
| aafDA | AAF-II chaperone and fimbrial subunits | 1.7 kb | EcoRI fragment from pJC2 | ||
| AAF-III | agg-3A | AAF-III fimbrial subunit | 462 bp | 5′-GTATCATTGCGAGTCTGGTATTCAG-3′ | This study |
| 5′-GGGCTGTTATAGAGTAACTTCCAG-3′ | |||||
| agg-3C | AAF-III usher subunit | 485 bp | 5′-GTTTGGAACCGGGAATTAACATTG-3′ | This study | |
| 5′-ATACTTTAGATACCCCTCACGCAG-3′ | |||||
| AggR | aggR | AAF-I and AAF-II tran- scriptional activator | 308 bp | 5′-CTAATTGTACAATCGATGTA-3′ | 11 |
| 5′-CTGAAGTAATTCTTGAAT-3′ | |||||
| DAEC factor, AFA | afaBC | AFA chaperone and usher subunits | 750 bp | 5′-GCTGGGCAGCAAACTGATAACTCTC-3′ | 24 |
| 5′-CATCAAGCTGTTTGTTCGTCCGCCG-3′ | |||||
| EPEC factors | |||||
| EAF plasmidb | pJN16 fragment | Unknown function | 1 kb | BamHI-SalI fragment | 33 |
| Bundle-forming pili (Bfp) | bfpA | Bfp major subunit | 324 bp | 5′-CAATGGTGCTTGCGCTTGCT-3′ | 42 |
| 5′-GCCGCTTTATCCAACCTGGT-3′ | |||||
| Intimin | eaeA | Structural gene | 494 bp | 5′-GCAAATTTAGGTGCGGGTCAGCGTT-3′ | 18 |
| 5′-GGCTCAATTTGCTGAGACCACGGTT-3′ | |||||
The CVD432 or pAA probe correlates with the presence of a pAA virulence plasmid (2).
Plasmid encoding Bfp.
DNA sequencing and sequence analysis.
We sequenced double-stranded DNA using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.). Sequence data were analyzed with ABI version 3.0.1b3 software. Sequences were screened for similarity to previously published sequences by using the BLASTN and BLASTX computer programs at the National Center for Biotechnology Information. Multiple sequence alignments were generated with the CLUSTAL W program.
Allelic replacement.
We carried out allelic exchange for the agg-3A gene, using pILL1268 as previously described (8). The strategy involved the use of recombination to replace a 419-bp agg-3A internal segment (bp 47 to 472) with a PCR product containing the nonpolar kanamycin resistance cassette and 80-nucleotide [nt] homology arms. We used the following primers for PCR: Agg3A/5′Km (5′-GAATACAATATTTAGGAGATTTTTCATATGAAAAAGATAAGTATCATTGCGAGTCTGGTATTCAGCTTGTACAGTGGGTTCTAACTAGGAGGAATAAATG-3′) and Agg3A/3′Km (5′-CATCCACTTATGCGATCAAAAAGCGCTAAGTGGGGCTTAGCGCACTGTTTTTATTTATGGTAAAACCATAACACCATGGGTTCCCTCCAGGTACTAAAAC-3′).
Electron microscopy.
For observation of cellular appendages, bacteria were negatively stained with 1% ammonium molybdate in water as previously described (25). Staining of some samples was enhanced by rotary platinum shadowing. Transmission electron microscopy of infected HeLa cells was carried out as previously described (20).
Adhesion and invasion assays.
Human HeLa (ECACC 84121901) and Caco-2 cells were maintained as previously described (20, 39). Intestine-407 (Int407) cells were maintained in Dulbecco's modified Eagle medium (GIBCO) supplemented with 1% glutamine and 10% fetal calf serum. They were cultured at 37°C in 5% CO2-95% air. Adhesion and invasion assays were performed as previously described (20).
Nucleotide sequence accession number.
The GenBank accession number for the 12,012-bp region of pILL1268 reported in this paper is AF411067.
RESULTS
Identification of a new variant AAF.
To identify AA factors (other than AAF-I and AAF-II) involved in the interaction of EAEC strains with epithelial cells, we constructed a cosmid library from isolate 55989. This isolate is a typical EAEC strain, containing both chromosomal and plasmid-borne genes associated with EAEC virulence, but it does not contain genes encoding AAF-I or AAF-II (Table 3). This strain displays a typical AA (AAt) pattern, with bacterial aggregates adhering to both cells and glass coverslips (Fig. 1A), and it forms aggregates in static broth cultures. A role for autoagglutination via lipopolysaccharide (26) in such phenomena has been ruled out because no macroscopically visible aggregates are observed if bacteria grown on agar plates are suspended in 2% NaCl and shaken on a glass slide at room temperature.
TABLE 3.
Characterization of the adhesion properties of diarrhea-associated EAEC strains isolated from HIV-positive patients
| Isolate | Adhesion phenotypeb | Detectiona of:
|
|||||
|---|---|---|---|---|---|---|---|
| EAEC factors
|
DAEC factor, AFA | ||||||
| pAA | Adhesins
|
AggR | |||||
| aggA | aafA | aag-3Ac | |||||
| 56390 | AAt | + | + | − | − | + | − |
| 645125 | AAt | + | + | − | − | + | − |
| 384P | AAt | + | +/− | − | − | − | − |
| 55989d | AAt | + | − | − | + | + | − |
| 387Ad | AAt | + | − | −c | + | + | − |
| 885254d | AAt | + | − | − | + | + | − |
| 11097 | AAt | + | − | − | − | + | − |
| 11006 | AAt | + | +/− | − | − | + | − |
| 11053 | AAII | + | − | − | − | + | − |
| 381A | AAt | − | − | −c | − | − | − |
| 911277 | AAt | − | − | − | − | − | − |
| 041344 | AAt | − | − | − | − | − | − |
| 11074 | AAt | − | − | − | − | − | − |
| 50A | AAll | − | − | −c | − | − | − |
| 113A | AAll | − | − | −c | − | − | − |
| 374P | AAll | − | − | − | − | − | − |
| 36197A | AAll | − | − | − | − | − | − |
| 100A | AAll | − | − | − | − | − | − |
| 647129 | AAll | − | − | − | − | − | − |
| 495P | AAll | − | − | − | − | − | − |
| 934290 | AAll | − | − | − | − | − | − |
| 650139 | AAll | − | − | − | − | − | − |
| 950305 | AA/DA | − | − | −c | − | − | + |
| 148A | AA/DA | − | − | − | − | − | + |
| 504P | AA/DA | − | − | − | − | − | − |
+, positive detection; −, absence of detection; +/−, the strain was positive by hybridization and negative by PCR. No strain was positive for EPEC factors.
AAll, low level of aggregative adhesion; AA/DA, mixed phenotype.
The same results were obtained with the agg-3C amplicon used as a probe.
Strain tested positive for shf, aspU, astA, pic, and irp2 sequences.
Strain negative with the aafA probe but positive with the aafDA probe.
FIG. 1.
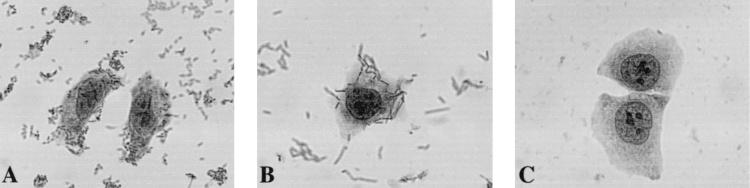
Photomicrographs of HeLa cells infected with AAF-III-producing strains: the wild-type strain 55989 (A) and the recombinant HB101(pILL1268) (B). Bacteria displayed an aggregative pattern of adhesion to the cells and, to a lesser extent, to glass coverslips. (C) HB101 (negative control).
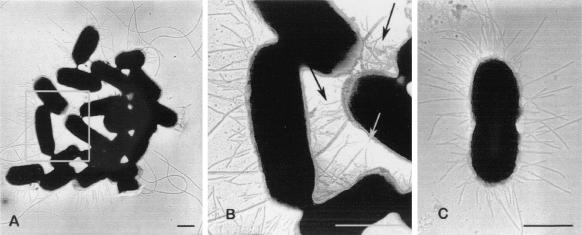
Bacteriophage lambda particles carrying recombinant cosmid molecules containing fragments of 55989 DNA were prepared and used to infect E. coli HB101. We screened 1,460 recombinant HB101 strains for the autoagglutination properties of the parental 55989 strain and identified 23 clones that formed aggregates in static broth cultures. The recombinant cosmid clone HB101(pILL1268) was selected for further study. HB101(pILL1268) displayed a pattern of adhesion to HeLa cells similar to that of strain 55989 (Fig. 1B). The pILL1268 cosmid carried an insert of about 45 kb. Detection of sequences specific to pAA virulence plasmids in this clone indicated that the pILL1268 insert corresponded to plasmid sequences. PCR experiments revealed that, like the parental strain 55989, HB101(pILL1268) carried aggR, aspU, and astA sequences, which were mapped to the cosmid insert by Southern hybridization (Fig. 2). Electron microscopy of the parental strain 55989 grown in static broth showed that the bacterial surface was surrounded by long, flexible fimbrial structures. These fimbriae seemed to be involved in autoagglutination, facilitating contact between bacteria for aggregate formation (Fig. 3A and B). Although no surface appendages were detected for strain AAEC185 under the same conditions (data not shown), long fimbriae (diameter, 3 to 5 nm) similar to those of the parental strain were observed on the surface of AAEC185 transformed with pILL1268 (Fig. 3C). Unlike AAF-I and AAF-II, which formed bundles (10, 34), the novel fimbrial structure encoded by pILL1268, designated AAF-III, was most commonly observed as individual filaments.
FIG. 2.
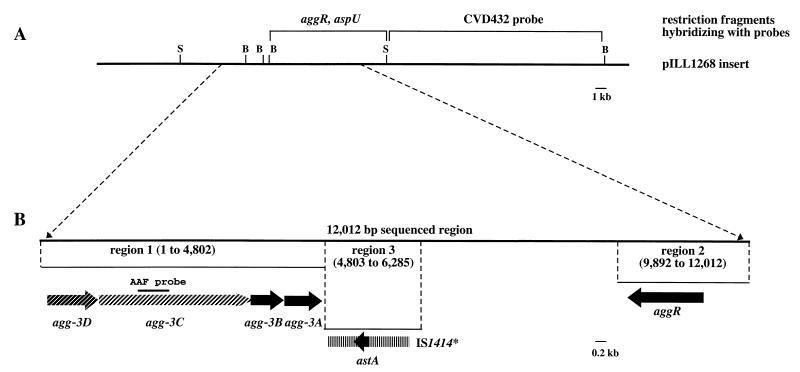
Structure of the agg-3 region. (A) Physical map of the pILL1268 insert. Restriction fragments hybridizing with the aggR, aspU, and CVD432 probes are indicated. (B) Genetic organization of the 12,012-bp sequenced region. The locations and directions of transcription of the ORFs described in this study are indicated by arrows. The truncated IS1414 element (IS1414∗) is indicated by a box. The region used as the AAF probe is also shown. Abbreviations: B, BamHI; S, SalI.
FIG. 3.
Electron micrographs of AAF-III-producing E. coli strain preparations negatively stained with 1% aqueous ammonium molybdate and shadowed with platinum. (A) Strain 55989; (B) detail of the region framed in panel A; (C) AAEC185(pILL1268). The wild-type 55989 isolate produces both flagella and long, flexible fimbriae, which are involved in the cohesion of bacterial aggregates, as indicated by arrows in panel B. AAEC185(pILL1268) produces fimbriae similar to those observed in 55989. These fimbriae are designated AAF-III. Bars, 0.5 μm.
Internalization of bacteria producing AAF-III.
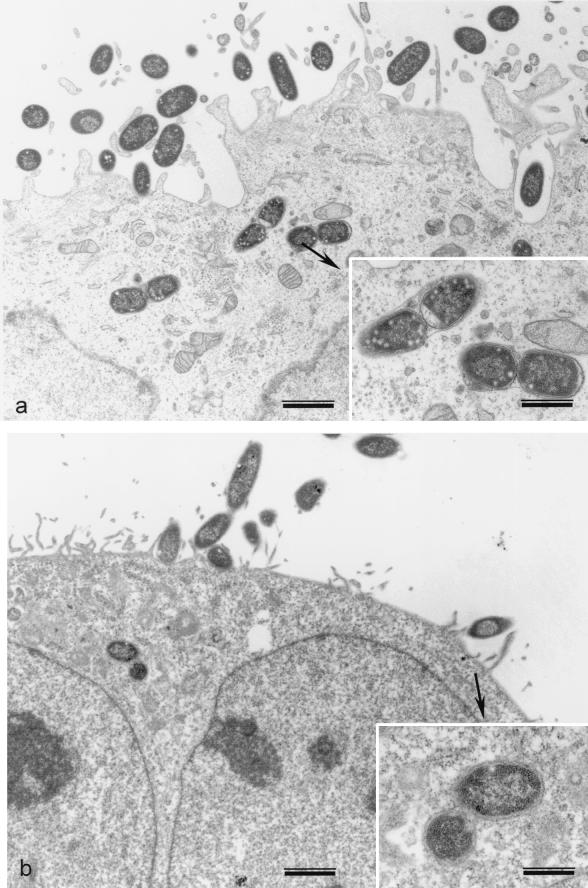
We investigated the invasion properties of strains producing AAF-III by comparing those of the wild-type strain 55989 and the recombinant strain AAEC185(pILL1279) with those of the EPEC strain E2348/69, used as a positive control, and strain AAEC185, used as a negative control. The results of invasion assays suggested a low level of internalization for AAF-III-producing strains (Table 4). Transmission electron microscopy of HeLa cells infected with strain 55989 or HB101(pILL1268) confirmed these data. For both strains, bacteria were observed within the cells. Internalized bacteria were present within membrane-bound vacuoles, frequently with 2 bacteria per vacuole for isolate 55989 (Fig. 4).
TABLE 4.
Invasion of HeLa cells by E. coli strains producing AAF-III
| Strain | Virulence factor(s) | % Invasiona (mean ± SD) | Relative % invasionb |
|---|---|---|---|
| E2348/69 (EPEC) | Bfp, Eaec | 0.22 ± 0.05 | 100 |
| 55989 | AAF-III | 0.05 ± 0.02 | 22.7 |
| AAEC185 (pILL1279) | AAF-III | 0.02 ± 0.005 | 9.1 |
| AAEC185d | None | 0.001 ± 0.0007 | 0.45 |
Bacteria resisting treatment with gentamicin as a percentage of the number of bacteria present in the original inoculum. Results are means ± standard deviations from two experiments, each performed in duplicate.
Invasion relative to that with E. coli E2348/69, which was defined as 100%.
Bfp, bundle-forming pili; Eae, intimin.
Similar results were obtained with AAEC185 harboring pBR322.
FIG. 4.
Transmission electron micrographs of HeLa cells infected with E. coli strains expressing the agg-3 operon. (a) Strain 55989; (b) HB101 carrying the recombinant plasmid pILL1268. Bars, 1 μm. (Insets) Intracellular bacteria. Bars, 0.5 μm.
Sequence analysis of agg-3 and associated loci.
We characterized the AAF-III-encoding genes by partial sequence determination of pILL1268, initiated from the aggR region (Fig. 2B). We sequenced a 12,012-bp region. Within this sequence, we identified regions displaying similarity to previously described AAF-encoding gene clusters (the agg and aaf operons, encoding AAF-I and AAF-II, respectively). One such region was the agg-3 gene cluster (region 1), which comprises four open reading frames (ORFs), transcribed in the same orientation, mapping to the same loci as the aggD, aggC, aggB, and aggA genes in the agg operon. We investigated whether agg-3 genes conferred adhesion properties by PCR amplification of a 4.7-kb agg-3-containing region of pILL1268. The amplicon was ligated into pBR322. The resulting plasmid, pILL1279, conferred on the host strains (MC1061 and AAEC185) the ability to produce fimbriae involved in bacterial aggregation and to bind HeLa cells. For three of the ORFs, agg-3D, agg-3C, and agg-3B, a BLASTX search predicted amino acid sequence similarities between the products of AAF-encoding operons and those of afa operons. The Agg-3D product displays significant sequence similarity to putative periplasmic chaperone proteins such as AggD, AafD, and AfaB (63 to 67% identity and 77 to 82% similarity), and the Agg-3C product is similar to the AggC, AafC, and AfaC putative usher proteins (65 to 67% identity and 79% similarity). Significant similarities were found between the Agg-3B protein and members of the invasin family, for which AfaD-III (from the afa-3 operon) is the prototype (16). Agg-3B is 58, 59, and 54% identical to AggB, AafB, and AfaD-III, respectively. The sequence similarities extend over the full length of the proteins, and Agg-3B contains the highly conserved 8-amino-acid (aa) region (ATGRXXCR) described for all proteins of the family (14). The agg-3A gene was located 174 bp downstream of agg-3B. A potential ribosome-binding site precedes the start codon, and a putative signal sequence was identified in the 165-aa polypeptide, which displays no significant sequence similarity to any protein sequence in the database. However, alignments of the Agg-3A, AggA, and AafA sequences highlighted the conservation of residues described as typical of the family of adhesins encoded by afa-related operons (10), including 2 cysteine residues separated by 31 to 33 aa in the N-terminal moiety of the mature product and a glycine near the C terminus (Fig. 5). Agg-3A was demonstrated to function as an adhesin by replacing part of agg-3A by a nonpolar kanamycin cassette in pILL1268. The resulting plasmid, pILL1282, did not confer on the host strain aggregate formation in static broth culture, fimbria production, or adhesion to HeLa cells (Table 1).
FIG. 5.
Sequence alignment of adhesins encoded by the agg-3, agg, and aaf operons. Gaps, indicated by dashes, have been inserted to optimize the alignment. Asterisks and points represent identical and similar residues, respectively, identified with the CLUSTAL W program. Numbers correspond to amino acid positions in the protein encoded by the agg-3 gene cluster. The predicted translational start site of Agg-3A is indicated by +1.
A 2,121-bp region of the pILL1268 insert (region 2) located 5 kb downstream of the stop codon of the aggA gene was found to be very similar (99%) to region 2, which is involved in the production of AAF-I. Sequence alignment showed that this region contained a gene 100% identical to the previously published aggR gene, which encodes a transcriptional activator (37).
A third interesting region (region 3) follows the agg-3 gene cluster (Fig. 2B). This region contains an ORF very similar to the astA gene, encoding the EAST1 toxin, from the enterotoxigenic E. coli (ETEC) strain 27D (31). A single base difference between the two nucleotide sequences, in codon 4, does not result in an amino acid substitution, and the products encoded by the two genes are 100% identical. This region is also very similar in sequence to IS1414 (1,106 bp identical over 1,111 bp), an insertion element identified in ETEC strain 27D. As in strain 27D, the EAST1-encoding gene is present within the insertion element. The IS1414-like element of strain 55989 is very unlikely to be functional, because it contains an incomplete transposase gene (with 211 bp deleted from the 3′ end) and the target site is not duplicated. Instead, sequences similar to parts of IS91 and IS2, respectively, were found on either side.
The 3,606-bp nucleotide sequence between regions 2 and 3 showed no significant similarity to sequences in the database, except for a region from nt 8889 to nt 9292, which displayed 97% identity (398 nt identical over 409 nt) to part of insertion sequence IS629 from Shigella flexneri (30).
Distribution of AAF-encoding operons in EAEC strains isolated from HIV patients.
We investigated 25 diarrhea-associated isolates, including isolate 55989, initially identified as EAEC strains on the basis of their AA patterns in HeLa cell adhesion assays in Bangui and Dakar. We first checked the adhesion properties of these strains on HeLa cells after storage. Various AA phenotypes were observed (Table 3). Twelve strains exhibited a typical AA pattern (AAt). Ten isolates showed a low-level AA pattern (AAll). Finally, three strains showed a mixed adhesion pattern (AA/DA) corresponding to AAt or diffuse adhesion (DA) depending on the HeLa cell examined. We also performed adhesion assays with human intestinal cell lines, such as undifferentiated Caco-2 cells and Int407 cells. For all isolates, the adhesion pattern was similar for intestinal and HeLa cells. We characterized these strains in more detail, using PCR and probes to detect genes for adhesion factors from the three pathotypes of diarrhea-associated enteroadherent E. coli (EAEC, DAEC, and EPEC) (Tables 2 and 3). None of the isolates tested positive for the genes involved in the localized adherence pattern of EPEC strains. Two of the AA/DA isolates tested positive for afa operons, which confer DA on DAEC (uropathogenic and diarrheal) strains. Eleven of the 25 strains tested negative for all the factors. These 11 strains comprised 3 AAt, 7 AAll, and 1 AA/DA strain.
Most of the AAt strains (8 of 12) and 1 of the AAll isolates tested positive by classical methods for the pAA virulence plasmid and the aggR gene.
The aggA gene, which encodes the AAF-I fimbrial subunit, was detected by both PCR and hybridization. Only four pAA-positive strains also tested positive for aggA. Two of these four isolates tested positive by both approaches and also tested positive for aggR; these two strains were classified as typical AAF-I strains. Two other isolates testing positive only for aggA, by hybridization, were described as carrying agg-related sequences.
No isolate hybridized with the aafA probe, corresponding to the AAF-II fimbrial subunit. However, five strains (two AAt, two AAll, and one AA/DA strain) hybridized with a probe corresponding to a larger region (aafDA) of the aaf operon, suggesting that they carried aaf-related sequences. Interestingly, four of these strains were negative for pAA and aggR. One of the three AA/DA isolates contained both an afa operon and an aaf-related operon.
We used PCR assays to evaluate the frequency of the agg-3 operon. Two sets of agg-3-specific primers were designed from the nucleotide sequences of the agg-3C and agg-3A genes for detection of the corresponding sequences (Table 2). The specificity of the oligonucleotide pairs was evaluated by testing representative strains. Amplicons were obtained from strain 55989, from which the agg-3 genes were cloned, and HB101(pILL1268). No amplification was observed with strain JM221, 042, or A30, carrying genes encoding the AAF-I, AAF-II, and AFA-III adhesins, respectively, or with the E. coli K-12 strain HB101, used as a negative control. Only 3 of the 25 EAEC strains in our collection tested positive by PCR for both agg-3C and agg-3A: isolates 55989, 885254, and 387A. All these strains were isolated from AIDS patients in the Central African Republic, and all carried the same genetic markers (CVD432, aggR, aspU, astA, shf, pic, and irp2). In our collection, the frequency of AAF-III strains (12%) was similar to that of AAF-I strains (16%) and of strains carrying aaf-related sequences (20%) (Table 3).
Development of a probe to detect AAF-encoding operons.
Characterization of the EAEC collection highlighted the variability between isolates. However, identification of the agg-3 operon demonstrated conservation of the AAF-encoding gene clusters involved in the adhesion phenotype. Moreover, the results of the adhesion and hybridization experiments presented in this study (Table 3) clearly indicated that, in addition to the agg, aaf, and agg-3 operons, other, as yet unidentified gene clusters must be associated with the expression of an AA pattern. Based on alignment of the three available sequences of AAF-encoding operons, we showed that the usher-encoding region (aggC, aafC, or agg-3C) was the best conserved region in these operons. We selected a 485-bp fragment of the agg-3C gene from strain 55989 as a potential probe for AAF-encoding genes (Fig. 2B; also see Table 2 for sequences of the oligonucleotides used as primers). The recombinant plasmid pILL1277 was produced by inserting the AAF probe sequence into pUC18. The specificity of the AAF probe (485-bp amplicon) was evaluated by testing representative strains by colony hybridization at 65°C (Table 5). Strains carrying the agg operon [JM221 and HB101(pILL1218)] and strains expressing the aaf operon [042 and HB101(cosmid D6)] tested positive with the probe. No hybridization was detected with strain HB101, used as a negative control. Similar results were obtained if hybridization was performed at 60°C.
TABLE 5.
Comparison of probes for detection of EAEC strains by hybridization
| Isolate and adhesion phenotype | No. (%) of strains positive with probe:
|
|
|---|---|---|
| CVD432 | AAF | |
| Diarrheagenic strains | ||
| AAt (n = 12) | 8a (66.6) | 10b (83.3) |
| AA/DA (n = 3) | 0 | 3c (100) |
| AAll (n = 10) | 1a (10) | 3d (30) |
| Nondiarrheagenic strains | ||
| AAt (n = 3) | 1a (33.3) | 2 (66.6) |
| AAll (n = 4) | 1a (25) | 3 (75) |
| Total (n = 32) | 11 (34.4) | 21 (65.6) |
| Control strains | ||
| AAt (JM221, 042) | + | + |
| + | + | |
| DA (A30) | − | + |
All of these strains were positive with the AAF probe.
One of the two strains negative with the CVD432 probe carries sequences related to that of the aaf gene cluster (it was positive with the aafDA probe and negative with the aafA probe).
Two strains were positive for AFA; one strain was positive for AAF-III.
The two strains negative with the CVD432 probe carry sequences related to that of the aaf gene cluster (they were positive with the aafDA probe and negative with the aafA probe).
This study and previous reports provide evidence for a high level of similarity between the AAF-encoding operons and the afa operons carried by DAEC strains (13, 14). Not surprisingly, clinical and recombinant strains carrying an afa operon also hybridized with the AAF probe. We obtained positive results with strains A30, AL845 (25), and HB101(pILL1151), carrying the afa-3 operon; strain AL851, carrying the afa-5 operon (25); and strain C1845, carrying the daa operon (which belongs to the afa family of operons) (3, 15). In contrast, seven DAEC strains that did not carry afa operons tested negative with the AAF probe. We evaluated the sensitivity of the probe by testing the 32 EAEC strains (25 diarrheagenic and 7 nondiarrheagenic isolates) of the collection. Once again, the results of hybridizations at 65 and 60°C were similar. However, the results were easier to interpret if hybridization was carried out at 60°C. All strains testing positive with the CVD432 probe also tested positive with the AAF probe (11 strains). However, 10 other strains also hybridized with the AAF probe, giving a total of 21 AAF probe-positive strains (65.6%). Only 20% of the AAt and 57% of the AAll strains were not identified with the AAF probe, which compares favorably with the 40 and 85.7%, respectively, not identified with the classical CVD432 probe.
DISCUSSION
EAEC strains may be considered to be a heterogeneous set of strains with factors in common, resulting in a common AA phenotype on HEp-2 and HeLa cells (35). Some EAEC isolates have been recognized as pathogenic strains associated with various intestinal diseases. However, the genetic bases of EAEC pathogenesis have not yet been fully elucidated, and EAEC strains may be pathogenic or nonpathogenic. Great progress toward the identification of pathogenic EAEC strains could be achieved by characterizing new adhesion factors and developing genetic tools for detection of EAEC clones in stools.
We identified the AAF-III adhesion factor in isolate 55989. The genetic system encoding this factor also encodes a fimbrial adhesin involved in the aggregation of bacteria and the adhesion of bacteria to cells. The genetic organization of the gene cluster, designated agg-3, is very similar to those of the agg and aaf operons from EAEC isolates. A gene encoding a product identical to AggR, a positive transcriptional regulator of both the agg and aaf gene clusters, was detected downstream of agg-3, but there is no evidence that it is involved in AAF-III production. Analysis of the sequence of agg-3 also confirmed that the operons encoding AA factors were similar to afa operons. The Agg-3B product is similar to AggB and AfaD, both of which are invasins. Thus, as suggested for AafB, to which it is also similar, Agg-3B is probably an invasin (14). HeLa cells may take up agg-3-expressing wild-type and recombinant strains. It is unknown whether invasion is specifically promoted by Agg-3B in AAF-III-producing bacteria. Like DA afa-expressing isolates, agg-3-expressing EAEC strains invade HeLa cells at low frequency. The ability of EAEC strains to invade epithelial cells may reflect an evolutionary strategy designed to establish bacterial reservoirs and persistence within the host. This notion is strongly supported by the isolation of these strains from patients presenting with persistent enteric infections. The present lack of clinical evidence of invasiveness may be accounted for by a low efficiency of internalization into epithelial cells both in vivo and in vitro.
Like the agg and aaf operons, agg-3 is plasmid borne and is genetically linked to the astA gene, which encodes the EAST1 toxin. The region carrying agg-3 displays a genetic organization similar to that reported for the agg and aaf regions: the adhesion-encoding operon is followed by the toxin gene, which in turn is followed by the regulator gene. These genes are also tightly linked to sequences reminiscent of the sequences of various insertion elements. Thus, multiple recombination events have been involved in the genetic linkage of adhesion- and toxin-encoding gene blocks on pAA plasmids, suggesting that such topological associations of virulence genes could play a role in determining the pathogenic potential of an isolate.
EAEC strains isolated from AIDS patients have never before been characterized. We investigated 25 EAEC strains isolated from HIV-infected patients presenting with persistent diarrhea. We characterized these strains phenotypically and genotypically on the basis of their adhesion properties. These strains were originally isolated in Bangui and Dakar, and were selected based on their AA patterns on cells. However, the adhesion phenotypes of these strains differed in some cases in our hands, suggesting that genotypic identification should greatly facilitate epidemiological studies, overcoming variation due to individual interpretation, laboratory growth conditions, and storage conditions. We studied the prevalences of the three AAF-encoding operons in these strains. To detect AAF-I- and AAF-II-encoding genes, we used internal segments of the aggA and aafA fimbrial subunit genes, which display no significant sequence similarity to each other, as probes, to avoid problems with cross-reactivity. The results obtained with the AAF-II probe were surprising. Although no positive hybridization signals were obtained with the aafA probe, five isolates (20%) hybridized with the aafDA probe, which carries the sequences of both the chaperone and fimbrial subunit genes. These data indicate a high level of heterogeneity for the aafA gene of the aaf operon, which has not previously been reported. Use of the aafDA probe has been suggested as a possible means of detecting AAF-II as a relevant marker of pathogenic EAEC (38). It would be useful to type the fimbrial subunits encoded by the various operons detected with this probe, as a means of identifying the subunits associated with pathogenicity. In previous studies, detection of aaf operons was reported to be strongly associated with detection of both the regulator-encoding aggR gene and the pAA plasmid. We found no such relationship in most of the strains carrying aaf-related sequences (four of five), indicating that the potential aaf operons detected here differ from the AAF-II-encoding gene cluster. Two PCR tests have been developed for the specific detection of agg-3-positive strains: they detect the agg-3C gene and the agg-3A adhesin-encoding gene, respectively. With the aid of these tests, we identified three AAF-III-producing strains among the diarrheagenic isolates from HIV-infected patients tested. Three isolates had a set of plasmid- and chromosome-borne virulence genes in common. According to our results, the frequencies of AAF-I, AAF-II-related, and AAF-III strains seem to be similar. Further studies should be performed to address the prevalence and potential clonality of AAF-III strains. Interestingly, we recently identified an AAF-III-positive isolate from the diarrheagenic stools of a child in New Caledonia, indicating that AAF-III production is not restricted to strains from HIV-infected patients. Although none of the 25 strains carried both AAF-I- and AAF-III-encoding sequences, one AAF-III-producing isolate also tested positive for aaf-related operon sequences, confirming that two AA factor-encoding gene clusters may be present in a single EAEC strain. Only 11 of the 25 EAEC isolates (44%) studied gave a positive result with at least one of the three AAF adhesion system probes, and the basis of AA remains unknown for 14 strains. The prevalence (36%) of strains testing positive with the CVD432 probe was very low, which is consistent with several previously published results (36). On the basis of our results, we were unable to differentiate strains isolated from HIV-infected patients from isolates of other origins.
Characterization of the collection of EAEC isolates studied provided evidence for variability between strains and demonstrated the existence of a new variant fimbria, AAF-III. Several other studies have also suggested that some EAEC strains carry virulence plasmids that do not hybridize with the CVD432 probe (11). However, sequence analysis of the agg-3 gene cluster showed a high degree of conservation between the various operons involved in the adhesion phenotype. We therefore decided to design a new EAEC probe for sequences directly involved in expression of the AA pattern. We report here that an internal fragment of the agg-3C gene functions efficiently as a probe for the identification of strains carrying AAF-encoding operons. This probe is sensitive for a family of operons widespread among enteroadherent E. coli. Indeed, in addition to AAF-encoding operons, this probe detects the afa operons carried by DAEC and uncharacterized operons involved in various adhesion phenotypes, such as AA/DA or AAll, described in this study. Use of this probe could make it possible to identify genetically a large proportion of enteroadherent pathogenic E. coli for which it is not always easy to determine an adhesion pattern.
EAEC strains are an emerging cause of diarrheal infections worldwide. The epidemiological significance of the virulence markers identified to date should be evaluated by testing large collections of EAEC isolated from diarrheagenic patients and healthy controls. One limitation to such studies is the lack of probes for detection of all adhesion factors. The probe described here should facilitate progress in this direction.
Acknowledgments
We thank A. Labigne, in whose unit this work was carried out, for her continuing interest and helpful discussions. We also thank Y. Germani (Pasteur Institute of Bangui, Central African Republic, and GEID) and A. Aïdara-Kane (Pasteur Institute of Dakar, Senegal, and GEID) for the EAEC isolates. We thank J. Nataro for the gift of EAEC reference strains, pCVD301, and Pic primers, J.-M. Ghigo for the gift of pKOBEG and help in allelic exchange by recombination with a PCR product, and C. Parsot for providing the nonpolar kanamycin resistance cassette. We also thank L. du Merle and H. Ohayon for expert technical assistance.
C. Bernier was supported by a doctoral fellowship from the Ministère de l'Education Nationale de la Recherche et de la Technologie. C. Le Bouguénec was supported in part by grant 99162 from l'Agence Nationale de Recherche sur le SIDA.
Editor: A. D. O'Brien
REFERENCES
- 1.Baldini, M. M., J. B. Kaper, M. M. Levine, D. C. Candy, and H. W. Moon. 1983. Plasmid-mediated adhesion in enteropathogenic Escherichia coli. J. Pediatr. Gastroenterol. Nutr. 2:534-538. [DOI] [PubMed] [Google Scholar]
- 2.Baudry, B., S. J. Savarino, P. Vial, J. B. Kaper, and M. M. Levine. 1990. A sensitive and specific DNA probe to identify enteroaggregative Escherichia coli, a recently discovered diarrheal pathogen. J. Infect. Dis. 161:1249-1251. [DOI] [PubMed] [Google Scholar]
- 3.Bilge, S. S., C. R. Clausen, W. Lau, and S. L. Moseley. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J. Bacteriol. 171:4281-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., M. S. McClain, J. A. Princ, P. J. Calie, and B. I. Eisenstein. 1991. Type 1 fimbriation and fimE mutants of Escherichia coli K-12. J. Bacteriol. 173:5298-5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bolivar, F., R. L. Rodriguez, P. J. Greene, M. C. Betlach, H. L. Heyneker, and H. W. Boyer. 1977. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene 2:95-113. [PubMed] [Google Scholar]
- 6.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 7.Chang, A. C. Y., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chaveroche, M. K., J. M. Ghigo, and C. d'Enfert. 2000. A rapid method for efficient gene replacement in the filamentous fungus Aspergillus nidulans. Nucleic Acids Res. 28:E97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins, J. 1979. Escherichia coli plasmids packageable in vitro in lambda bacteriophage particles. Methods Enzymol. 68:309-326. [DOI] [PubMed] [Google Scholar]
- 10.Czeczulin, J. R., S. Balepur, S. Hicks, A. Phillips, R. Hall, M. H. Kothary, F. Navarro-Garcia, and J. P. Nataro. 1997. Aggregative adherence fimbria II, a second fimbrial antigen mediating aggregative adherence in enteroaggregative Escherichia coli. Infect. Immun. 65:4135-4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Czeczulin, J. R., T. S. Whittam, I. R. Henderson, F. Navarro-Garcia, and J. P. Nataro. 1999. Phylogenetic analysis of enteroaggregative and diffusely adherent Escherichia coli. Infect. Immun. 67:2692-2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Durrer, P., R. Zbinden, F. Fleisch, M. Altwegg, B. Ledergerber, H. Karch, and R. Weber. 2000. Intestinal infection due to enteroaggregative Escherichia coli among human immunodeficiency virus-infected persons. J. Infect. Dis. 182:1540-1544. [DOI] [PubMed] [Google Scholar]
- 13.Elias, W. P., Jr., J. R. Czeczulin, I. R. Henderson, L. R. Trabulsi, and J. P. Nataro. 1999. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J. Bacteriol. 181:1779-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garcia, M.-I., M. Jouve, J. P. Nataro, P. Gounon, and C. Le Bouguénec. 2000. Characterization of the AfaD-like family of invasins encoded by pathogenic Escherichia coli associated with intestinal and extra-intestinal infections. FEBS Lett. 479:111-117. [DOI] [PubMed] [Google Scholar]
- 15.Garcia, M. I., P. Gounon, P. Courcoux, A. Labigne, and C. Le Bouguénec. 1996. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol. Microbiol. 19:683-693. [DOI] [PubMed] [Google Scholar]
- 16.Garcia, M. I., A. Labigne, and C. Le Bouguénec. 1994. Nucleotide sequence of the afimbrial-adhesin-encoding afa-3 gene cluster and its translocation via flanking IS1 insertion sequences. J. Bacteriol. 176:7601-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gassama, A., P. S. Sow, F. Fall, P. Camara, A. Gueye-N'diaye, R. Seng, B. Samb, S. M'Boup, and A. Aïdara-Kane. 2001. Ordinary and opportunistic enteropathogens associated with diarrhea in Senegalese adults in relation to human immunodeficiency virus serostatus. Int. J. Infect. Dis. 5:192-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Germani, Y., E. Begaud, and C. Le Bouguénec. 1997. Detection of the Escherichia coli attaching and effacing gene (eaeA) in enteropathogenic strains by polymerase chain reaction. Res. Microbiol. 148:177-181. [DOI] [PubMed] [Google Scholar]
- 19.Germani, Y., P. Minssart, M. Vohito, S. Yassibanda, P. Glaziou, D. Hocquet, P. Berthelemy, and J. Morvan. 1998. Etiologies of acute, persistent, and dysenteric diarrheas in adults in Bangui, Central African Republic, in relation to human immunodeficiency virus serostatus. Am. J. Trop. Med. Hyg. 59:1008-1014. [DOI] [PubMed] [Google Scholar]
- 20.Jouve, M., M. I. Garcia, P. Courcoux, A. Labigne, P. Gounon, and C. Le Bouguénec. 1997. Adhesion to and invasion of HeLa cells by pathogenic Escherichia coli carrying the afa-3 gene cluster are mediated by the AfaE and AfaD proteins, respectively. Infect. Immun. 65:4082-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotler, D. P., T. T. Giang, M. Thiim, J. P. Nataro, E. M. Sordillo, and J. M. Orenstein. 1995. Chronic bacterial enteropathy in patients with AIDS. J. Infect. Dis. 171:552-558. [DOI] [PubMed] [Google Scholar]
- 22.Labigne-Roussel, A., and S. Falkow. 1988. Distribution and degree of heterogeneity of the afimbrial-adhesin-encoding operon (afa) among uropathogenic Escherichia coli isolates. Infect. Immun. 56:640-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lalioui, L., M. Jouve, P. Gounon, and C. Le Bouguénec. 1999. Molecular cloning and characterization of the afa-7 and afa-8 gene clusters encoding afimbrial adhesins in Escherichia coli strains associated with diarrhea or septicemia in calves. Infect. Immun. 67:5048-5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Bouguénec, C., M. Archambaud, and A. Labigne. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 30:1189-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Bouguénec, C., M. I. Garcia, V. Ouin, J. M. Desperrier, P. Gounon, and A. Labigne. 1993. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect. Immun. 61:5106-5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Minor, L., and C. Richard. 1993. Méthodes de laboratoire pour l'identification des entérobactéries. Institut Pasteur, Paris, France.
- 27.Maniatis, T., E. F. Fritsh, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 28.Mathewson, J. J., Z. D. Jiang, A. Zumla, C. Chintu, N. Luo, S. R. Calamari, R. M. Genta, A. Steephen, P. Schwartz, and H. L. DuPont. 1995. HEp-2 cell-adherent Escherichia coli in patients with human immunodeficiency virus-associated diarrhea. J. Infect. Dis. 171:1636-1639. [DOI] [PubMed] [Google Scholar]
- 29.Mathewson, J. J., R. A. Oberhelman, H. L. Dupont, F. Javier de la Cabada, and E. V. Garibay. 1987. Enteroadherent Escherichia coli as a cause of diarrhea among children in Mexico. J. Clin. Microbiol. 25:1917-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matsutani, S., H. Ohtsubo, Y. Maeda, and E. Ohtsubo. 1987. Isolation and characterization of IS elements repeated in the bacterial chromosome. J. Mol. Biol. 196:445-455. [DOI] [PubMed] [Google Scholar]
- 31.McVeigh, A., A. Fasano, D. A. Scott, S. Jelacic, S. L. Moseley, D. C. Robertson, and S. J. Savarino. 2000. IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect. Immun. 68:5710-5715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mwachari, C., B. I. Batchelor, J. Paul, P. G. Waiyaki, and C. F. Gilks. 1998. Chronic diarrhoea among HIV-infected adult patients in Nairobi, Kenya. J. Infect. 37:48-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nataro, J. P., M. M. Baldini, J. B. Kaper, R. E. Black, N. Bravo, and M. M. Levine. 1985. Detection of an adherence factor of enteropathogenic Escherichia coli with a DNA probe. J. Infect. Dis. 152:560-565. [DOI] [PubMed] [Google Scholar]
- 34.Nataro, J. P., Y. Deng, D. R. Maneval, A. L. German, W. C. Martin, and M. M. Levine. 1992. Aggregative adherence fimbriae I of enteroaggregative Escherichia coli mediate adherence to HEp-2 cells and hemagglutination of human erythrocytes. Infect. Immun. 60:2297-2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nataro, J. P., J. B. Kaper, R. Robins-Browne, V. Prado, P. Vial, and M. M. Levine. 1987. Patterns of adherence of diarrheagenic Escherichia coli to HEp-2 cells. Pediatr. Infect. Dis. J. 6:829-831. [DOI] [PubMed] [Google Scholar]
- 36.Nataro, J. P., T. Steiner, and R. L. Guerrant. 1998. Enteroaggregative Escherichia coli. Emerg Infect. Dis. 4:251-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nataro, J. P., D. Yikang, D. Yingkang, and K. Walker. 1994. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 176:4691-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okeke, I. N., A. Lamikanra, J. Czeczulin, F. Dubovsky, J. B. Kaper, and J. P. Nataro. 2000. Heterogeneous virulence of enteroaggregative Escherichia coli strains isolated from children in Southwest Nigeria. J. Infect. Dis. 181:252-260. [DOI] [PubMed] [Google Scholar]
- 39.Pinto, M., S. Robine-Léon, M.-D. Appay, M. Kedinger, N. Triadou, E. Dussaulx, B. Lacroix, P. Simon-Assmann, K. Haffen, J. Fogh, and A. Zweibaum. 1983. Enterocyte-like differentiation and polarization of the human colon carcinoma cell line Caco-2 in culture. Biol. Cell 47:323-330. [Google Scholar]
- 40.Schmidt, H., C. Knop, S. Franke, S. Aleksic, J. Heesemann, and H. Karch. 1995. Development of PCR for screening of enteroaggregative Escherichia coli. J. Clin. Microbiol. 33:701-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzart, S., B. E. Guth, M. Z. Pedroso, U. M. Okafor, and T. A. Gomes. 2001. Diversity of surface structures and virulence genetic markers among enteroaggregative Escherichia coli (EAEC) strains with and without the EAEC DNA probe sequence. FEMS Microbiol. Lett. 201:163-168. [DOI] [PubMed] [Google Scholar]
- 42.Tornieporth, N. G., J. John, K. Salgado, P. de Jesus, E. Latham, M. C. Melo, S. T. Gunzburg, and L. W. Riley. 1995. Differentiation of pathogenic Escherichia coli strains in Brazilian children by PCR. J. Clin. Microbiol. 33:1371-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wanke, C. A., J. Gerrior, V. Blais, H. Mayer, and D. Acheson. 1998. Successful treatment of diarrheal disease associated with enteroaggregative Escherichia coli in adults infected with human immunodeficiency virus. J. Infect. Dis. 178:1369-1372. [DOI] [PubMed] [Google Scholar]
- 44.Wanke, C. A., H. Mayer, R. Weber, R. Zbinden, D. A. Watson, and D. Acheson. 1998. Enteroaggregative Escherichia coli as a potential cause of diarrheal disease in adults infected with human immunodeficiency virus. J. Infect. Dis. 178:185-190. [DOI] [PubMed] [Google Scholar]
- 45.Yamamoto, T., and P. Echeverria. 1996. Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64:1441-1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]