Abstract
In order to assess the role of polyphosphate kinase (PPK) in the physiology of Porphyromonas gingivalis, a ppk gene mutant, CW120, was constructed and characterized. P. gingivalis was demonstrated to synthesize short-chain polyphosphate (polyP) but not long-chain polyP. CW120 failed to survive in the stationary phase as well as the parental cell did, and it was attenuated in biofilm formation on polyvinylchloride and glass surfaces. Furthermore, the complementation by insertion of an intact copy of the ppk gene into the mutant CW120 restored its biofilm formation and stationary-phase survival. These results suggest that PPK may be important for incorporation of these organisms into subgingival plaque in the human oral cavity.
Periodontitis is one of the most common infectious diseases afflicting mankind (35). These infections appear to result from the inflammatory response of the host to mixed anaerobic bacterial infections of the gingival margin. Among the organisms implicated in these diseases, Porphyromonas gingivalis has attracted much attention (22). These organisms appear to express a variety of virulence factors that may be important in the etiology of periodontitis (15). Since P. gingivalis, along with other pathogenic bacteria in the oral cavity, appears to reside primarily in biofilm structures commonly termed dental plaque, it is important to delineate the molecular basis for biofilm formation by these organisms.
Investigations with several gram-negative bacteria have suggested that the polyphosphate kinase (PPK) gene ppk may be a potentially important virulence factor in these organisms (29, 30). This gene is highly conserved among both gram-positive and gram-negative bacteria and appears to play a role in adaptation to nutritional and other environmental stresses as well as stationary-phase survival (26, 34). In addition, ppk mutations in Escherichia coli, Salmonella enterica serovar Dublin, and Pseudomonas aeruginosa attenuate swimming, swarming, and twitching motility in these organisms (20, 28-30). Likewise, recent results have suggested an important role for PPK activity in biofilm formation and virulence in P. aeruginosa (30). It was therefore of interest to examine the potential role of the ppk gene in the properties of P. gingivalis. The construction of a specific ppk mutant of P. gingivalis 381 in the present study demonstrated that this gene plays an important role in stationary-phase survival as well as in biofilm formation in vitro.
Construction and identification of a ppk-deficient mutant of P. gingivalis 381 (CW120).
P. gingivalis strains were maintained anaerobically on blood agar plates containing tryptic soy broth (TSB; Difco Laboratory, Detroit, Mich.) supplemented with 10% sheep blood, hemin (5.0 μg/ml), menadione (1.0 μg/ml), and gentamicin (25 μg/ml). E. coli strains MG1655 and CF5802 were kindly provided by A. Kornberg, Stanford University School of Medicine (Stanford, Calif.). Plasmid prtT:Em was constructed in our laboratory previously (unpublished results). Plasmids pUC19, Topo/PCR vector, and prtT:Em were maintained in E. coli DH5α in the presence of 50 μg of ampicillin per ml. A ppk homologous sequence of P. gingivalis W83 was identified by searching The Institute for Genomic Research database (http://www.ncbi.nlm.nih.gov) with the amino acid sequence of E. coli. A pair of primers, 5′-AAC GAT CAG TAG CAC TGT GG-3′ and 5′-TTA TTT TGC AGC AGG AGT GGC-3′, were designed based upon the sequence of the ppk gene of P. gingivalis W83 and used for amplifying and cloning a 2.1-kb fragment of the ppk gene from strain 381. Inactivation of the P. gingivalis 381 ppk gene was accomplished following electroporation (36) with an Em cassette inserted into the gene following homologous recombination (Fig. 1). The Em cassette was introduced into the BglII sites, which are 703 and 871 bp downstream from the ATG initiation codon and are present within the conserved regions of ppk. Since the plasmid pCW111 was linearized, erythromycin-resistant transformants would grow only as a result of a double-crossover event between the regions flanking the Em cassette and the ppk gene on the chromosome. In order to confirm that the Em cassette was inserted into the predicted sites within the ppk gene on the chromosome of strain 381, Southern blot analysis (6) was carried out (Fig. 1). Chromosomal DNA from P. gingivalis strains was prepared with a Puregene isolation kit (Gentra System, Inc., Minneapolis, Minn.) by following the supplier's protocol. DNA was digested with the restriction enzymes indicated and loaded onto 1% agarose gels for electrophoresis, and the DNA fragments were transferred to nylon membranes (Amersham Corp., Arlington Heights, Ill.) after alkaline denaturation. The labeling of the probes, hybridization, and detection with an enhanced chemiluminescence system were performed as recommended by the supplier (Amersham). Thirty-three erythromycin-resistant colonies were obtained. One of the ppk mutants with erythromycin resistance was chosen for further study and designated CW120. The growth rate of CW120 was similar to that of wild-type 381 in TSB medium.
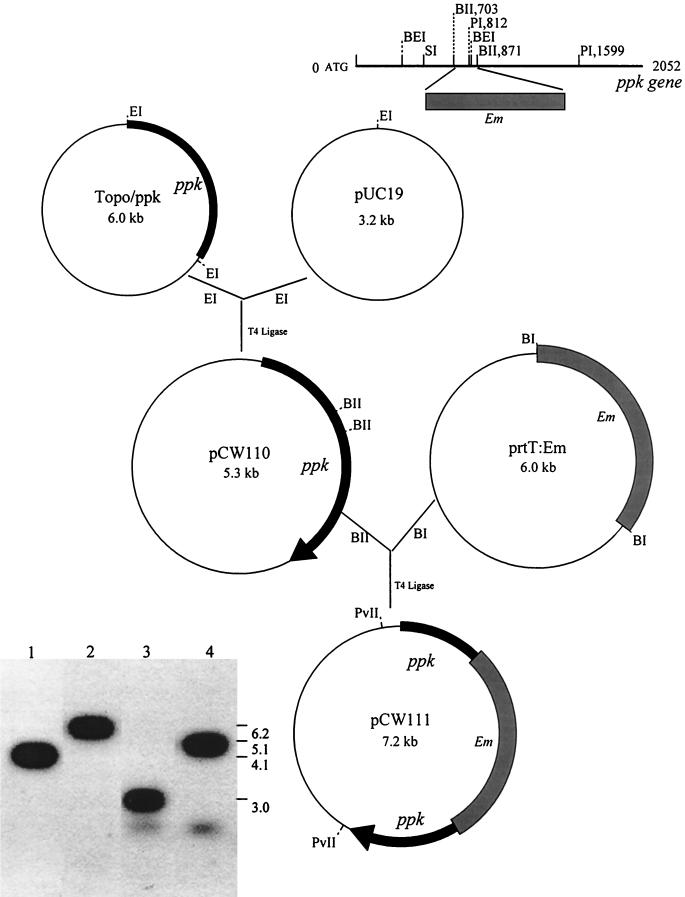
FIG. 1.
Construction of the ppk-deficient mutant CW120. A 2.1-kb ppk fragment amplified by PCR from P. gingivalis 381 was ligated into the pCR2.1-TOPO vector. The 2.1-kb fragment was then isolated and inserted into pUC19 following cleavage of plasmid TOPO/ppk and pUC19 with EcoRI, producing plasmid pCW110. A 2.1-kb ermF-ermAM BamHI-digested cassette from plasmid prtT:Em was next inserted into the ppk gene at the BglII site of pCW110. The resulting plasmid, pCW111, was linearized with PvuII and electroporated into P. gingivalis 381. The resulting mutant, CW120, was identified by Southern blot analysis of the genomic DNA of P. gingivalis (bottom left). The chromosomal DNA of strains 381 (lanes 1 and 3) and CW120 (lanes 2 and 4) was digested with BspEI (lanes 1 and 2) or StuI (lanes 3 and 4), respectively, and probed with a 787-bp fragment of the ppk gene digested with PstI. BEI, BspEI; BI, BamHI; EI, EcoRI; BII, BglII; PI, PstI; PvII, PvuII; SI, StuI; Em, erythromycin cassette.
Estimation of polyP in P. gingivalis.
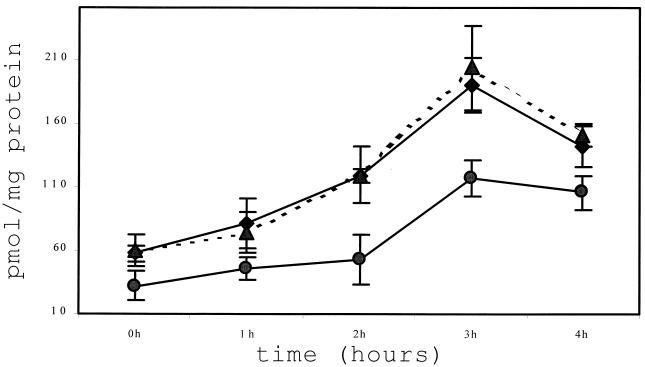
A radioactive assay confirmed the expression of the ppk gene in P. gingivalis 381, although PPK activity was low (data not shown). In order to compare the PPK activity between the wild-type 381 and mutant CW120, polyphosphate (polyP), the product of PPK, was examined. Bacterial cells (107 and ≈108 CFU) were pelleted for long-chain and short-chain polyP extraction, respectively. PolyP was assayed with the nonradioactive two-enzymes method described by Ault-Riche at al. (4). Bioluminescence was measured by using a 1450 MicroBeta TriLux counter (Wallac Oy, Turku, Finland). Long-chain polyP (with 60 to several hundred Pi residues) extraction was achieved with milkglass as described by Ault-Riche et al. (4). As positive and negative controls, E. coli strains MG1655 and CF5802 (ppk and ppx deficient), respectively, were cultured to mid-log phase in Luria-Bertani medium and then transferred into morpholinepropanesulfonic acid (MOPS) medium with 4% glucose and limiting phosphate (0.1 mM Pi) without amino acids. P. gingivalis 381 and CW120 were cultured to mid-log phase in TSB medium, transferred to the same prereduced MOPS medium supplemented with hemin and menadione and with or without 0.01% bovine serum albumin (BSA), and incubated anaerobically at 37°C. PolyP was isolated from the samples at different intervals (0 to ≈4 h) and measured (4). PolyP accumulation was observed in MG1655 (350 nmol/mg of protein at 3 h) but not in CF5802. P. gingivalis 381 and CW120 were incubated anaerobically in MOPS medium for prolonged time periods to collect samples for up to 24 h, since P. gingivalis grew much slower than E. coli. However, long-chain polyP was not detectable in P. gingivalis 381 or CW120 at any time points (data not shown). The P. gingivalis strains were also stimulated with osmotic shock, changes in pH, temperature upshifts, and oxidative stress in TSB. None of these stress conditions could induce detectable accumulation of long-chain polyP. Since bacteria can produce polyP of various chain lengths (7, 17, 32), an assay to detect short-chain polyP (<60 Pi residues) was then carried out. The isolation of short-chain polyP was performed as described by Ruiz et al. (32). Picomolar levels of short-chain polyP were detected from the same bacterial samples. The levels of short-chain polyP from mutant CW120 were decreased to about 50% of that from WT381. The peak of short-chain accumulation (200 pmol/mg of protein) was at 3 h for strain 381. Mutant CW120 also exhibited lower detectable short-chain polyP accumulation (100 pmol/mg of protein). Furthermore, the ppk-deficient complemented strain, CW120C (described below), displayed a normal level of short-chain polyP relative to that of the wild-type 381 (Fig. 2).
FIG. 2.
Short-chain polyP assay of P. gingivalis 381, ppk-deficient mutant CW120, and complemented strain CW120C. P. gingivalis 381 (♦), CW120 (•), and CW120C (▴) were grown to mid-log phase in TSB medium with hemin and menadione. The cells were pelleted, resuspended, and incubated anaerobically in MOPS defined medium containing 0.1 mM Pi, 0.01% BSA, and 4 mg of glucose per ml. The samples were collected at 0, 1, 2, 3, and 4 h. Short-chain polyP was extracted and analyzed as described in the text. The results are averages of quadruple samples and their standard deviations are shown.
The ppk gene of P. gingivalis 381 is essential for stationary-phase survival.
There was no significant difference in the growth rate between strain 381 and mutant CW120 when cultured anaerobically in TSB medium at 37°C. In a long-term survival assay (8), strains 381 and CW120 were grown to the early stationary phase and further incubated in TSB medium anaerobically at 37°C for several days. Viable cell counts were then determined in triplicate for each day. After plating, the plates were routinely incubated anaerobically at 37°C for 7 days before the viable colonies were counted. The results at days 1 to ≈3 indicated an indistinguishable loss in viability between strains 381 and CW120. However, after the third day, the viability of CW120 was significantly reduced. The viable cells of CW120 were approximately 10, 1, 0.1, and 0.001% of that of wild-type 381 at days 4, 5, 6, and 7, respectively. After 7 days, a number of small-colony variants of CW120 arose, and only a few normal-size colonies could be observed on the plates, as with other bacteria (8, 12, 26). These small colonies from CW120 were lighter in color and could not be subsequently passaged. Characterization of these variants was not further explored. The heat shock survival and sensitivity to H2O2 (8) were also evaluated but no differences were observed between 381 and mutant CW120 (data not shown).
The ppk gene of P. gingivalis 381 is involved in biofilm formation.
Static biofilm formation of P. gingivalis strains was first examined on 96-well polyvinyl chloride (PVC) plates as described previously (24). Briefly, the stationary-phase cultures of P. gingivalis were inoculated into the diluted TSB medium (TSB/phosphate-buffered saline (PBS) ratio, 1:2). The cells were added to 96-well PVC plates (100 μl/well) and incubated anaerobically at 37°C for 12 h. Four wells of each sample were used for measuring total growth while another four identical wells were assayed for biofilms. Following incubation, the biofilm wells were stained with crystal violet (CV) and quantitated (24). Biofilm formation was scored as the absorbance of CV-stained biofilms at an optical density at 570 nm (OD570) divided by the absorbance of total growth (including biofilm cells and planktonic cells) at OD 570. Attachment, as an initial step of biofilm formation, of 381 and CW120 to KB cells (9, 10, 38) and PVC abiotic surfaces (24) was also evaluated. There was no significant difference detected in attachment between the wild-type 381 and mutant CW120 (data not shown). However, with static continued incubation in the diluted TSB medium (TSB/PBS ratio, 1:2), the ppk mutant was shown to be attenuated in biofilm formation (Fig. 3).
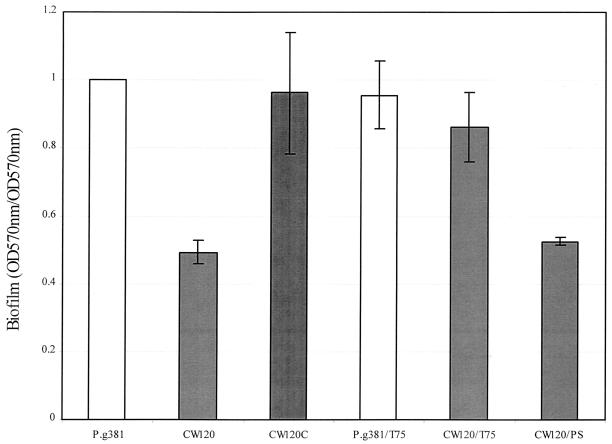
FIG. 3.
Biofilm formation assay in PVC plates. P. gingivalis strains were incubated overnight in TSB medium diluted with PBS (TSB/PBS ratio, 1:2) with supplementation of hemin, vitamin K in the wells of 96-well PVC microtiter dishes (100 μl/well). The resulting biofilms were analyzed as described previously (24). Biofilm formation was calculated as follows: (OD570 for the biofilm)/(OD570 of total cell growth). The data are averages of triplicate assays with the standard errors of the means. T75, 75 phosphate polymer of polyP (100 μg/ml); PS, polyvinyl sulfate (100 μg/ml).
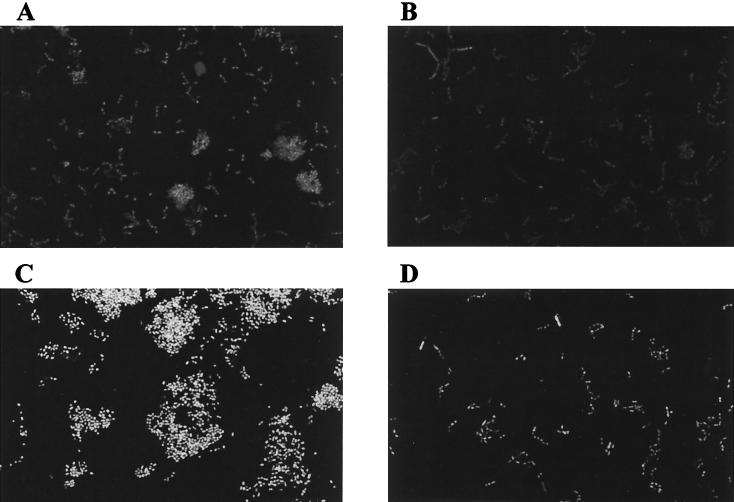
Continuous biofilm formation in the flow-cell system was also performed as described previously (13), except that the experiment was performed in an anaerobic glove box. Briefly, two-track flow cells were constructed using a microscope slide as the bottom and a no. 1.5 coverglass as the top. The flow cells were washed overnight with 0.1 M HCl, rinsed with several changes of distilled water over a period of 3 h, and then autoclaved. Overnight cultures (0.5 ml of each strain) of P. gingivalis 381 and mutant CW120 grown on porphyromonas broth (PB; Todd Hewitt broth [Difco] supplemented with 1 g of yeast extract, 5 mg of hemin, and 1 mg of vitamin K per liter) or PB plus 5 μg of erythromycin/ml (CW120) were spun down, washed with fresh PB, and resuspended in 8 ml of fresh PB without any antibiotic. After 3 h of growth, the cultures were adjusted to 10 Klett units and the suspensions of strain 381 and CW120 were injected (0.5 ml) into separate flow-cell tracks. The flow cells were inverted and bacteria were permitted to attach for 20 min. After that, the flow cells were returned to their original orientations and a flow of PB diluted 1:10 at 0.0125 mm/s (200 μl/min) was begun. After 4 h of flow, one flow cell for each strain was removed from the incubator, stained with BacLight Live/Dead (Molecular Probes, Eugene, Oreg.) (0.5 ml of a 1:1 mixture of the two dyes diluted 1,000-fold in PBS), and observed with a Leica TCS 4D confocal microscope (Leica Laser Technik, Heidelberg, Germany). Three randomly selected fields were imaged in each track. After an overnight (18-h) period of flow, the remaining flow cells were removed from the incubator, stained, and imaged. This experiment was repeated twice. In this continuous flow-cell system (25, 31), it was demonstrated that the attached monolayer of the ppk mutant CW120 is similar to that of wild-type 381 on glass surfaces at 4 h (Fig. 4). A few cell clusters on the glass surfaces were seen for strain 381 but not for CW120. After 18 h, characteristic mound-shaped cell clusters were formed by strain 381. By contrast, no cell clusters could be observed for mutant CW120. Mutant CW120 only formed a monolayer biofilm whose architecture differed dramatically from that of the wild-type biofilm. Thus, biofilm maturation appears to be strongly affected in the ppk mutant under these conditions. This defect does not result from attenuated interactions between the mutant cells, since autoaggregation of the wild-type strain 381 and ppk mutant are similar (data not shown) (39).
FIG. 4.
Confocal images of P. gingivalis 381 and CW120 biofilms in a flow cell. (A) Strain 381 at 4 h; (B) CW120 at 4 h; (C) strain 381 at 18 h; (D) CW120 at 18 h.
Complementation of the ppk gene mutation.
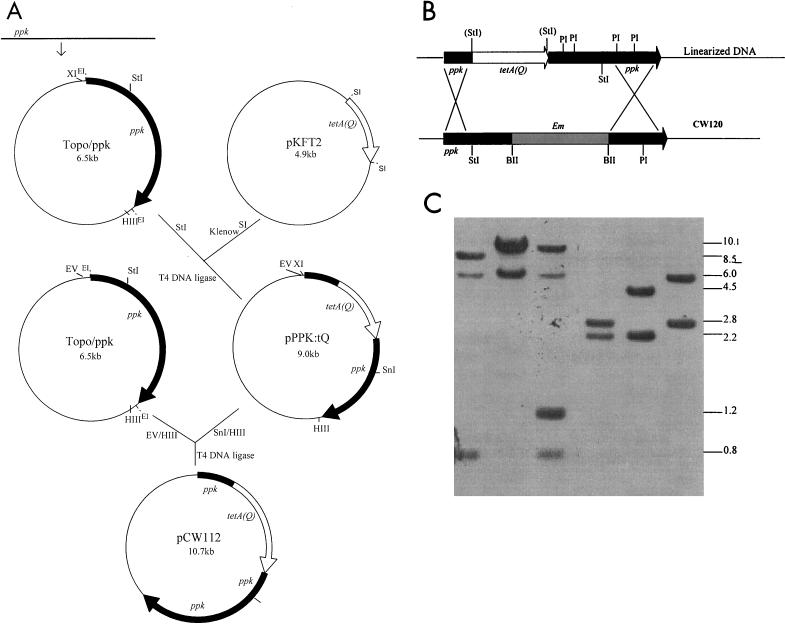
Since multiple transposons have been detected in several strains of P. gingivalis, it was important to confirm that the altered phenotypes of a constructed P. gingivalis mutant resulted from inactivation of the gene and not from a secondary spontaneous mutation (6). Therefore, complementation of the PPK defect was carried out. The tetA(Q) gene was chosen for use as a selection marker for the complementation. The tetA(Q) cassette was ligated either upstream or downstream of the ppk gene in the suicide vectors (pACYC:PPK and pKFT2, respectively), and the suicide plasmids were linearized for recombination into the P. gingivalis chromosome by electroporation. Neither approach resulted in generation of tetracycline-resistant transformants in strains 381 and CW120. Therefore, a strategy was devised to add a strain 381 promoter upstream of the tetA(Q) gene by inserting a 5′-end fragment of the ppk gene, including the 450-bp upstream flanking region (Fig. 5A). Plasmid pKFT2 containing the tetracycline resistance gene [tetA(Q)] was kindly provided by M. Curtis (Royal London School of Medicine and Dentistry, London, United Kingdom) and maintained in E. coli DH5α in the presence of 50 μg of ampicillin/ml. The resulting plasmid, pCW112, was linearized with HindIII and electroporated into strain CW120 (36) to restore PPK activity. Following 16 h of incubation, the cell cultures were plated onto TSB agar plates containing tetracycline (1 μg/ml) and incubated anaerobically at 37°C for 7 to 10 days. The transformants that grew only on tetracycline plates and not on erythromycin plates were potential target colonies which were then grown anaerobically in TSB with tetracycline (0.25 μg/ml) for characterization. One of the 12 resulting purified transformants, which were demonstrated to have lost their erythromycin resistance and were tetracycline resistant, was named CW120C. In addition, Southern blot analysis confirmed the integration of the target gene from plasmid CW112 into the CW120 chromosome (Fig. 5B and C). The complemented mutants exhibited normal levels of stationary-phase survival ability, similar to that of parental strain 381 (data not shown). Significantly, the ppk gene-complemented strain CW120C produced biofilms at levels similar to those produced by wild-type 381 (Fig. 3). In addition, the complemented strain, CW120C, produced wild-type levels of short-chain polyP that were elevated relative to the levels produced by the CW120 mutant (Fig. 2). Furthermore, when commercial polyP (Type 75; Sigma Chemical Co., St. Louis, Mo.) was added to the cultures of 381 and the ppk mutant CW120, CW120 formed biofilms at a level similar to that of 381, but CW120 stationary-phase survival ability could not be rescued. PolyP (Type 75) did not affect biofilm formation of the wild-type strain 381. When another polyanion, polyvinyl sulfate, or orthophosphate was added to cultures of the ppk mutant CW120, biofilm formation of CW120 was not augmented (Fig. 3). This suggested that the complementing effects of polyP addition were not due to a nonspecific polyanionic effect or to phosphate limitation. Taken together, these results suggested that the defects exhibited by CW120 resulted from the loss of PPK activity and not from a secondary spontaneous mutation.
FIG. 5.
Strategy for complementation of the ppk-deficient mutant CW120. (A) A 2.4-kb ppk gene fragment, including the 400-bp upstream flanking region, amplified by PCR from P. gingivalis 381 was ligated into the pCR 2.1-TOPO vector. A 2.2-kb tetA(Q) cassette digested with SstI from pKFT2 was blunted and inserted into the ppk gene at the StuI site in the same transcription direction as the ppk gene. The portion of the ppk gene downstream from tetA(Q) was cut out with SnaB I and HindIII. An intact copy of the ppk gene was ligated downstream of tetA(Q) at the SnaB I and HindIII sites of pPPK:tQ. The resulting plasmid, designated pCW112, was linearized with HindIII and electroporated into the ppk-null mutant CW120. (B) The predicted integration of the resulting plasmid pCW112 into CW120. (C) Southern blot analysis of the genomic DNA of P. gingivalis wild-type 381, ppk mutant CW120, and the ppk-complemented strain CW120C. The chromosomal DNA of 381(lanes 1 and 4), CW120 (lanes 2 and 5), and CW120C (lanes 3 and 6) was digested with StuI (lanes 1, 2, and 3) and PstI (lanes 4, 5, and 6), respectively, and probed with a 2.4-kb ppk fragment.
In several bacteria species, ppk genes have been mutated to determine the role of polyP production in virulence, including some of the major pathogenic species (1, 3, 11, 16, 18, 41). The PPK of P. gingivalis exhibited good homology with the PPK of E. coli (identity, 37%; similarity, 59%). However, there is no detectable ppk/ppx (exopolyphosphatase, ppx) operon in P. gingivalis as observed in E. coli or some other microorganisms (2, 5, 8, 19, 33). Therefore, this suggests that polyP metabolism in P. gingivalis may be somewhat different from that in E. coli. PolyP accumulation may vary in different microorganisms (4, 14, 17, 21, 23, 32, 37, 40, 43). Long-chain polyP could not be detected in P. gingivalis and may require specific stress conditions (4). The inability to detect polyP accumulation under these stress conditions may also indicate that another factor(s) is involved in these responses in P. gingivalis (45) or that P. gingivalis might degrade long-chain polyP during isolation (7). PPK may not be the only enzyme capable of catalyzing the production of short-chain polyP. A second gene may be involved in short-chain polyP synthesis, as suggested for other bacteria (45). The ppk mutant CW120 displayed lower, but significant, levels of short-chain polyP accumulation at 3 h, providing a mechanism for short-chain polyP accumulation independent of PPK activity.
It appears that the ppk gene is essential for stationary-phase long-term survival in P. gingivalis, although ppk may not be the only enzyme involved in the production of polyP. However, unlike E. coli, the ppk mutant CW120 of P. gingivalis still remained sensitive to heat and oxidants, as did parental strain 381. Some studies have reported that long-chain polyP, even at relatively low levels, is essential for adaptation to various stresses and for survival of bacteria in the stationary phase (4, 26, 27).
As with P. aeruginosa (30), the present results suggest that the ppk gene does not affect the initial attachment of P. gingivalis to abiotic surfaces. Therefore, the ppk gene appears to be involved in biofilm maturation of P. gingivalis. The molecular mechanisms involved in biofilm maturation still remain to be elucidated. However, since the metabolism of biofilm bacteria is similar to that of stationary-phase cells (44) it is of interest that the ppk mutant CW120 was attenuated in both biofilm formation and stationary-phase survival. The present results suggest that the mutant could be altered in colonizing the subgingival margin and subsequently periodontal inflammation. Therefore, the ppk gene of P. gingivalis, as well as of other periodontopathogens, might be targeted for the development of specific inhibitors of subgingival plaque formation and periodontitis. Such a strategy has been suggested for other virulent bacteria (42).
Acknowledgments
We gratefully acknowledge the assistance in the bioluminescence assays by A. Thakur and L. Zhao. We also thank F. Scannapieco and A. Sharma for critically reviewing the manuscript.
This study was supported in part by NIH grant DE08293.
Editor: E. I. Tuomanen
REFERENCES
- 1.Ahn, K., and A. Kornberg. 1990. Polyphosphate kinase from Escherichia coli. Purification and demonstration of a phosphoenzyme intermediate. J. Biol. Chem. 265:11734-11739. [PubMed] [Google Scholar]
- 2.Akiyama, M., E. Crooke, and A. Kornberg. 1993. An exopolyphosphatase of Escherichia coli. The enzyme and its ppx gene in a polyphosphate operon. J. Biol. Chem. 268:633-639. [PubMed] [Google Scholar]
- 3.Akiyama, M., E. Crooke, and A. Kornberg. 1992. The polyphosphate kinase gene of Escherichia coli. Isolation and sequence of the ppk gene and membrane location of the protein. J. Biol. Chem. 267:22556-22561. [PubMed] [Google Scholar]
- 4.Ault-Riche, D., C. D. Fraley, C. M. Tzeng, and A. Kornberg. 1998. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J. Bacteriol. 180:1841-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Booth, J. W., and G. Guidotti. 1995. An alleged yeast polyphosphate kinase is actually diadenosine-5′,5″-P1, P4-tetraphosphate alpha,beta-phosphorylase. J. Biol. Chem. 270:19377-19382. [DOI] [PubMed] [Google Scholar]
- 6.Chen, W., and H. K. Kuramitsu. 1999. Molecular mechanism for the spontaneous generation of pigmentless Porphyromonas gingivalis mutants. Infect. Immun. 67:4926-4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark, J. E., H. Beegen, and H. G. Wood. 1986. Isolation of intact chains of polyphosphate from “Propionibacterium shermanii” grown on glucose or lactate. J. Bacteriol. 168:1212-1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crooke, E., M. Akiyama, N. N. Rao, and A. Kornberg. 1994. Genetically altered levels of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 269:6290-6295. [PubMed] [Google Scholar]
- 9.de Jong, M. H., and J. S. van der Hoeven. 1987. The growth of bacteria in saliva. J. Dent. Res. 66:498-505. [DOI] [PubMed] [Google Scholar]
- 10.Duncan, M. J., S. A. Emory, and E. C. Almira. 1996. Porphyromonas gingivalis genes isolated by screening for epithelial cell attachment. Infect. Immun. 64:3624-3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissdorfer, W., A. Ratajczak, and W. Hillen. 1998. Transcription of ppk from Acinetobacter sp. strain ADP1, encoding a putative polyphosphate kinase, is induced by phosphate starvation. Appl. Environ. Microbiol. 64:896-901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottesman, S., and M. R. Maurizi. 2001. Cell biology. Surviving starvation. Science 293:614-615. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, M. C., R. J. Palmer, and D. C. White. 2000. Flow-cell culture of Porphyromonas gingivalis biofilm under anaerobic condition. J. Microbiol. Methods. 40:233-239. [DOI] [PubMed] [Google Scholar]
- 14.Hardoyo, K. Yamada, H. Shinjo, J. Kato, and H. Ohtake. 1994. Production and release of polyphosphate by a genetically engineered strain of Escherichia coli. Appl. Environ. Microbiol. 60:3485-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt, S. C., and T. E. Brammanti. 1991. Factors in virulence expression and their role in periodontal disease pathogenesis. Crit. Rev. Oral Biol. Med. 2:177-281. [DOI] [PubMed] [Google Scholar]
- 16.Ishige, K., A. Kameda, T. Noguchi, and T. Shiba. 1998. The polyphosphate kinase gene of Pseudomonas aeruginosa. DNA Res. 5:157-162. [DOI] [PubMed] [Google Scholar]
- 17.Ishige, K., and T. Noguchi. 2001. Polyphosphate:AMP phosphotransferase and polyphosphate:ADP phosphotransferase activities of Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 281:821-826. [DOI] [PubMed] [Google Scholar]
- 18.Kato, J., T. Yamamoto, K. Yamada, and H. Ohtake. 1993. Cloning, sequence and characterization of the polyphosphate kinase-encoding gene (ppk) of Klebsiella aerogenes. Gene 137:237-242. [DOI] [PubMed] [Google Scholar]
- 19.Keasling, J. D., and G. A. Hupf. 1996. Genetic manipulation of polyphosphate metabolism affects cadmium tolerance in Escherichia coli. Appl. Environ. Microbiol. 62:743-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornberg, A., N. N. Rao, and D. Ault-Riche. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda, A., and H. Ohtake. 2000. Molecular analysis of polyphosphate accumulation in bacteria. Biochemistry (Moscow) 65:304-308. [PubMed] [Google Scholar]
- 22.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 62:1244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McGrath, J. W., and J. P. Quinn. 2000. Intracellular accumulation of polyphosphate by the yeast Candida humicola G-1 in response to acid pH. Appl. Environ. Microbiol. 66:4068-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, R. J., K. Kazmerzak, M. Hansen, and P. Kolenbrander. 2001. Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69:5794-5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao, N. N., and A. Kornberg. 1996. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J. Bacteriol. 178:1394-1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rao, N. N., S. Liu, and A. Kornberg. 1998. Inorganic polyphosphate in Escherichia coli: the phosphate regulon and the stringent response. J. Bacteriol. 180:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rashid, M. H., N. N. Rao, and A. Kornberg. 2000. Inorganic polyphosphate is required for motility of bacterial pathogens. J. Bacteriol. 182:225-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rashid, M. H., K. Rumbaugh, L. Passador, D. G. Davies, A. N. Hamood, B. H. Iglewski, and A. Kornberg. 2000. Polyphosphate kinase is essential for biofilm development, quorum sensing, and virulence of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:9636-9641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogers, J. D., R. J. Palmer, P. E. Kolenbrander, and F. A. Scannapieco. 2001. Role of Streptococcus gordonii amylase-binding protein A in adhesion to hydroxyapatite, starch metabolism, and biofilm formation. Infect. Immun. 69:7046-7056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz, F. A., C. O. Rodrigues, and R. Docampo. 2001. Rapid changes in polyphosphate content within acidocalcisomes in response to cell growth, differentiation, and environmental stress in Trypanosoma cruzi. J. Biol. Chem. 276:26114-26121. [DOI] [PubMed] [Google Scholar]
- 33.Sethuraman, A., N. N. Rao, and A. Kornberg. 2001. The endopolyphosphatase gene: essential in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 98:8542-8547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiba, T., K. Tsutsumi, K. Ishige, and T. Noguchi. 2000. Inorganic polyphosphate and polyphosphate kinase: their novel biological functions and applications. Biochemistry (Moscow) 65:315-323. [PubMed] [Google Scholar]
- 35.Slots, J., and R. J. Genco. 1984. Black-pigmented Bacteriodes species, Capnocytophaga species, and Actinobacillus actinomycetemcomitans in human periodontal diseases: virulence factors in colonization, survival and tissue destruction. J. Dent. Res. 63:412-421. [DOI] [PubMed] [Google Scholar]
- 36.Smith, C. J., A. Parker, and M. B. Rogers. 1990. Plasmid transformation of Bacteroides spp. by electroporation. Plasmid 24:100-109. [DOI] [PubMed] [Google Scholar]
- 37.Tinsley, C. R., and E. C. Gotschlich. 1995. Cloning and characterization of the meningococcal polyphosphate kinase gene: production of polyphosphate synthesis mutants. Infect. Immun. 63:1624-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tokuda, M., M. Duncan, M.-I. Cho, and H. K. Kuramitsu. 1996. Role of Porphyromonas gingivalis protease activity in colonization of oral surfaces. Infect. Immun. 64:4067-4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tokuda, M., T. Karunakaran, M. Duncan, N. Hamada, and H. K. Kuramitsu. 1998. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect. Immun. 66:1159-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trelstad, P. L., P. Purdhani, W. Geissdorfer, W. Hillen, and J. D. Keasling. 1999. Polyphosphate kinase of Acinetobacter sp. strain ADP1: purification and characterization of the enzyme and its role during changes in extracellular phosphate levels. Appl. Environ. Microbiol. 65:3780-3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tzeng, C. M., and A. Kornberg. 2000. The multiple activities of polyphosphate kinase of Escherichia coli and their subunit structure determined by radiation target analysis. J. Biol. Chem. 275:3977-3983. [DOI] [PubMed] [Google Scholar]
- 42.Tzeng, C. M., and A. Kornberg. 1998. Polyphosphate kinase is highly conserved in many bacterial pathogens. Mol. Microbiol. 29:381-382. [DOI] [PubMed] [Google Scholar]
- 43.Van Niel, E. W., K. J. Appeldoorn, A. J. Zehnder, and G. J. Kortstee. 1998. Inhibition of anaerobic phosphate release by nitric oxide in activated sludge. Appl. Environ. Microbiol. 64:2925-2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu, K. D., M. J. Franklin, C. H. Park, G. A. McFeters, and P. S. Stewart. 2001. Gene expression and protein levels of the stationary phase sigma factor, RpoS, in continuously fed Pseudomonas aeruginosa biofilms. FEMS Microbiol. Lett. 199:67-71. [DOI] [PubMed] [Google Scholar]
- 45.Zago, A., S. Chugani, and A. M. Chakrabarty. 1999. Cloning and characterization of polyphosphate kinase and exopolyphosphatase genes from Pseudomonas aeruginosa 8830. Appl. Environ. Microbiol. 65:2065-2071. [DOI] [PMC free article] [PubMed] [Google Scholar]