Abstract
A non-O1 non-O139 Vibrio cholerae strain, 10259, belonging to the serogroup O53 was shown to harbor genes related to the vibrio pathogenicity island (VPI) and a cholera toxin (CT) genetic element called CTX. While the nucleotide sequence of the strain 10259 tcpA gene differed significantly (26 and 28%) from those of O1 classical and El Tor biotype strains, respectively, partial sequence analysis data of certain other VPI-associated genes (aldA, tagA, tcpP/H, toxT, acfB/C, and int) and intergenic regions (tcpF to toxT and tcpH to tcpA) of the strain showed only minor variations (0.4 to 4.8%) from corresponding sequences in O1 strains. Strain 10259 also contained CTX element-associated toxin genes with sequences almost identical to those of O1 strains. Growth of the organism in Luria broth (LB) under ToxR inducing conditions (30°C and pH 6.5) led to transcriptional activation of tcpP/H, toxR, toxT, and tcpA genes, but not of ctxA, as determined by reverse transcription-PCR (RT-PCR). Subsequent analysis revealed that strain 10259 possessed only two copies (instead of three or more copies found in epidemic-causing O1 or O139 strains) of the heptanucleotide (TTTTGAT) repeats in the intergenic region upstream of ctxAB. Therefore, a strain 10259 mutant was generated by replacement of this region with a homologous region (1.4 kb) derived from a V. cholerae O1 classical biotype strain (O395) that contained seven such repeats. The resultant recombinant strain (10259R) was found to be capable of coordinately regulated expression of toxT, ctxA, and tcpA when grown under the ToxR inducing conditions. Serological studies also demonstrated that the recombinant strain produced TcpA and a significantly (∼1,000-fold) higher level of CT in vitro compared to that of the parent strain. Virulence gene expression in two other non-O1 non-O139 strains (serogroup O37) containing VPI and the CTX element was studied by RT-PCR and serological assay. One strain (S7, which was involved in an epidemic in Sudan in 1968) showed coordinately regulated expression of virulence genes leading to the production of both CT and TcpA in LB medium. However, the other strain, V2, produced RT-PCR-detectable transcripts of toxT, ctxA, or tcpA genes in the early phase (6 h), but not in the late phase (16 h) of growth in LB medium. These results are consistent with the low levels of production of CT and TcpA by the strain that were serologically detectable. The significance of these results is discussed in relation to the role of virulence genes and their expression to the pathogenic potential of V. cholerae strains belonging to non-O1 serogroups.
In humans, the disease cholera is caused by strains of the gram-negative bacterium Vibrio cholerae that belong to the O1 or O139 serogroup. The organism enters into the host during ingestion of contaminated water or food material, colonizes the small intestine, and produces an enterotoxin (cholera toxin [CT]) that is primarily responsible for the induction of massive loss of salt and water in the form of diarrhea (18). Colonization of the gut is facilitated through the expression of bundle-forming pilus structures (toxin-coregulated pilus [TCP]) on the surface of the bacterium (48). The expression of both CT and TCP is coordinately regulated at the transcriptional level by a cascade of signaling pathways that involve several transmembrane and cytosolic regulatory proteins (23, 45). Briefly, in response to appropriate environmental stimuli, the transmembrane protein ToxR, in association with ToxS of the toxRS operon, activates toxT, the gene encoding the cytosolic protein ToxT, which in turn activates transcription of genes for CT (ctxAB), the major structural protein subunit gene tcpA, and several others involved in TCP biosynthesis and secretion (23, 45, 54). More recently, membrane-associated proteins (e.g., TcpP and TcpH of the tcpPH operon) have been shown to play an additional regulatory role in the transcriptional activation of toxT (6, 15). Furthermore, activation of tcpPH is controlled, either positively (46) or negatively (2), by several regulatory elements. The ctxAB operon is located within a larger genetic element called CTX, which also encodes genes for several other toxins and accessory virulence factors (38). CTX was likely to be integrated into the host chromosome following lysogenic conversion of a filamentous bacteriophage, CTXφ (51). Similarly, the tcp gene cluster has been shown to be a part of a 39-kb DNA region, referred to as a vibrio pathogenicity island (VPI), which contains a gene for the ToxT regulatory protein as well as several other clusters of genes of known and unknown function (19). Like CTX, the VPI has been proposed to be of another lysogenic bacteriophage origin (20). The acquisition of VPI by V. cholerae endows the organism with the ability to express TCP, which acts as a receptor for CTXφ (20). The VPI and CTX genetic elements are primarily found in V. cholerae strains of O1 and O139 serogroups, which are associated with epidemic cholera (18, 19). The majority of strains belonging to about 200 other non-O1 non-O139 serogroups (53) do not contain genes for CT and/or TCP (18, 31, 47), although the gene for ToxR is ubiquitously present in these strains (34). The non-O1 non-O139 strains, which are predominantly isolated from an aquatic environment, are largely nonpathogenic in nature, although some of these are known to cause sporadic cases or occasional outbreaks of diarrhea in humans (30). Recently, however, ctxAB- and tcp-related genes have been shown to be present in certain strains of non-O1 non-O139 V. cholerae of both clinical and environmental origins (7, 14, 35, 36, 40, 42). This has raised important issues related to their evolution as well as relevance from the public health point of view. The point assumes considerable significance in view of the fact that strains belonging to non-O1 non-O139 serogroups have recently been implicated as the causative agents of a large number of cases of diarrhea in various parts of the world (1, 9, 41, 43). Evidently, documentation of the mere presence of the virulence-associated genes is not likely to provide sufficient information on the pathogenic potential of these strains, which is likely to depend on the presence of complex signaling pathways to couple appropriate environmental signals to virulence gene expression. Although some of the non-O1 non-O139 strains described earlier were shown to express detectable amounts of CT and/or TcpA protein in vitro (10, 14, 31, 32, 35), only limited information about their VPI and CTX is available so far (7, 14, 20), and no data on the regulation of virulence gene expression are available. In an earlier study, we characterized a new type of TcpA protein in a toxigenic V. cholerae strain, 10259, belonging to serogroup O53 (35). In the present communication, we describe partial characterization of VPI and CTX genetic elements of strain 10259 and provide information on its pathogenic potential by determining its ability to express virulence genes when grown in vitro under conditions that favor the expression of the same genes in V. cholerae O1 strains. We have extended this study by including data obtained with two other non-O1 non-O139 strains harboring VPI and CTX-related genes.
The bacterial strains and plasmids used in this study are listed in Table 1. The presence of virulence-associated genes in V. cholerae strains was determined by PCR amplification experiments with the primers listed in Table 2. All of these target genes, except toxR, are located in the VPI or CTX genetic element of the V. cholerae chromosome. Bacterial cells were grown overnight in Luria broth (LB) at 37°C, and chromosomal DNA was isolated from the harvested bacteria by a standard protocol. PCR amplification of target DNA was carried out in a thermal cycler (Perkin-Elmer) by essentially following the methodology described earlier (34). The reaction mixture was subjected to 30 cycles of amplification. Each cycle consisted of three successive steps in the following order: denaturation at 94°C for 30 s, annealing at 55°C for 50 s, and extension at 72°C for 50 s.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Serogroup or characteristica | Source or reference |
|---|---|---|
| Strains | ||
| V. cholerae | ||
| O395 | O1 (classical/Ogawa), Smr | 25 |
| AD48 | O1 (E1 Tor/Ogawa) | 14 |
| Co900 | O1 (E1 Tor/Inaba) | Clinical isolate (local) |
| V1228 | O1 (E1 Tor/Ogawa), ΔtcpA | 49 |
| 10259 | O53 | 10 |
| V2 | O37 | 10 |
| S7 | O37 | 3 |
| E. coli SM10λ pir | thi thr leu tonA lacA supE recA::RP4-Tc::Muλ pir R6K | 29 |
| Plasmid | ||
| pCVD 442 | sacB gene containing suicide vector, Apr | 12 |
Smr, streptomycin resistant; Apr, ampicillin resistant.
TABLE 2.
List of primers used in this study
| Primera | Primer sequence (5′→3′) | Reference |
|---|---|---|
| VPI | ||
| aldA-(F) | TGTGTTTCTCGTCCAATGCC | This study |
| aldA-(R) | TCGTCTTCGCAAGATGTCGAACTTGC | |
| tagA-(F) | GGTGGTAAGATATTCACTC | 33 |
| tagA-(R) | GAGACATCTATAGAATACTGGCTG | |
| tcpP-(F) | ACTCTGTGAATATCATCCTGCC | This study |
| tcpH-(R) | CTGGGTAAGCCAAACATTGG | |
| tcpI-(F) | GCCGTCTCCGCATTAAGCTCTGCAC | 33 |
| tcpA-(F) | CACGATAAGAAAACCGGTCAAGAG | 33 |
| tcpA-(R) | ACCAAATGCACGCCGAATGGAGC | |
| tcpQ-(R) | GAGGACTGTTCTGCAATCTGCTCAT | This study |
| toxT-(F) | ACTGTATAGCAAAGCATATTCAGAGA | This study |
| toxT-(R) | CGCGGATCCATACAATCGAAAATAGGA | |
| tcpF-(F) | CTGTCAAACCATATCAGC | This study |
| tcpJ-(R) | TAAAGTAAAGCCCGAGC | |
| acfB-F | AATGTCAGACTTTGGCG | This study |
| acfC-R | CGAGATCGATAAGTCTTCC | |
| orfZ-(F) | CCATCATTCACGCCTGGGACTTCAT | 7 |
| orfW-(R) | TTCGTAATATGGCTGAGGATCATCTG | |
| int-(F) | GATAAAGAGATCAAAGCC | 33 |
| int-(R) | ATCTGCTTCCATGTGGG | |
| LJ-(F) | GTGAATCTTGATGAGACGCTCTG | 33 |
| LJ-(R) | GGTGAGCCAGGCTTATTTGGG | |
| RJ-(F) | TCGTTAGCGTGTTCGGTTCGCAGG | 33 |
| RJ-(R) | TGCTTTGTACCAGTCACAGATAG | |
| CTX | ||
| cep-(F) | TCGTTAGCGTGTCGGTTCGCAGG | This study |
| cep-(R) | TGCTTTGTACCAGTCACAGATAG | |
| orfU-(F) | GCTACATGTTTAGCTCACTG | This study |
| orfU-(R) | AGGTGCGTTAGTCATCAGCG | |
| ace-(F) | GGCGTATTGTATCTATTAAA | This study |
| ace(R) | GGTGTTATTTGATGGCTGCATG | |
| zot-(R) | GGCGGTACGAGTAAAACAAATCC | This study |
| zot-(F) | GCTTATGATGGACACCCTTTA | |
| ctxA-(F) | TTTAACGCTCGCAGGGC | 35 |
| ctxA-(R) | GGGCGAGAAAGGACGC | |
| ctxB-(F) | GGTTGCTTCTCATCATCGAACCAC | 7 |
| ctxB-(R) | GATACACATAATAGAATTAAGGAT | |
| toxR | ||
| toxR-(F) | ATGTTCGGATTAGGACAC | 13 |
| toxR-(R) | TACTCACACACTTTGATGGC |
F, forward primer; R, reverse primer.
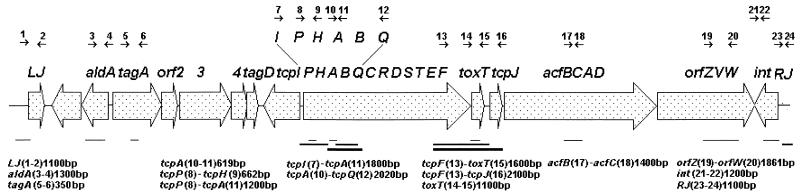
Our earlier study (34) demonstrated that the toxR gene was present in about 98% of V. cholerae strains tested, and strain 10259 was found to be no exception to this rule. The toxR gene nucleotide sequence of 10259 differed at only 3 base positions and 1 base position, respectively, from those of classical and El Tor strains (data not shown). Furthermore, PCR amplification of VPI-associated genes in the non-O1 non-O139 V. cholerae strain 10259 produced amplicons with all of the combinations of primers as shown in Fig. 1. Amplicon sizes were verified by comparison of the products with those generated with the V. cholerae O1 strain (O395). Some of the amplicons generated with strain 10259 were purified and sequenced with an automated DNA sequencer (Applied Biosystems) by using the end or internal primers. Partial nucleotide sequences of the genes aldA, tagA, tcpP/H, tcpA, toxT, acfB/C, and int of strain 10259 were determined and compared with those already available in the literature for V. cholerae O1 strains of both the classical and El Tor biotypes (GenBank accession no. X64098 and X74730 and AE004168 and AE004169, respectively). The summarized results presented in Table 3 show only minor variations (0.3 to 2.8%) between the 10259 sequence and the corresponding sequences from either classical or El Tor strains. In contrast, the nucleotide sequence of 10259 tcpA differed from those of classical and El Tor tcpA genes by 26 and 28%, respectively (35). Partial sequence analysis of the intergenic region between tcpF-toxT of 10259 revealed only minor (1%) variation from sequence of the corresponding region of V. cholerae O1 strains. However, the tcpH-tcpA intergenic sequence covering the promoter proximal region (302 bp) of tcpA showed 3.3 and 4.3% changes with the corresponding region sequences of classical and El Tor strains, respectively. No difference, however, was detectable at −10 and −35 regions upstream of tcpA. The GC content of the partially sequenced region of 10259 VPI was found to be 34.6%, which is quite comparable to those (35.5%) of VPIs of O1 strains (19), but differs from the overall GC content (47%) of the V. cholerae genome (16). It is therefore reasonable to conclude that, as is the case with V. cholerae O1, the 10259 VPI was also acquired from an outside donor. Generation of amplicons with right and left junction primers (Fig. 1) suggests that the VPI was likely to be integrated in the 10259 chromosome at sites identical to those of O1 strains (19, 22).
FIG. 1.
Schematic representation of V. cholerae VPI with its right (RJ) and left (LJ) junctions. Amplicons generated with strain 10259 by using various primer pairs are shown with their respective sizes. Arrows represent forward and reverse primer pairs.
TABLE 3.
Sequence divergence of VPI-associated genes of non-O1 non-O139 V. cholerae strain 10259 from those of O1 strains
| Target gene(s) or intergenic region | Size (bp) of:
|
Divergence from O1 strains in bp (%)
|
||
|---|---|---|---|---|
| Target sequence | Sequence compared | Classicala | E1 Torb | |
| aldA | 1,300 | 1,180 | 8 (0.6) | 9 (0.7) |
| tagA | 350 | 315 | 1 (0.3) | 2 (0.6) |
| tcpP-tcpH | 662 | 610 | 6 (0.9) | 7 (1.1) |
| Intergenic region (tcpH-tcpA) | 598 | 302 | 10 (3.3)c | 13 (4.3) |
| tcpA | 619 | 590d | 157 (26.0)d | 169 (28.0)d |
| Intergenic region (tcpF-toxT) | 207 | 187 | 2 (1.0) | 2 (1.0) |
| toxT | 1,100 | 588 | 7 (1.2) | 17 (2.8) |
| acfB-acfC | 1,400 | 963 | 5 (0.5) | 9 (0.9) |
| int | 1,200 | 955 | 3 (0.3) | 9 (0.9) |
Sequence analysis data obtained so far suggest that the 10259 VPI differs from the VPIs of O1 strains primarily with respect to the tcpA gene, although minor variations could also be detected in the adjoining regions. In a recent study, Karaolis et al. (21) demonstrated that most of the divergence between the VPIs of 6th (classical) and 7th (El Tor) pandemic strains were located in or around the tcpA gene constituting the central region of VPI, but not its left or right segments. The authors hypothesized that this could arise as a result of a recombinational event involving genes located in the central segment. A similar mechanism might have contributed to generate the 10259 VPI, which shows an equal degree of divergence from both classical and El Tor biotype strains with respect to its tcpA and adjacent region sequences. If true, a crucial factor leading to such a recombinational event would be the generation of new variants of TcpA different from both classical or El Tor TcpA proteins. A comparative analysis (35) of four different TcpA variants (including the 10259 TcpA) revealed that the sequence variations were primarily in the carboxyl-terminal half of the 20-kDa protein with the majority of the variable or hypervariable residues located along the surface of TcpA, leading to alterations in their surface structures or epitopes (8). Such alterations are likely to influence the reactivity of the pilus or pilus protein to antibodies and/or ligands or receptors of biological importance.
The CTX genetic element of the strain 10259 was also probed with the primers for ctxA, ctxB, zot, ace, orfU, and cep genes, as listed in Table 2. All of these primers produced amplicons of the desired sizes, thereby documenting the presence of these genes in this non-O1 non-O139 strain. Amplicons generated with ctxA and orfU primers were subjected to sequencing analysis, and data were compared with those of O1 strains. The ctxA of 10259 showed only minimum changes from those of classical (0 bp) and El Tor (2 bp) strains. On the other hand, the 10259 orfU sequence diverged significantly from that of the classical strain by 56 bp (9.1% with 14 synonymous and 42 nonsynonymous changes), although it differed from the El Tor orfU by only 8 bp (1.3% with 6 synonymous and 2 nonsynonymous changes).
Recently, orfU has been proposed to be involved in the interaction of CTXφ to its receptor TCP on the V. cholerae surface (5). A significant difference between the orfU sequences of classical and El Tor biotype strains was postulated to be responsible for the specific recognition of CTXφs by biotype-specific TcpA proteins, the major structural unit of TCP. Therefore, the fact that the orfU sequence of 10259 shows close similarity to that of El Tor (but not of classical orfU), appears to be somewhat at variance to this concept, since 10259 TcpA was predicted to differ equally from both classical and El Tor TcpA proteins (Table 3). Our results are, however, in agreement with those obtained with certain other non-O1 non-O139 strains, the orfU sequences of which were shown to be more similar to those of El Tor than to those of classical strains (5). At least, two of these strains, 158 and 208, possessed a new variant of TcpA with significant differences from classical, El Tor, as well as 10259 TcpA (35). All of these considerations would suggest that the drift in the TcpA sequence in non-O1 non-O139 V. cholerae strains may not necessarily be related to the need to bind to different OrfU proteins for the acquisition of new type of CTXφ. As a matter of fact, TcpA-independent acquisition of CTXφ by V. cholerae under selective conditions has been documented recently (4, 13).
Documentation of the presence of VPI and CTX elements in strain 10259 and partial characterization of its virulence-associated genes have prompted us to address the question of their expression in relation to pathogenesis. Therefore, the expression of toxR, tcpP/H, toxT, ctxA, and tcpA genes was studied by reverse transcription-PCR (RT-PCR) with the set of primers listed in Table 1. Briefly, bacterial RNA was extracted with TRIZOL reagent (GIBCO-BRL) from cells grown under the appropriate culture conditions, and the extracted material was treated with RNase-free DNase (Ambion). Purified RNA was used to obtain cDNA. For this, 1 μg of RNA was mixed with 0.1 M dithiothreitol (DTT), 2 pmol of each primer of the primer pair, 10 mM deoxynucleotide triphosphate (dNTP), and 5× first-strand buffer in 9.5 μl of reaction volume, and the mixture was incubated at 42°C for 2 min followed by immediate cooling. Next, 100 U of the enzyme reverse transcriptase (GIBCO-BRL) was added to this mixture, which was incubated at 42°C for 50 min. The reaction was terminated by incubating the mixture at 70°C for 15 min. The cDNA preparation thus obtained was amplified by PCR by the methodology described earlier.
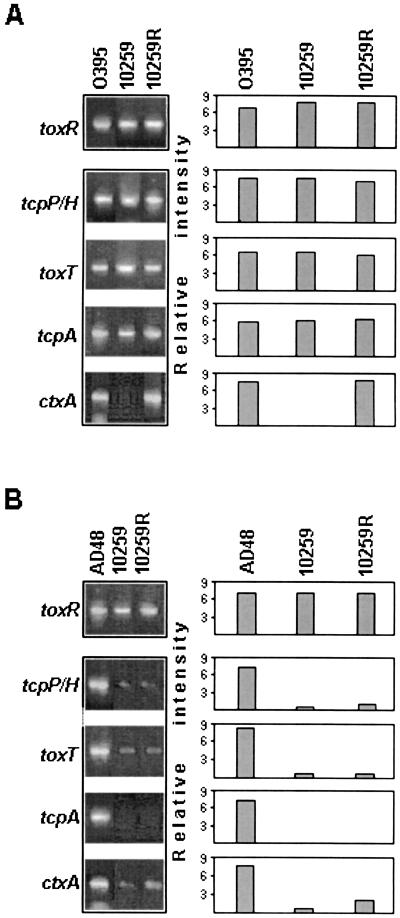
The results obtained in the RT-PCR experiments demonstrate that strain 10259, when grown in LB at pH 6.5 for 16 h at 30°C under mild shaking conditions, expressed transcripts for toxR, tcpP/H, toxT, and tcpA, but not for ctxA (Fig. 2A). Similar results were obtained when the organism was grown in the same LB medium under static conditions or in colonization factor antigen (CFA) agar plates (data not shown). In all of these experiments, the V. cholerae O1 classical strain (O395) was used as the positive control to document the expression of virulence gene-associated transcripts as mentioned above (Fig. 2A). The expression of these genes was also tested by growing strain 10259 in modified AKI medium (52), which is known to favor the expression of ctxAB in El Tor biotype strains. While the control El Tor strain AD48 expressed toxR, tcpP/H, toxT, ctxA, and tcpA when grown in AKI medium at 37°C at pH 7.8 for 4 h under static conditions followed by 2 h of shaking conditions (24), strain 10259 showed expression of toxR with only weakly detectable transcripts of toxT and ctxA, but none for tcpP/H or tcpA (Fig. 2B). Culture of strain 10259 for a longer period (4 h of static conditions plus 6 h of shaking conditions) also failed to produce elevated levels of transcripts for these genes (data not shown).
FIG. 2.
Detection of toxR, tcpP/H, toxT, tcpA, and ctxA transcripts by RT-PCR in V. cholerae strains O395, AD48, 10259, and 10259R. Organisms were grown in LB medium at pH 6.5 at 30°C for 16 h under mild shaking conditions (A) and AKI medium at pH 7.8 at 37°C for 4 h under static conditions followed by 2 h of shaking conditions (B). Columns in the right-hand panel show the relative intensity of the bands (left-hand panel) determined by the densitometric analysis with the Molecular Analyst (version 1.5) software package.
Failure to detect ctxA expression of 10259 in LB medium (which activated the transcription of both toxR and toxT genes) prompted us to examine the promoter proximal region located upstream of ctxAB, which contains the binding sites for ToxR and/or ToxT proteins (11, 27, 28, 39). Therefore, the region between the zot and ctxA genes in strain 10259 was amplified with the forward primer of the zot gene and a reverse one from the ctxA gene (Table 2). The amplified product of 1.4 kb, which contained the zot-ctxA intergenic region (124 bp) and parts of the ctxA and zot genes (434 and 923 bp, respectively), was sequenced, and the data were compared with the corresponding region sequence of V. cholerae O1 strain O395. The comparison revealed that while strain O395 contained seven copies of a heptanucleotide (TTTTGAT) repeat, strain 10259 contained only two such copies (Fig. 3). This difference appeared to be of considerable interest, since earlier studies (26) already demonstrated that the repeated sequence is required for the activation of the ctxAB promoter and a higher number of such tandem repeats produces higher levels of toxin. Apart from this, the zot-ctxA intergenic region sequence of 10259 did not show any other difference from that of the strain O395. However, two nucleotide changes were noted between these two strains in the partial (443 bp) zot sequences.
FIG. 3.
Partial nucleotide sequence comparison of the ctxAB upstream regions of V. cholerae strains O395 and 10259. The sequences are aligned to show differences in the numbers of heptanucleotide repeats (in boxes).
Based on the results presented so far, it is tempting to speculate that the observed lack of ctxA expression by strain 10259 under toxR- and toxT-expressing conditions could be attributed to the presence of only two copies of the heptanucleotide repeats, instead of the three to eight such copies that are reported to be present in the toxigenic O1 strains tested so far (26). To test this hypothesis, a recombinant of strain 10259 was generated through the replacement of its zot-ctxA intergenic region with the homologous region derived from V. cholerae O1 strain O395, which has seven copies of the TTTTGAT repeats. For this, the chromosomal DNA of O395 was amplified with the zot forward and ctxA reverse primers, and the resultant amplified product of 1.4 kb was directly cloned into a suicide vector, pCVD442 (12). Briefly, the plasmid pCVD442 was digested with SmaI, and a single 3′-T overhang (44) of the linearized product was generated by incubating 2 μg of the purified digested material with 10 mM dTTP, 50 mM MgCl2, 5 U of Taq DNA polymerase, and 10× reaction buffer in a final reaction volume of 50 μl. An aliquot (20 μl) of the mixture was purified and mixed with 1.5 μg of the amplified product, and the mixture was then incubated at 23°C for 16 h in the presence of T4 DNA ligase (GIBCO-BRL). The ligated product (pCAS442) was used to transform Escherichia coli SM10λpir (29), and ampicillin-resistant transformants were selected. Transformed E. coli cells harboring the recombinant vector pCAS442 were allowed to conjugate with V. cholerae strain 10259 at a donor/recipient ratio of 1:10. Transconjugants of V. cholerae (harboring the chromosomally integrated vector) were selected on thiosulfate-citrate-bile-sucrose (TCBS) agar plates containing ampicillin (50 μg/ml) and checked for their reactivity to the antisera to the O53 serogroup. Next, selected transconjugant colonies were grown on Luria agar (LA) plates containing 10% (wt/vol) sucrose to select organisms to undergo a second recombinational event resulting in the deletion of the suicide vector sequence from the host chromosome. The recombinant colonies thus obtained (10259R) were further checked by their growth requirements, reactivity to O53-specific antisera, and sensitivity to ampicillin. Finally, the chromosomal DNA of 10259R was amplified with the zot-ctxA primer pair, and the product was sequenced to ensure that it contained seven repeats of the heptanucleotide sequence (TTTTGAT) at the proper position.
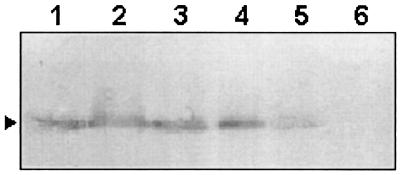
Following characterization, the recombinant strain 10259R was tested for expression of virulence genes by RT-PCR as described earlier. When grown in LB at pH 6.5 at 30°C, the organism was shown to express ctxA in addition to tcpP/H, toxR, toxT, and tcpA (Fig. 2A). However, transcription of tcpP/H, toxT, and ctxA was detectable at low levels only in AKI medium-grown cells of strain 10259R (Fig. 2B). The RT-PCR data were extended through determination of the translated products CT and TcpA. Production of CT was estimated by GM1 enzyme-linked immunosorbent assay (ELISA) (17) with culture supernatants of cells grown under the appropriate culture conditions. While the wild-type strain 10259 produced a barely detectable level of CT in the LB-grown culture supernatant, the recombinant strain 10259R produced a significantly (about 1,000-fold) larger amount of CT in cultures grown under the same conditions (Table 4). In fact, CT produced by 10259R was quite comparable to that produced by the classical strain O395 in LB medium. Production of TcpA was also tested by immunoblotting experiments with a rabbit polyclonal antiserum to the protein. Both the wild-type and recombinant strains expressed TcpA when grown in LB medium, and the level of expression was quite comparable to that of the classical strain O395 grown under the same culture conditions (Fig. 4).
TABLE 4.
CT production by V. cholerae strains grown under two different cultural conditions
| Strain | Amt of CTa produced in medium (ng/ml/unit of opacity)
|
|
|---|---|---|
| LBb | AKIc | |
| O395 | 2,700 | NDd |
| 10259 | 2.8 | 16 |
| 10259R | 2,650 | 53 |
| AD48 | ND | 663 |
| Co900 | ND | 1,035 |
| V2 | 216 | 24 |
| S7 | 1,800 | 80 |
Assayed by the GM1 ELISA method (17) and expressed as nanograms of toxin per milliliter of culture supernatant per unit of opacity of bacterial suspension measured at 540 nm.
LB medium, pH 6.5, incubation temperature of 30°C, 16 h of culture.
AKI medium, pH 7.8, incubation temperature of 37°C, 4 h of static culture with limited aeration followed by 16 h of mild shaking culture with adequate aeration.
ND, not determined.
FIG. 4.
Expression of TcpA by V. cholerae strains grown in LB medium (pH 6.5 at 30°C for 16 h) as determined by immunoblotting experiments with anti-TcpA serum. Whole-cell lysates of O395 (lane 1), 10259 (lane 2), 10259R (lane 3), S7 (lane 4), V2 (lane 5), and a ΔtcpA strain, V1228 (lane 6), were used as antigens. An arrowhead indicates 20-kDa TcpA bands.
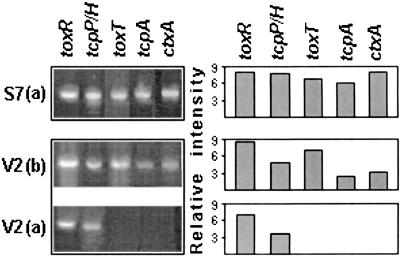
Expression of virulence genes in two other non-O1 non-O139 strains (V2 and S7) was also studied by RT-PCR. The strains, which were obtained from different sources, belonged to the same serogroup, O37 (Table 1), and possessed similar restriction fragment length polymorphism pattern on pulsed-field gel electrophoresis. Both of the strains contained VPI (with classical type of tcpA) and CTX-associated genes that were demonstrable by the PCR and/or DNA probe assay (14, 19; A. Sarkar and A. C. Ghose, unpublished data). When grown in LB medium, the strain S7 produced tcpP/H, toxR, toxT, tcpA, and ctxA transcripts (Fig. 5), which could be corroborated through the determination of its ability to produce CT and TcpA by ELISA (Table 4) and immunoblotting experiments (Fig. 4), respectively. On the other hand, only tcpP/H and toxR transcripts were detectable with the strain V2 grown for 16 h in LB (Fig. 5). Interestingly enough, transcripts of toxT, ctxA, and tcpA (in addition to those of tcpP/H and toxR) could be detected in V2 when grown for 6 h under the same culture conditions. These results are in apparent agreement with the low-level production of CT (Table 4) and TcpA (Fig. 4) by the strain as demonstrable by serological assays.
FIG. 5.
Detection of toxR, tcpP/H, toxT, tcpA, and ctxA transcripts by RT-PCR in V. cholerae strains V2 and S7. Organisms were grown in LB medium at pH 6.5 at 30°C for 16 h (a) or 6 h (b) under mild shaking conditions. Columns in the right-hand panel show the relative intensity of the bands (left-hand panel) determined by densitometric analysis.
Non-O1 non-O139 Vibrio cholerae strains harboring CT genes have been shown to produce considerably smaller amounts of CT than those produced by their O1 counterparts (10, 14, 32, 33). Several possible explanations may be provided for this, including (i) the absence of ToxT protein (due to the absence of VPI or toxT gene), which is known to be responsible for enhanced production of CT in V. cholerae; (ii) the presence of a toxT allele functionally deficient from the canonical toxT; (iii) poor production or a lack of production of ToxT as a result of defects or deviations in the signaling pathway that couple appropriate environmental signals to toxT activation; and (iv) failure to activate ctxAB operon (despite the production of an adequate amount of functional ToxT) due to the lack of a sufficient number of the heptanucleotide repeats in its promoter region.
Expression of ctxA in absence of any detectable transcripts of tcpA by strain 10259R in AKI medium raises an interesting question regarding the relative efficiency with which these two genes can be transcriptionally activated in V. cholerae. In a recent study, Yu and DiRita (55) have demonstrated that, although ToxT is the direct activator of both ctxA and tcpA, ctxA transcription regulation is much more complex than that of tcpA. It is also suggested that the ctxA is under the control of a more efficient ToxT-dependent promoter compared to that of tcpA. Thus, under ToxT-limiting conditions, as is the case for strain 10259R grown in AKI medium, expression of ctxA may be achieved even in the absence of a detectable level of tcpA transcripts.
Coordinately regulated expression of CT and TcpA is an essential (although perhaps not sufficient) feature of epidemic-causing strains of V. cholerae, which so far has differentiated them from strains that are not associated with epidemics. Evidently, the genetic background of these strains plays a crucial role in this effect. The emergence of the O139 strain with epidemic potential from an O1 El Tor strain demonstrates that this feature may be retained by a non-O1 strain as well, provided it has a genetic makeup otherwise similar to that of O1 strains (50). Coordinate expression of CT and TcpA in the non-O1 non-O139 V. cholerae strain S7 described here also supports this concept, since this strain was likely to be derived from an O1 classical strain (3) and known to be involved in an epidemic in Sudan in 1968. Results obtained with the genetically engineered strain 10259R, however, demonstrate that coordinate expression of high levels of CT and TcpA is also achievable with a non-O1 non-O139 strain that may not be directly related to an epidemic strain as its origin. Although strain 10259R has shown its pathogenic potential in in vitro experiments, preliminary data generated in our laboratory have demonstrated considerable enhancement of its ability to produce toxin in vivo.
Acknowledgments
This work was supported by a grant [37(1008)/99-EMR-II] from the Council of Scientific and Industrial Research, Government of India.
The helpful technical assistance of Prabal Gupta and Debashis Mazumder is acknowledged.
Editor: V. J. DiRita
REFERENCES
- 1.Bagchi, K., P. Echeverria, J. D. Arthur, O. Sethabutr, O. Serichantalergs, and C. W. Hoge. 1993. Epidemic diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J. Clin. Microbiol. 31:1315-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behari, J., L. Stagon, and S. B. Calderwood. 2001. pepA, a gene mediating pH regulation of virulence genes in Vibrio cholerae. J. Bacteriol. 183:178-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bik, E. M., R. D. Gouw, and F. R. Mooi. 1996. DNA fingerprinting of Vibrio cholerae strains with a novel insertion sequence element: a tool to identify epidemic strains. J. Clin. Microbiol. 34:1453-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd, E. F., and M. K. Waldor. 1999. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: generalized transduction of CTXΦ by bacteriophage CP-T1. Infect. Immun. 67:5898-5905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd, E. F., A. J. Heilpern, and M. K. Waldor. 2000. Molecular analysis of a putative CTXφ precursor and evidence for independent acquisition of distinct CTXφs by toxigenic Vibrio cholerae. J. Bacteriol. 182:5530-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll, P. A., K. T. Tashima, M. B. Rogers, V. J. DiRita, and S. B. Calderwood. 1997. Phase variation in tcpH modulates expression of the ToxR regulon in Vibrio cholerae. Mol. Microbiol. 25:1099-1111. [DOI] [PubMed] [Google Scholar]
- 7.Chakraborty, S., A. K. Mukhopadhyay, R. K. Bhadra, A. N. Ghosh, R. Mitra, T. Shimada, S. Yamasaki, S. M. Faruque, Y. Takeda, R. R. Colwell, and G. B. Nair. 2000. Virulence genes in environmental strains of Vibrio cholerae. Appl. Environ. Microbiol. 66:4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chattopadhyaya, R., and A. C. Ghose. 2002. Model of Vibrio cholerae toxin coregulated pilin capable of filament formation. Protein Eng. 15:297-304. [DOI] [PubMed] [Google Scholar]
- 9.Dalsgaard, A., M. J. Albert, D. N. Taylor, T. Shimada, R. Meza, O. Serichantalergs, and P. Echeverria. 1995. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J. Clin. Microbiol. 33:2715-2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datta-Roy, K., K. Banerjee, S. P. De, and A. C. Ghose. 1986. Comparative study of expression of hemagglutinins, hemolysins, and enterotoxins by clinical and environmental isolates of non-O1 Vibrio cholerae in relation to their enteropathogenicity. Appl. Environ. Microbiol. 52:875-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DiRita, V. J., C. Parsot, G. Jander, and J. J. Mekalanos. 1991. Regulatory cascade controls virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 88:5403-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eae deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faruque, S. M., Asadulghani, A. R. M. A. Alim, M. J. Albert, K. M. N. Islam, and J. J. Mekalanos.1998. Induction of the lysogenic phage encoding cholera toxin in naturally occurring strains of toxigenic Vibrio cholerae O1 and O139. Infect. Immun. 66:3752-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ghosh, C., R. K. Nandy, S. K. Dasgupta, G. B. Nair, R. H. Hall, and A. C. Ghose. 1997. A search for cholera toxin (CT), toxin coregulated pilus (TCP), the regulatory element ToxR and other virulence factors in non-O1/non-O139 Vibrio cholerae. Microb. Pathog. 22:199-208. [DOI] [PubMed] [Google Scholar]
- 15.Hase, C. C., and J. J. Mekalanos. 1998. TcpP protein is a positive regulator of virulence gene expression in Vibrio cholerae. Proc. Natl. Acad. Sci. USA 95:730-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolavea, J. Vamathevan, S. Bass, H. Qin, I. Daragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosome of the cholera pathogen Vibrio cholerae. Nature 406:477-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holmgren, J. 1973. Comparison of the tissue receptors for Vibrio cholerae and Escherichia coli enterotoxins by means of gangliosides and natural cholera toxoid. Infect. Immun. 8:851-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaper, J. B., J. G. Morris, Jr., and M. M. Levine. 1995. Cholera. Clin. Microbiol. Rev. 8:48-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karaolis, D. K. R., J. A. Johnson, C. C. Bailey, E. C. Boedeker, J. B. Kaper, and P. R. Reeves. 1998. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc. Natl. Acad. Sci. USA 95:3134-3139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karaolis, D. K. R., S. Somara, D. R. Manaeval, Jr., J. A. Johnson, and J. B. Kaper. 1999. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature 399:375-379. [DOI] [PubMed] [Google Scholar]
- 21.Karaolis, D. K. R., R. Lan, J. B. Kaper, and P. R. Reeves. 2001. Comparison of Vibrio cholerae pathogenicity islands in sixth and seventh pandemic strains. Infect. Immun. 69:1947-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kovach, M. E., M. D. Shaffer, and K. M. Peterson. 1996. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology 142:2165-2174. [DOI] [PubMed] [Google Scholar]
- 23.Krukonis, S. E., R. R. Yu, and V. J. DiRita. 2000. The Vibrio cholerae ToxR/TcpP/ToxT virulence cascade: distinct roles for two membrane-localized transcriptional activators on a single promoter. Mol. Microbiol. 38:67-84. [DOI] [PubMed] [Google Scholar]
- 24.Medrano, A. I., V. J. DiRita, G. Castillo, and J. Sanchez. 1999. Transient transcriptional activation of the Vibrio cholerae El Tor virulence regulator ToxT in response to culture conditions. Infect. Immun. 67:2178-2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mekalanos, J. J. 1983. Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35:253-263. [DOI] [PubMed] [Google Scholar]
- 26.Mekalanos, J. J., D. J. Swartz, G. D. N. Pearson, N. Harford, F. Groyne, and M. de Wilde. 1983. Cholera toxin genes: nucleotide sequence, deletion analysis and vaccine development. Nature 306:551-557. [DOI] [PubMed] [Google Scholar]
- 27.Miller, V. L., and J. J. Mekalanos. 1984. Synthesis of cholera toxin is positively regulated at the transcriptional level by toxR. Proc. Natl. Acad. Sci. USA 81:3471-3475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, V. L., R. K. Taylor, and J. J. Mekalanos. 1987. Cholera toxin transcriptional activator ToxR is a transmembrane DNA binding protein. Cell 48:271-279. [DOI] [PubMed] [Google Scholar]
- 29.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morris, J. G., Jr. 1990. Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol. Rev. 12:179-191. [DOI] [PubMed] [Google Scholar]
- 31.Morris, J. G., Jr. 1994. Non-O group 1 Vibrio cholerae strains not associated with epidemic disease, p. 103-115. In I. K. Wachsmuth, P. A. Blake, and Ø. Olsvik (ed.), Vibrio cholerae and cholera: molecular to global perspectives. ASM Press, Washington, D.C.
- 32.Mukhopadhyay, A. K., P. K. Saha, S. Garg, S. K. Bhattacharya, T. Shimada, T. Takeda, Y. Takeda, and G. B. Nair. 1995. Distribution and virulence of Vibrio cholerae belonging to serogroups other than O1 and O139: a nation survey. Epidemiol. Infect. 114:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mukhopadhyay, A. K., S. Chakraborty, Y. Takeda, G. B. Nair, and D. E. Berg. 2001. Characterization of VPI pathogenicity island and CTXφ prophage in environmental strains of Vibrio cholerae. J. Bacteriol. 183:4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nandi, B., R. K. Nandy, S. Mukhopadhyay, G. B. Nair, T. Shimada, and A. C. Ghose. 2000. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J. Clin. Microbiol. 38:4145-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nandi, B., R. K. Nandy, A. C. P. Vicente, and A. C. Ghose. 2000. Molecular characterization of a new variant of toxin-coregulated pilus protein (TcpA) in a toxigenic non-O1/non-O139 strain of Vibrio cholerae. Infect. Immun. 68:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novais, R. C., A. Coelho, C. A. Salles, and A. C. Vicente. 1999. Toxin-co-regulated pilus cluster in non-O1, non toxigenic Vibrio cholerae: evidence of a third allele of pilin gene. FEMS Microbiol. Lett. 171:49-55. [DOI] [PubMed] [Google Scholar]
- 37.Ogierman, M. A., E. Voss, C. Meaney, R. Faast, R. Attridge, and P. A. Manning. 1996. Comparison of the promoter proximal regions of the toxin coregulated tcp gene cluster in classical and El Tor strains of Vibrio cholerae O1. Gene 170:9-16. [DOI] [PubMed] [Google Scholar]
- 38.Pearson, G. D. N., A. Woods, S. L. Chiang, and J. J. Mekalanos. 1993. CTX genetic element encodes a site specific recombination system and an intestinal colonization factor. Proc. Natl. Acad. Sci. USA 90:3750-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pfau, J. D., and R. K. Taylor. 1996. Genetic footprint of the ToxR-binding site in the promoter for cholera toxin. Mol. Microbiol. 20:213-222. [DOI] [PubMed] [Google Scholar]
- 40.Rivera, I. N. G., J. Chun, A. Huq, R. B. Sack, and R. R. Colwell. 2001. Genotypes associated with virulence in environmental isolates of Vibrio cholerae. Appl. Environ. Microbiol. 67:2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudra, S., R. Mahajan, M. Mathur, K. Kathuria, and V. Talwar. 1996. Cluster of cases of clinical cholera due to Vibrio cholerae O10 in east Delhi. Indian J. Med. Res. 103:71-73. [PubMed] [Google Scholar]
- 42.Said, B., H. R. Smith, S. M. Scotland, and B. Rowe. 1995. Detection and differentiation of the gene for toxin co-regulated pili (tcpA) in Vibrio cholerae non-O1 using the polymerase chain reaction. FEMS Microbiol. Lett. 125:205-210. [DOI] [PubMed] [Google Scholar]
- 43.Sharma, C., M. Thungapathra, A. Ghosh, A. K. Mukhopadhyay, A. Basu, R. Mitra, I. Basu, S. K. Bhattacharya, T. Shimada, T. Ramamurthy, T. Takeda, S. Yamasaki, Y. Takeda, and G. B. Nair. 1998. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera like disease in Calcutta, India. J. Clin. Microbiol. 36:756-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shuman, S. 1994. Novel approach to molecular cloning and polynucleotide synthesis. J. Biol. Chem. 269:32678-32684. [PubMed] [Google Scholar]
- 45.Skorupski, K., and R. K. Taylor. 1997. Control of the ToxR virulence regulon in Vibrio cholerae by environmental stimuli. Mol. Microbiol. 25:1003-1009. [DOI] [PubMed] [Google Scholar]
- 46.Skorupski, K., and R. K. Taylor. 1999. A new level in the Vibrio cholerae ToxR virulence cascade: AphA is required for transcriptional activation of the tcpPH operon. Mol. Microbiol. 31:763-771. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, R., C. Shaw, K. Peterson, P. Spears, and J. Mekalanos. 1988. Safe, live Vibrio cholerae vaccines? Vaccine 6:151-154. [DOI] [PubMed] [Google Scholar]
- 48.Taylor, R. K., V. L. Miller, D. B. Furlong, and J. J. Mekalanos. 1987. Use of phoA gene fusions to identify a pilus colonization factor coordinately regulated with cholera toxin. Proc. Natl. Acad. Sci. USA 84:2833-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voss, E., P. A. Manning, and S. T. Attridge. 1996. The toxin-coregulated pilus is a colonization factor and protective antigen of Vibrio cholerae El Tor. Microb. Pathog. 20:141-153. [DOI] [PubMed] [Google Scholar]
- 50.Waldor, M. K., and J. J. Mekalanos. 1994. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect. Immun. 62:72-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waldor, M. K., and J. J. Mekalanos. 1996. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science 272:1910-1914. [DOI] [PubMed] [Google Scholar]
- 52.Waldor, M. K., E. J. Rubin, G. D. N. Pearson, H. Kimsey, and J. J. Mekalanos. 1997. Regulation, replication, and integration functions of the Vibrio cholerae CTX are encoded by region RS2. Mol. Microbiol. 24:917-926. [DOI] [PubMed] [Google Scholar]
- 53.Yamai, S., T. Okitsu, T. Shimada, and Y. Katsube. 1997. Distribution of serogroups of Vibrio cholerae nonO1/nonO139 with specific reference to their ability to produce cholera toxin and addition of novel serogroups. Jpn. J. Assoc. Infect. Dis. 71:1037-1045. [DOI] [PubMed] [Google Scholar]
- 54.Yu, R. R., and V. J. DiRita. 1999. Analysis of an autoregulatory loop controlling ToxT, cholera toxin, and toxin-coregulated pilus production in Vibrio cholerae. J. Bacteriol. 181:2584-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, R. R., and V. J. DiRita. 2002. Regulation of gene expression in Vibrio cholerae by ToxT involves both the antirepression and RNA polymerase stimulation. Mol. Microbiol. 43:119-134. [DOI] [PubMed] [Google Scholar]