Abstract
Klebsiella pneumoniae is an opportunistic pathogen responsible for nosocomial infections that initially colonize the intestinal tract of patients. Signature-tagged mutagenesis was used to identify genes required for this function. A library of 2,200 mutants was analyzed for the inability of the mutants to survive in a murine model of intestinal colonization and to adhere to human intestinal cells (Int-407) in vitro. Twenty-nine attenuated mutants were selected for further analyses after competition assays against the wild-type strain. Whatever the screening model, most of the transposon insertions occurred in genes involved in metabolic pathways, membrane transport, DNA metabolism, transcriptional regulation, and unknown functions. Only one mutant was attenuated in both the murine colonization and the in vitro adhesion models, and the sequence disrupted by the transposon had homology to adhesin-encoding genes of Haemophilus sp.
Nosocomial infections are a major public health problem. Among the gram-negative bacilli most frequently implicated in such infections, the ubiquitous bacterium Klebsiella pneumoniae plays an important role. In humans, K. pneumoniae is a common saprophyte of the nasopharynx and the intestinal tract. It is an opportunistic pathogen that occasionally causes suppurative infections, bacteremia, and septicemia. Epidemiological studies have shown that, whatever the infection site, the first stage in nosocomial infections due to K. pneumoniae consists of colonization of the patient's gastrointestinal (GI) tract (6, 19). The factors that allow bacteria to be established and maintained inside the complex microbial ecosystem of the human intestine are largely unknown. Disruption of this open ecosystem by antibiotics probably contributes greatly to colonization by K. pneumoniae, since most of the strains involved are highly resistant to antibiotics (31). However, bacterial strains can only persist inside the GI tract if they are firmly established on the intestinal mucous surface and hence are able to resist the strong waves of peristalsis.
As observed with pathogens causing enteritis, colonization of the mucous surfaces of the host requires specialized factors encoded by the microorganisms that specifically bind to host epithelial cells (9). Previous results from our laboratory showed that several adhesins were involved in the interactions between K. pneumoniae and human intestinal cell lines (4, 7, 8). It has also been shown by use of a murine model that the K. pneumoniae capsule plays an active role during the intestinal colonization process (10, 11). Since bacteria must survive host defense mechanisms and acquire nutrients from the surrounding environment, often competing with the host microbial flora, other factors, probably including metabolic properties, are likely to be required. Signature-tagged mutagenesis (STM), a negative-selection strategy (13), has recently been used to identify genes important for in vivo virulence in a variety of gram-negative and gram-positive bacteria (2, 5, 17, 20, 22, 23, 28, 35). We adapted the STM method to K. pneumoniae to identify bacterial factors that are required for the colonization of the colon in a mouse infection model and for adhesion to intestinal epithelial cells in a culture cell model.
Generation of the transposon mutant library.
A pool of signature-tagged mini-Tn5 Km2 transposons in the pUT delivery vector was generously supplied by David Holden (London, United Kingdom). The pool was used to transform Escherichia coli strain CC118 λpir to ampicillin (50 μg/ml) and kanamycin (50 μg/ml) resistance. Approximately 10,000 transformants were grown in Luria-Bertani (LB) broth supplemented with ampicillin and kanamycin, and plasmid DNA was isolated with a plasmid purification kit (Qiagen, Courtaboeuf, France). An aliquot of this sample was used to electroporate E. coli strain S17-1 λpir to ampicillin and kanamycin resistance. About 50,000 transformants were pooled, and plasmids were transferred to K. pneumoniae LM21 (Rifr) by conjugation. Since K. pneumoniae is naturally resistant to ampicillin, all 5,000 transconjugants selected were then screened for the loss of the suicide vector by colony hybridization with a nucleic acid probe specific for the pGP704 plasmid. About 2,200 (44%) clones had lost the plasmid and were selected for further studies in the animal and culture cell models. For determination of the diversity of the insertion sites of mini-Tn5 in K. pneumoniae LM21, DNA from 30 randomly selected mutants was subjected to Southern analysis. Genomic DNA was digested with XhoI and SalI, and the blots were hybridized with the gene encoding kanamycin resistance in mini-Tn5 Km2. The results (data not shown) demonstrated that each mutant arose from the single integration of the transposon at a distinct site.
Screening for attenuated mutants in a mouse intestinal colonization model.
Female IOPS mice were used (OF1 strain, 8 to 18 weeks old, average weight of 26 g; Iffa Credo, L'Arbresle, France). To facilitate the proliferation of naturally ampicillin-resistant Klebsiella, the mice received the antibiotic (50 μg/ml) in drinking water 24 h before inoculation and throughout the time of infection. The 2,200 mutants were orally given in pools of 48 to the mice at a total challenge dose of 106 CFU per mouse in 50 μl of 20% sucrose solution, and two mice were tested for each pool of mutants. After 48 h, the mice were killed and their colons harvested. The entire contents (mucus and ceca) of the colons were removed and plated onto kanamycin-containing LB agar plates and incubated for 18 h at 37°C. The generation of PCR probes and hybridizations were performed as follows. Portions of the input pool and the output pool were used to make [α-32P]dCTP-radiolabeled tags by PCR with primers P2 (5′-TACCTACAACCTCAAGCT-3′) and P4 (5′-TACCCATTCTAACCAAGC-3′) in a two-step process (13). First, chromosomal DNA was amplified in a 50-μl PCR mixture (a 1 μM concentration of primers, 0.2 μg of DNA, a 40 μM concentration of each deoxynucleoside triphosphate, 1.2 mM MgC12) with the following PCR program: 95°C for 4 min, 30 cycles of 95°C for 30 s, 50°C for 1 min, and 72°C for 10 s, and an extension at 72°C for 4 min. The 80-bp PCR product was then purified from a 4% agarose gel following the Qia ExII procedure for agarose gel extraction (Qiagen) and subjected to a second amplification step under the same conditions. The resulting PCR product was digested by HindIII to isolate the internal specific 40-bp fragments which were purified from a 4% agarose gel by use of a Qia ExII kit (Qiagen) and used as a DNA probe. The probes were hybridized to filters containing replicas from the mutant library made by transferring the strains from a 96-well plate onto a nylon SandS Nytran membrane (Schleicher & Schuell) on an LB agar plate containing kanamycin. Colonies formed after 4 h of incubation at 37°C were lysed as previously described (21). A total of 44 mutants gave positive hybridization signals in the inoculum but were not recovered from the intestines of the two animals tested.
Because STM mutants are outcompeted by many other strains during a mixed infection, the attenuation of mutants was confirmed and quantified by an in vivo competition assay. Each of the 44 mutant strains (Kmr) and the wild-type strain LM21 (Kms) were grown separately in LB broth at 37°C for about 18 h. The mutants were independently mixed with the wild-type strain in equal amounts in 50 μl of 20% sucrose solution and administered orally to five mice. In parallel, dilutions of this suspension were plated onto LB agar only (to measure parent CFU) and LB agar with kanamycin (to determine mutant CFU). After 5 days of incubation, the entire contents of the colons were removed and plated onto LB agar plates containing rifampin (to measure total CFU) and onto LB agar plates containing rifampin and kanamycin (to measure mutant CFU). Only those bacterial concentrations (CFU/milliliter) with CFU values between 30 and 300 were preserved for calculations. The exact ratio of mutant to wild-type CFU was then calculated for both the inocula and the intestinal contents. For analysis of results, the averages were compared by studying variance with SPSS software (version 10.0). The competition index (CI) was defined as the output ratio (mutant/wild-type CFU) divided by the input ratio (mutant/wild-type CFU). Only 13 mutants with a statistically significant colonization defect compared with the wild type (CI < 1) were selected.
To determine whether the reduction in colonization was caused by a general growth defect, an in vitro competition assay was performed with the attenuated mutants and the parent strain by mixing equal amounts of bacteria (105 to 106 CFU of both wild-type and mutant bacteria) in LB broth. The cultures were incubated at 37°C for 6 h, after which serial dilutions were plated with medium with or without kanamycin. Experiments were performed in triplicate, and CIs were determined as described above. Thirteen mutants with an in vivo CI below 1 and an in vitro CI in excess of 1 were selected (Table 1).
TABLE 1.
Characterization of mutants
| Mutant type | Putative product of the disrupted gene | Strain | Homologous sequence, organism | % Similarity (% identity)a | Sequence length (bp) | CI(s)b | In vitro CIc |
|---|---|---|---|---|---|---|---|
| Colonization and adhesion attenuated | Adhesin | ||||||
| HMWf surface-exposed protein | A-13/C-65 | hmw1A, H. influenzae | 39 (24) | 479 | 0.81d, 0.21e | 1.38 | |
| Colonization attenuated | Metabolic enzymes | ||||||
| O-sialoglycoprotein endo- peptidase | C-79 | ygjD, E. coli | 89 (87) | 530 | 0.21 | 1.19 | |
| Lactose metabolic enzyme | C-80 | lacI-lacZ intergenic region, E. coli | 99 (98) | 161 | 0.78 | 1.17 | |
| Cyclohexadienyl dehydratase | C-107 | pheC, Pseudomonas aeruginosa | 55 (39) | 521 | 0.83 | 1.08 | |
| Alpha-glucan phosphorylase | C-106 | glgP, E. coli | 96 (91) | 173 | 0.43 | 1.79 | |
| Hydrolase | C-91 | PA2698, Pseudomonas aeruginosa | 73 (63) | 413 | 0.57 | 1.21 | |
| Transporters | |||||||
| Amide-urea-binding protein | C-74 | AF315580, A. tumefaciens | 57 (42) | 659 | 0.45 | 1.20 | |
| Harpin type III secretion system | C-81 | hrcU, Erwinia amylovora | 52 (35) | 227 | 0.24 | 1.27 | |
| DNA-related enzymes | |||||||
| DNA primase | C-75 | ECs0303, E. coli | 52 (32) | 416 | 0.51 | 2.31 | |
| Adenine-specific methylase | C-95 | BAB36637, E. coli | 89 (86) | 575 | 0.66 | 1.51 | |
| Transcriptional regulators | |||||||
| Nitrogen metabolism regulator | C-51 | ntrC, E. coli | 68 (42) | 680 | 0.27 | 1.33 | |
| Glycine metabolism regulator | C-85 | gcvR, E. coli | 88 (81) | 431 | 0.36 | 1.61 | |
| Protein of unknown function | C-93 | yebE, hypothetical protein in the ptrB-purT intergenic region, E. coli | 63 (51) | 377 | 0.56 | 1.10 | |
| Adhesion attenuated | Metabolic enzymes | ||||||
| l-Fucose metabolic enzyme | A-20 | fucA-fucP region, E. coli | 93 (84) | 152 | 0.85 | 2.01 | |
| l-Fucose metabolic enzyme | A-37 | fucA-fucP region, E. coli | 93 (84) | 152 | 0.70 | 1.47 | |
| l-Fucose metabolic enzyme | A-46 | fucA-fucP region, E. coli | 93 (84) | 152 | 0.87 | 1.85 | |
| 2-Dehydro-3-deoxygalactokinase | A-29 | EG11717, E. coli | 57 (45) | 365 | 0.68 | 1.09 | |
| Short-chain dehydrogenase/ reductase | A-14 | m115766, Rhizobium sp. | 73 (58) | 566 | 0.74 | 1.08 | |
| Transporters | |||||||
| Pullulanase secretion envelope | A-16 | pulC, K. pneumoniae | 70 (61) | 281 | 0.83 | 1.54 | |
| Maltose/maltodextrin transport ATP-binding protein | A-2 | malK, Salmonella serovar Typhimurium | 98 (93) | 347 | 0.60 | 1.05 | |
| Maltose-inducible porin | A-18 | lamB, Aeromonas salmonicida | 59 (41) | 613 | 0.60 | 1.46 | |
| Fructose permease | A-30 | fruB, E. coli | 57 (37) | 389 | 0.86 | 1.29 | |
| DNA-related enzymes | |||||||
| Type 1 restriction-modification endonuclease | A-3 | XF2721, Xylella fastidiosa | 56 (41) | 272 | 0.86 | 1.27 | |
| Transposase | A-45 | 177546, IS903 E. coli | 95 (90) | 593 | 0.88 | 1.43 | |
| Transcriptional regulators | |||||||
| Regulator | A-5 | mlr1983, Mesorhizobium loti | 56 (35) | 332 | 0.81 | 1.10 | |
| Propanediol metabolism regulator | A-21 | pocR, Salmonella serovar Typhimurium | 91 (85) | 575 | 0.59 | 1.35 | |
| Protein of unknown function | |||||||
| A-8 | CC1834 hypothetical protein, Caulobacter crescentus | 42 (29) | 341 | 0.81 | 1.73 | ||
| A-55 | Hypothetical 60.8-kDa protein in the ssb-soxS intergenic region, E. coli | 44 (65) | 575 | 0.86 | 1.16 | ||
| A-43 | No homologous sequence in the nonredundant data bank | 0.89 | 1.99 |
Similarity at the amino acid level over the open reading frame, as determined by BLASTX.
CIs are the averages of results from five animals and/or of adhesion CIs from three adhesion assays.
CIs obtained from static cultures.
CI for adhesion to Int-407 cells.
Mouse colonization CI.
HMW, high molecular weight.
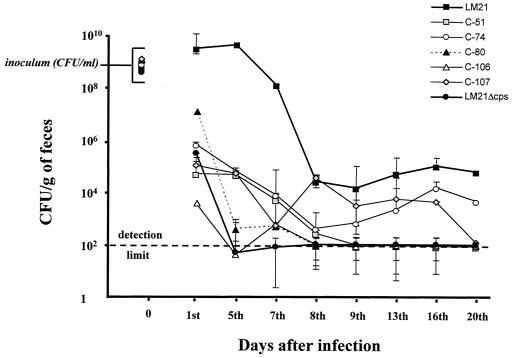
Each attenuated mutant was then tested individually for its ability to colonize the intestinal tract. Similar experiments were performed in parallel with both the wild-type strain LM21 and its capsule-deficient mutant (LM21Δcps) (10). For each strain, two mice were orally infected with 108 to 109 CFU from an overnight culture and stools were recovered at several times over a 3-week period. The stools were weighted and then homogenized in 20 ml of saline, and the resulting dilutions were plated onto selective LB agar plates. Determination of the CFU was performed after 24 h of incubation. The results are expressed in terms of CFU per gram of feces and represent the average of two determinations. Five mutants (C-51, C-74, C-80, C-106, and C-107) showed a remarkable decrease in their colonization ability compared to that of the wild-type strain, which is similar to the result obtained with the K. pneumoniae capsule-deficient mutant (Fig. 1). Important variations were observed in the results (CFU per gram of feces) obtained for these five mutants in individual colonizations on day 5 (Fig. 1) compared with the data from competitive assays (0.27 < CI < 0.83) (Table 1). These variations could be due to partial complementation of the mutants when they were tested in competition with the wild-type strain. Further analysis of these mutants, including determination of the exact function impaired, should elucidate this point. The other eight mutants showed a decrease in colonization ability during the first week (at least 1 log lower than what was obtained with the wild type), but the levels of colonization during the last days of the experiment were either similar to or slightly lower than those obtained with the wild-type strain (data not shown).
FIG. 1.
Individual colonization ability of five STM mutants (C-51, C-74, C-80, C-106, and C-107), the wild-type K. pneumoniae LM21, and the capsule-deficient mutant K. pneumoniae LM21Δcps. The strains were fed separately to two mice. The symbols for day 0 indicate the size of the inoculum. At the indicated times, fecal samples were plated as described in the text. The data are the mean values of two determinations, the error bars represent the values from the two different mice, and the detection limit (dotted line) was 102 CFU per gram of feces.
Screening for attenuated mutants in an intestinal-cell adhesion model.
We used a procedure similar to that described for the mouse intestinal colonization model to detect mutants attenuated in their ability to adhere to Intestine-407 (Int-407) cells (ATCC CCL6) in vitro. These cells were derived from human embryonic jejunum and ileum and were maintained in Eagle minimal essential medium (Seromed; Poly Labo, Strasbourg, France) supplemented with heat-inactivated fetal calf serum (Seromed), 1% l-glutamine, 1% nonessential amino acids, 10,000 U of penicillin per ml, 10 μg of streptomycin per ml, and 25 μg of amphotericin B per ml in an atmosphere of 5% CO2. Confluent monolayers (106 epithelial cells) in tissue culture plate wells were rinsed twice with 1 ml of phosphate-buffered saline (pH 7.4). The mutants were tested in pools of 48 at a total challenge dose of 108 CFU (multiplicity of infection, 100). After 3 h of incubation at 37°C in an atmosphere of 5% CO2, each well was rinsed three times with phosphate-buffered saline. Adherent bacteria were released by the addition of distilled water and were plated on LB agar plates. The generation of PCR probes and hybridizations were performed as described above with genomic DNA from colonies obtained after 18 h of incubation at 37°C. Individual cell adhesion competition and in vitro culture competition (each mutant versus the wild-type strain) assays were performed following a procedure similar to that used with the in vivo colonization experiments. Fifty-nine mutants were initially isolated, but only 17 exhibited CIs lower than 1 when tested in competition adhesion assays against the wild-type strain. It is likely that a longer competition assay (>3 h) would have resulted in lower CIs; however, alteration of cells by the bacterial products of metabolism prevented longer incubation periods. None of these 17 mutants showed an in vitro CI below 1; thus, they were all retained for further study (Table 1). Only one mutant (A-13/C-65) was attenuated in both the adhesion and colonization models.
Molecular analysis of the mutants.
The insertion sites of mini-Tn5 in the colonization- and adhesion-defective mutants were determined. The 29 mutants underwent Southern analysis with several endonucleases. SalI-digested genomic DNA gave DNA fragments containing the gene encoding kanamycin resistance that are compatible with cloning experiments. This endonuclease was therefore selected. All but three of the resulting hybridization patterns were different, confirming that most of the mutants were unique. Genomic DNA containing the cassette encoding kanamycin resistance was then cloned in the pUC18 vector. E. coli JM109 was then transformed with the recombinant plasmids by use of the kanamycin resistance marker of mini-Tn5 Km2 as a selective agent. Recombinant plasmid DNA was isolated with a QiaFilter plasmid isolation kit (Qiagen), and the sequences of inserts were obtained from ESGS Ltd. (Evry, France) by use of the mini-Tn5 Km2-specific primer, P6 (5′-CCTAGGCGGCCAGATCTGAT-3′). Sequence homologies were determined with the BLAST 2.0 search algorithm at the National Center for Biotechnology Information.
Whatever the screening method used (colonization or adhesion), the mutants fell into six categories, with transposon insertions in genes encoding (i) enzymes in metabolic pathways, (ii) proteins with membrane transport functions, (iii) DNA-related enzymes, (iv) transcriptional regulators, (v) adhesin, or (vi) proteins of unknown functions (Table 1).
The largest class of mutants affected metabolic functions, although no auxotrophic mutant was detected when bacteria were grown on M9 minimal medium (data not shown).
Two mutants (C-51 and C-74) with colonization defects had insertions in genes encoding products involved in nitrogen metabolism. In mutant C-51, the transposon was inserted in a sequence showing homology to the nitrogen regulatory protein C (ntrC) of E. coli (36). NtrC is the response regulator of a two-component regulatory system that includes a modulator (kinase), NtrB, and has also been described in Klebsiella species (18). This regulatory system controls, among other genes, the expression of the urease-encoding operon in K. pneumoniae (3). Our observation that mutant C-51 was also urease-negative after phenol red urease testing (data not shown) is consistent with these findings. A second mutant, clone C-74, also had a urease-negative phenotype (data not shown); the insertion event occurred in a gene whose product shows homology to an amide-urea-binding FmdD-like protein of Agrobacterium tumefaciens (33). FmdD is a periplasmic binding protein which is part of a high-affinity, binding-protein-dependent, active transport system for short-chain amides and urea (25). Many species of bacteria, including K. pneumoniae, are able to use urea as a source of nitrogen for growth by virtue of their ability to hydrolyze it to ammonia and carbon dioxide by using cytoplasmic ureases (26). The inability to metabolize urea could therefore impair the growth capacity of K. pneumoniae within the GI tract, where urea is abundant. Indeed, the impairment of these two mutants in colonizing the murine GI tract was corroborated in individual assays (Fig. 1). Similar observations have been made with other pathogens. Intestinal colonization defects were detected in Vibrio cholerae (15) and E. coli K1 (20), both mutated in the gene encoding the nitrogen starvation sigma factor 54, which controls many cellular functions related to nitrogen assimilation and urea utilization (24).
Another mutant falls into the group of those with a transposon insertion within a gene encoding enzymes of metabolic pathways. In clone C-106, the insertion occurred in a sequence related to glgP of E. coli encoding the catabolic glycogen phosphorylase (34). Alpha-glucan phosphorylases catalyze the reversible cleavage of polysaccharides in α-d-glucose-1-phosphate and hence play a central role in the mobilization of storage polysaccharides such as endogenous glycogen (30). Since clone C-106 was the most attenuated mutant in our study (Fig. 1), we postulate that the use of this endogenous source of energy is crucial for K. pneumoniae during intestinal colonization, i.e., in stress response or for swift adaptation to changing environments.
In three independent clones resulting from the adhesion screening, A-20, A-37, and A-46, the transposon had inserted in a region homologous to the E. coli fucA-fucP intergenic region containing the promoters of both the fucose aldolase- and fucose permease-encoding genes. All of these mutants grew more quickly than the wild-type strain (in vitro CIs of >1.4), indicating a possible disruption of the cell cycle and therefore suggesting that the expression of the l-fucose metabolism enzymes was affected. The occurrence of three insertion events in the same DNA region could be linked to the presence of a hot spot for transposon insertion. Conversely, there could be a relationship between l-fucose metabolism and the adhesion phenotype. In Bacteroides thetaiotaomicron, a component of the intestinal microflora, the l-fucose metabolic pathway is coordinated with the production of fucosylated glycans in enterocytes (14). This host-microbial interaction is mediated by an as-yet-unidentified bacterial signal and is thought to provide a nutrient foundation that can be used by the bacterium when l-fucose supplies are low (14). Since extracellular glycoconjugates also mediate attachment of bacteria to epithelial surfaces (27), it is tempting to speculate that K. pneumoniae uses this cell surface structure as a receptor and is able to promote its synthesis by a mechanism similar to the one seen with the commensal B. thetaiotaomicron.
In mutant A-30, attenuation of adhesion was correlated with the potential alteration of a sugar metabolic pathway; this clone had a transposon insertion in fruB, a gene potentially encoding a subunit of the fructose permease, one component of a phosphotransferase system. Phosphotransferases were initially described as sugar-phophorylating systems (29). It was later shown that these systems play a role in a wide variety of cellular metabolic processes and control the expression of numerous genes, including virulence genes in Salmonella enterica serovar Typhimurium (12, 21). Using in vivo expression technology (IVET) in a murine model of infection with K. pneumoniae after intraperitoneal administration, Lai et al. recently isolated several genes, including fruB (pftA), involved in the infection of mouse spleens (16). Mutations occurring in the V. cholerae homologue of fruB were also isolated when colonization-attenuated mutants were screened in an infant-mouse model (2). Thus, this metabolic pathway may play a crucial role in the interactions between bacteria and the host organism by interacting with cellular regulatory processes.
Of the 29 attenuated mutants, only 1 was impaired in both colonization and adhesion functions. In this mutant (A-13/C-65), the transposon had inserted in a region showing homology to the gene encoding Haemophilus influenzae adhesins, high-molecular-weight surface-exposed proteins (1). These proteins are major adhesins of noncapsulated (nontypeable) H. influenzae and mediate attachment to human epithelial cells, a key step in the colonization of the upper respiratory tract (32). This suggests that as-yet-unidentified surface-exposed proteins are involved in the adhesion of K. pneumoniae to intestinal cells and are also required for intestinal colonization.
Conclusion.
The purpose of this study was to identify genes encoding factors required for the in vivo colonization of the GI tract by K. pneumoniae. Using STM, we were able to examine the behavior of mutants in the context of a host's normal microbial flora (murine intestinal colonization model) and in an in vitro assay of adhesion to human intestinal cells. To our knowledge, the only other STM screenings that have examined intestinal colonization in animal models were performed on V. cholerae (2), Yersinia pseudotuberculosis (22), and E. coli K1 (20) and permit the identification of novel genes required for colonization.
In our study, 103 mutants were initially identified as attenuated in either model but only 29 were retained for competition assays with the wild-type colonizer. It is likely that the 74 remaining strains would show attenuation in vivo if inoculated in lower numbers, since they seemed to be attenuated when administered in pools of 48 mutants but nonattenuated when administered as one of two competing strains. Moreover, during the competition assays with the wild-type strain, we obtained in vivo CIs that were much higher than those obtained in studies with V. cholerae and Y. pseudotuberculosis but close to those observed in the E. coli K1 study. This suggests that commensal Enterobacteriaceae are better able to adapt metabolically to the GI tract than noncommensal bacteria or non-Enterobacteriaceae.
In a previous study (10), it was reported that capsular polysaccharides are involved in the colonization process, but surprisingly, no capsule-encoding gene was identified in the present study. The mutagenesis was not saturating, since only 2,200 insertional mutants were screened and we might have failed to isolate capsule-deficient mutants. Using STM to look for virulence genes in Streptococcus pneumoniae, Polissi et al. (28) and Lau et al. (17) did not identify capsule-deficient mutants despite the fact that this is one of the main virulence factors of this pathogen. This suggests that the STM technique, although powerful, is not sufficient and should be performed in parallel with other in vivo techniques such as IVET.
However, using STM, we were able to identify not only genes required for adhesion to intestinal cells but also genes with a previously unrecognized role in colonization of the GI tract. Surprisingly, only one mutant was affected in both its colonization and adhesion functions. This suggests that adhesion to intestinal cells in vitro poorly reflects the process in vivo. Intestinal cell adhesion may not be essential for colonization of the GI tract or represents only one late stage in the process: bacteria first have to counteract several barriers such as the mucous layer and peristalsis and to metabolically adapt to the environment. We anticipate that the information gathered in this study as well as future work based upon it will lead to an increased understanding of the processes by which pathogenic and nonpathogenic strains establish a successful intestinal colonization in the host.
Acknowledgments
We are grateful to David Holden for the generous gift of the pool of tagged transposons and for helpful advice. We thank Jean Pierre Girardeau for his help in analyzing the sequence data and Christophe De Champs for helpful discussions.
This work was supported by the “Contrat Quadriennal de Recherche, Equipe d'Accueil 2148” and by the program “Microbiologie et Maladies Infectieuses, Réseau Infections Nosocomiales” of the Ministère de l'Education Nationale, de la Recherche et des Technologies.
Editor: V. J. DiRita
REFERENCES
- 1.Barenkamp, S. J., and E. Leininger. 1992. Cloning, expression, and DNA sequence analysis of genes encoding nontypeable Haemophilus influenzae high-molecular-weight surface-exposed proteins related to filamentous hemagglutinin of Bordetella pertussis. Infect. Immun. 60:1302-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chiang, S. L., and J. J. Mekalanos. 1998. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol. Microbiol. 27:797-805. [DOI] [PubMed] [Google Scholar]
- 3.Collins, C. M., D. M. Gutman, and H. Laman. 1993. Identification of a nitrogen-regulated promoter controlling expression of Klebsiella pneumoniae urease genes. Mol. Microbiol. 8:187-198. [DOI] [PubMed] [Google Scholar]
- 4.Darfeuille-Michaud, A., C. Jallat, D. Aubel, D. Sirot, C. Rich, J. Sirot, and B. Joly. 1992. R-plasmid-encoded adhesive factor in Klebsiella pneumoniae strains responsible for human nosocomial infections. Infect. Immun. 60:44-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darwin, A. J., and V. L. Miller. 1999. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol. Microbiol. 32:51-62. [DOI] [PubMed] [Google Scholar]
- 6.De Champs, C., M. P. Sauvant, C. Chanal, D. Sirot, N. Gazui, R. Malhuret, J. C. Baguet, and J. Sirot. 1989. Prospective survey of colonization and infection caused by expanded-spectrum-β-lactamase-producing members of the family Enterobacteriaceae in an intensive care unit. J. Clin. Microbiol. 27:2887-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Martino, P., Y. Bertin, J. P. Girardeau, V. Livrelli, B. Joly, and A. Darfeuille-Michaud. 1995. Molecular characterization and adhesive properties of CF29K, an adhesin of Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 63:4336-4344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Di Martino, P., V. Livrelli, D. Sirot, B. Joly, and A. Darfeuille-Michaud. 1996. A new fimbrial antigen harbored by CAZ-5/SHV-4-producing Klebsiella pneumoniae strains involved in nosocomial infections. Infect. Immun. 64:2266-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donnenberg, M. S., and J. B. Kaper. 1992. Enteropathogenic Escherichia coli. Infect. Immun. 60:3953-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favre-Bonte, S., B. Joly, and C. Forestier. 1999. Consequences of reduction of Klebsiella pneumoniae capsule expression on interactions of this bacterium with epithelial cells. Infect. Immun. 67:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favre-Bonte, S., T. Rasklicht, C. Forestier, and K. A. Krogfelt. 1999. Klebsiella pneumoniae capsule expression is necessary for colonization of large intestines of streptomycin-treated mice. Infect. Immun. 67:6152-6156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Groisman, E. A., and M. H. J. Saier. 1990. Salmonella virulence: new clues to intramacrophage survival. Trends Biochem. Sci. 15:30-33. [DOI] [PubMed] [Google Scholar]
- 13.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269:400-403. [DOI] [PubMed] [Google Scholar]
- 14.Hooper, L. V., P. G. Falk, and J. I. Gordon. 2000. Analyzing the molecular foundations of commensalism in the mouse intestine. Curr. Opin. Microbiol. 3:79-85. [DOI] [PubMed] [Google Scholar]
- 15.Klose, K. E., and J. J. Mekalanos. 1998. Distinct roles of an alternative sigma factor during both free-swimming and colonizing phases of the Vibrio cholerae pathogenic cycle. Mol. Microbiol. 28:501-520. [DOI] [PubMed] [Google Scholar]
- 16.Lai, Y.-C., H.-L. Peng, and H.-Y. Chang. 2001. Identification of genes induced in vivo during Klebsiella pneumoniae CG43 infection. Infect. Immun. 69:7140-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lau, G. W., S. Haataja, M. Lonetto, S. E. Kensit, A. Marra, A. P. Bryant, D. McDevitt, D. A. Morrison, and D. W. Holden. 2001. A functional genomic of type 3 Streptococcus pneumoniae virulence. Mol. Microbiol. 40:555-571. [DOI] [PubMed] [Google Scholar]
- 18.Magasanik, B. 1993. The regulation of nitrogen utilization in enteric bacteria. J. Cell. Biochem. 51:34-40. [DOI] [PubMed] [Google Scholar]
- 19.Markowitz, S. M., J. M. Veazey, F. L. Macrino, C. G. Mayhall, and V. A. Lamb. 1980. Sequential outbreaks of infection due to Klebsiella pneumoniae in a neonatal intensive care unit: implication of a conjugative R plasmid. J. Infect. Dis. 142:106-112. [DOI] [PubMed] [Google Scholar]
- 20.Martindale, J., D. Stroud, E. R. Moxon, and C. M. Tang. 2000. Genetic analysis of Escherichia coli K1 gastrointestinal colonization. Mol. Microbiol. 37:1293-1305. [DOI] [PubMed] [Google Scholar]
- 21.Mass, R. 1983. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid 10:296-298. [DOI] [PubMed] [Google Scholar]
- 22.Mecsas, J., I. Bilis, and S. Falkow. 2001. Identification of attenuated Yersinia pseudotuberculosis strains and characterization of an orogastric infection in BALB/c mice on day 5 postinfection by signature-tagged mutagenesis. Infect. Immun. 69:2779-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mei, J. M., F. Nourbakhsh, C. W. Ford, and D. W. Holden. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol. Microbiol. 26:399-407. [DOI] [PubMed] [Google Scholar]
- 24.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mills, J., N. R. Wyborn, J. A. Greenwood, S. G. Williams, and C. W. Jones. 1998. Characterisation of a binding-protein-dependent, active transport system for short-chain amides and urea in the methylotrophic bacterium Methylophilus methylotrophus. Eur. J. Biochem. 251:45-53. [DOI] [PubMed] [Google Scholar]
- 26.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouricout, M. 1997. Interactions between the enteric pathogen and the host. An assortment of bacterial lectins and a set of glycoconjugate receptors. Adv. Exp. Med. Biol. 412:109-123. [PubMed] [Google Scholar]
- 28.Polissi, A., A. Ponttigia, G. Fager, M. Altieri, H. Mottl, L. Ferrari, and D. Simon. 1998. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect. Immun. 66:5620-5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saier, M. H., and J. Reizer. 1994. The bacterial phosphotransferase system: new frontiers 30 years later. Mol. Microbiol. 13:755-764. [DOI] [PubMed] [Google Scholar]
- 30.Schinzel, R., and B. Nidetzky. 1999. Bacterial alpha-glucan phosphorylases. FEMS Microbiol. Lett. 171:73-79. [DOI] [PubMed] [Google Scholar]
- 31.Sirot, J., C. Chanal, A. Petit, D. Sirot, R. Labia, and G. Gerbaud. 1988. Klebsiella pneumoniae and other Enterobacteriaceae producing novel plasmid-mediated β-lactamases markedly active against third-generation cephalosporins: epidemiologic studies. Rev. Infect. Dis. 10:850-859. [DOI] [PubMed] [Google Scholar]
- 32.St. Geme, J. W., III, S. Falkow, and S. J. Barenkamp. 1993. High-molecular-weight proteins of nontypable Haemophilus influenzae mediate attachment to human epithelial cells. Proc. Natl. Acad. Sci. USA 90:2875-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Trott, S., R. Bauer, H. J. Knackmuss, and A. Stolz. 2001. Genetic and biochemical characterization of an enantioselective amidase from Agrobacterium tumefaciens strain d3. Microbiology 147:1815-1824. [DOI] [PubMed] [Google Scholar]
- 34.Yu, F., Y. Jen, E. Takeuchi, M. Inouye, H. Nakayama, M. Tagaya, and T. Fukui. 1988. Alpha-glucan phosphorylase from Escherichia coli: cloning of the gene, and purification and characterization of the protein. J. Biol. Chem. 263:13706-13711. [PubMed] [Google Scholar]
- 35.Zhao, H., X. Li, D. E. Johnson, and H. L. T. Mobley. 1999. Identification of protease and rpoN-associated genes of uropathogenic Proteus mirabilis by negative selection in a mouse model of ascending urinary tract infection. Microbiology 145:185-195. [DOI] [PubMed] [Google Scholar]
- 36.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khordursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]