Abstract
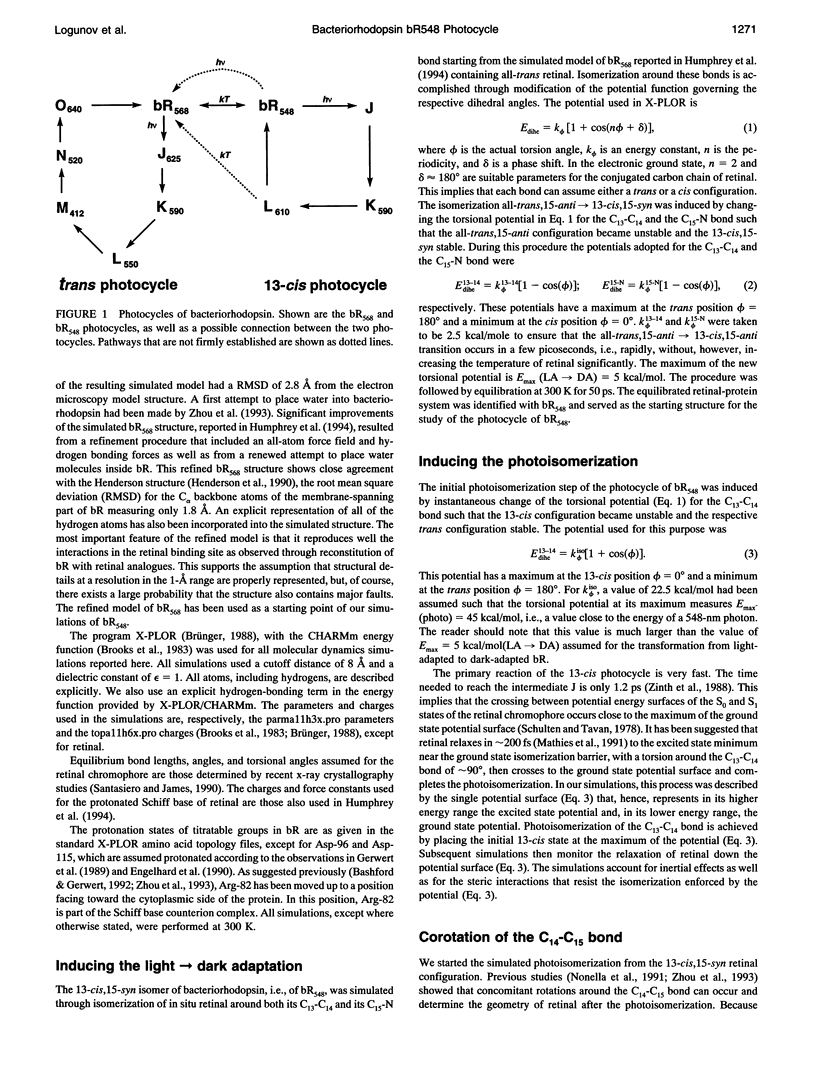
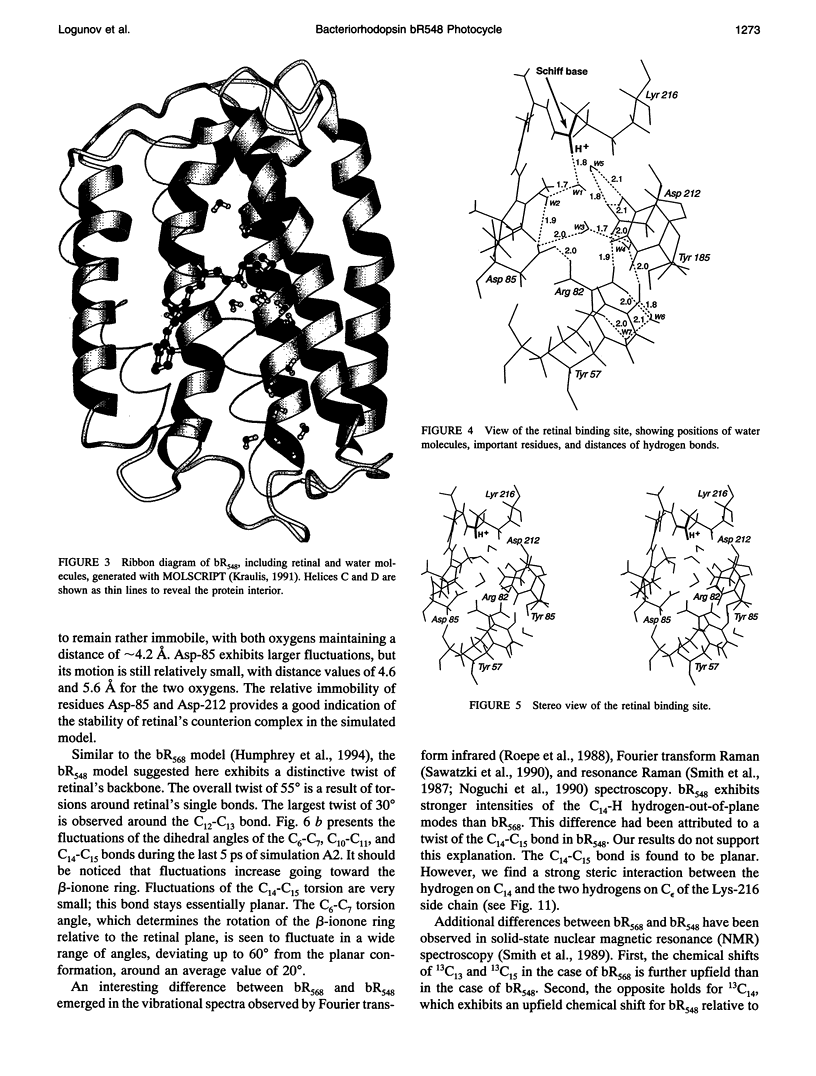
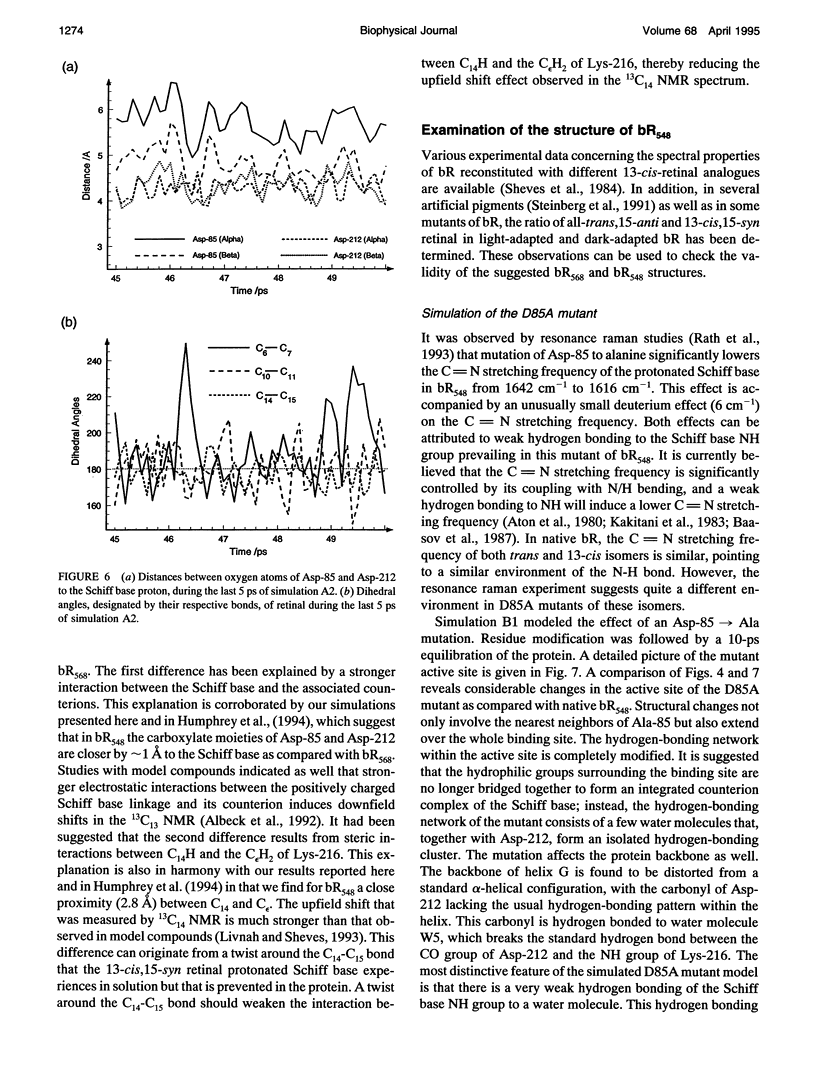
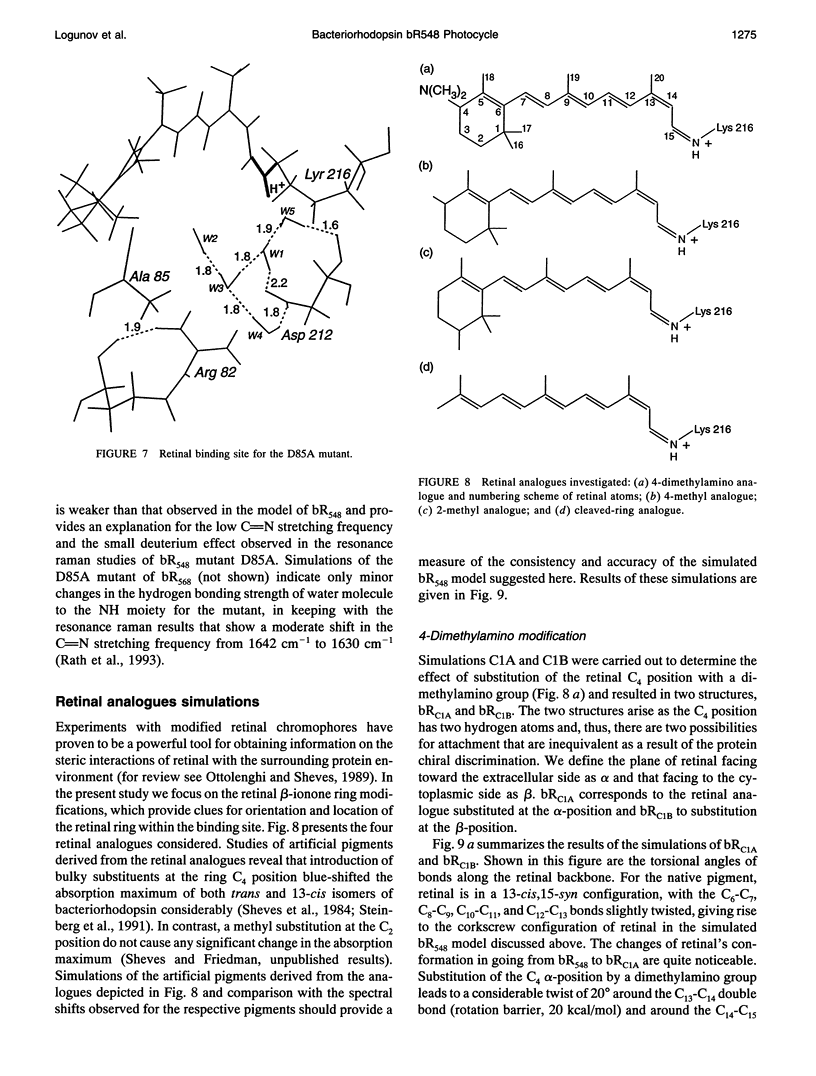
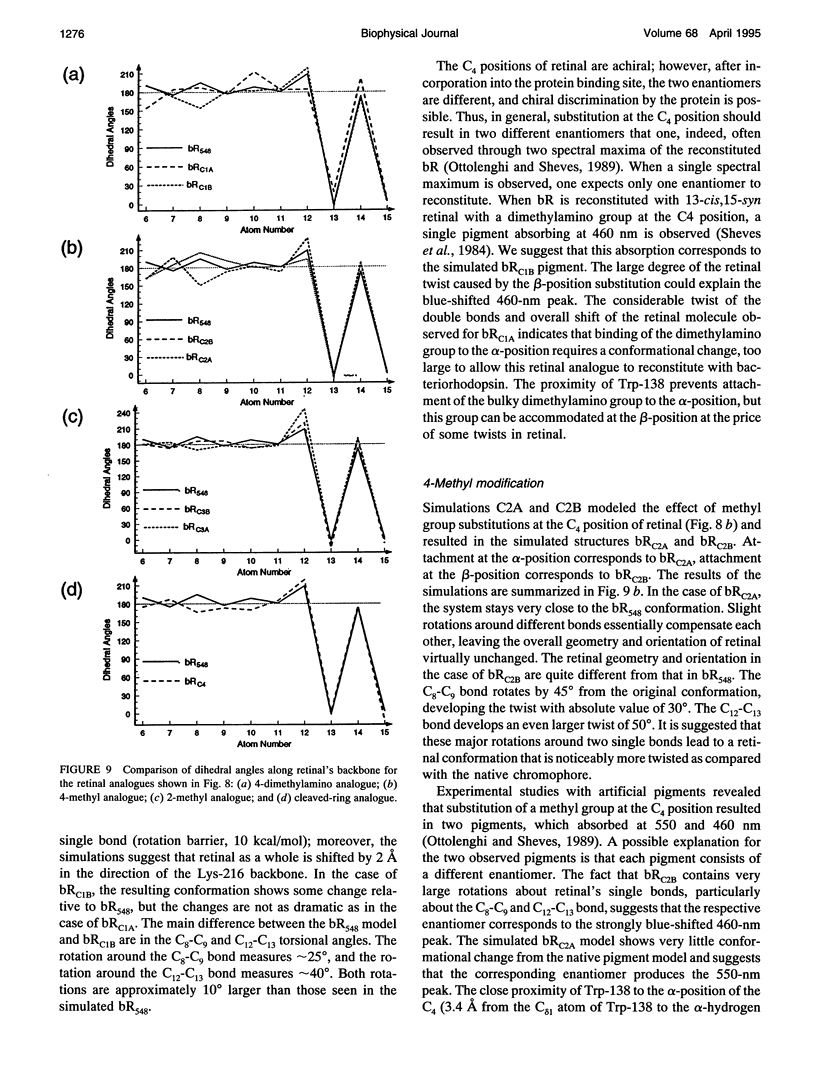
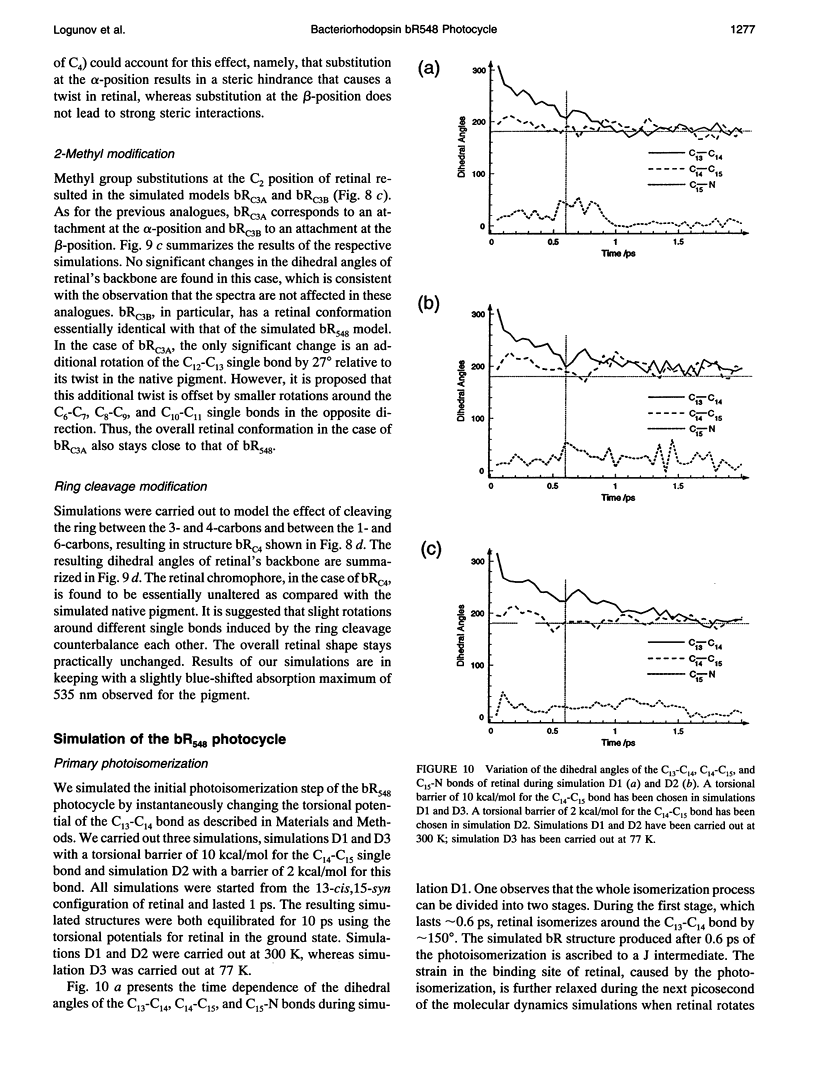
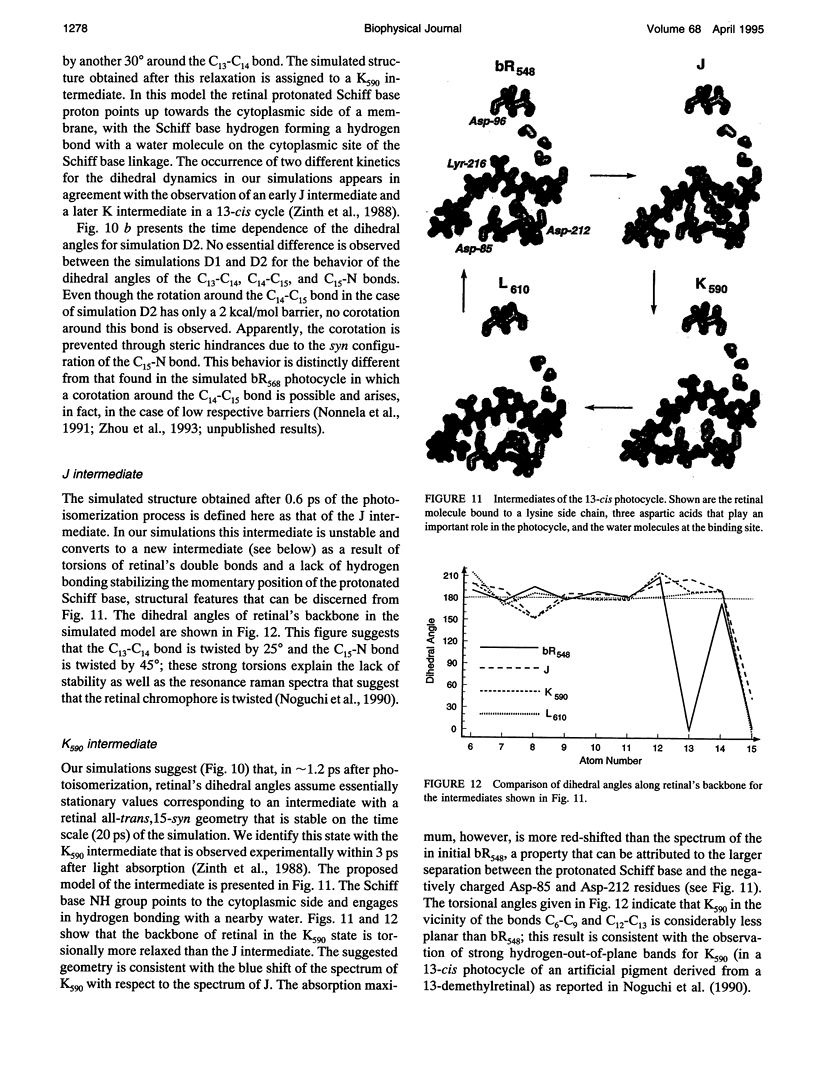
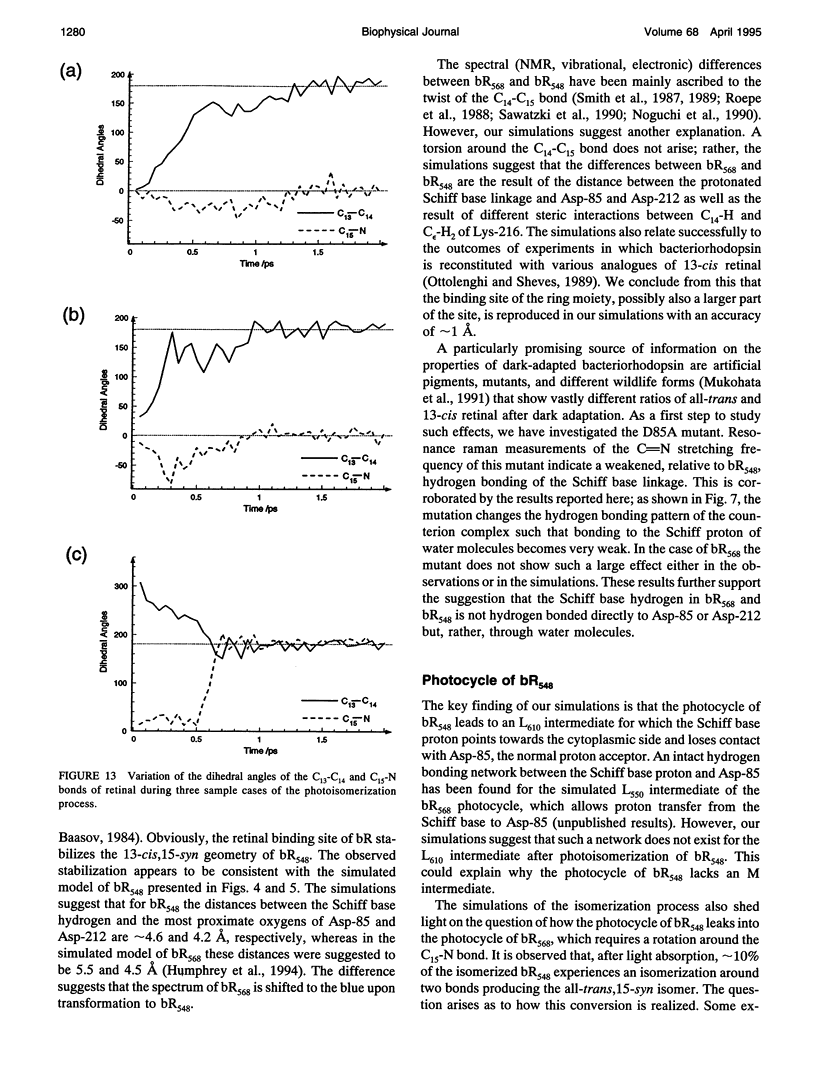
The structure and the photocycle of bacteriorhodopsin (bR) containing 13-cis,15-syn retinal, so-called bR548, has been studied by means of molecular dynamics simulations performed on the complete protein. The simulated structure of bR548 was obtained through isomerization of in situ retinal around both its C13-C14 and its C15-N bond starting from the simulated structure of bR568 described previously, containing all-trans,15-anti retinal. After a 50-ps equilibration, the resulting structure of bR548 was examined by replacing retinal by analogues with modified beta-ionone rings and comparing with respective observations. The photocycle of bR548 was simulated by inducing a rapid 13-cis,15-anti-->all-trans,15-syn isomerization through a 1-ps application of a potential that destabilizes the 13-cis isomer. The simulation resulted in structures consistent with the J, K, and L intermediates observed in the photocycle of bR548. The results offer an explanation of why an unprotonated retinal Schiff base intermediate, i.e., an M state, is not formed in the bR548 photocycle. The Schiff base nitrogen after photoisomerization of bR548 points to the intracellular rather than to the extracellular site. The simulations suggest also that leakage from the bR548 to the bR568 cycle arises due to an initial 13-cis,15-anti-->all-trans,15-anti photoisomerization.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aton B., Doukas A. G., Narva D., Callender R. H., Dinur U., Honig B. Resonance Raman studies of the primary photochemical event in visual pigments. Biophys J. 1980 Jan;29(1):79–94. doi: 10.1016/S0006-3495(80)85119-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baasov T., Friedman N., Sheves M. Factors affecting the C = N stretching in protonated retinal Schiff base: a model study for bacteriorhodopsin and visual pigments. Biochemistry. 1987 Jun 2;26(11):3210–3217. doi: 10.1021/bi00385a041. [DOI] [PubMed] [Google Scholar]
- Balashov S. P., Govindjee R., Kono M., Imasheva E., Lukashev E., Ebrey T. G., Crouch R. K., Menick D. R., Feng Y. Effect of the arginine-82 to alanine mutation in bacteriorhodopsin on dark adaptation, proton release, and the photochemical cycle. Biochemistry. 1993 Oct 5;32(39):10331–10343. doi: 10.1021/bi00090a008. [DOI] [PubMed] [Google Scholar]
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Birge R. R. Photophysics and molecular electronic applications of the rhodopsins. Annu Rev Phys Chem. 1990;41:683–733. doi: 10.1146/annurev.pc.41.100190.003343. [DOI] [PubMed] [Google Scholar]
- Drachev L. A., Dracheva S. V., Kaulen A. D. pH dependence of the formation of an M-type intermediate in the photocycle of 13-cis-bacteriorhodopsin. FEBS Lett. 1993 Oct 11;332(1-2):67–70. doi: 10.1016/0014-5793(93)80486-e. [DOI] [PubMed] [Google Scholar]
- Engelhard M., Hess B., Metz G., Kreutz W., Siebert F., Soppa J., Oesterhelt D. High resolution 13C-solid state NMR of bacteriorhodopsin: assignment of specific aspartic acids and structural implications of single site mutations. Eur Biophys J. 1990;18(1):17–24. doi: 10.1007/BF00185416. [DOI] [PubMed] [Google Scholar]
- Gergely C., Ganea C., Váró G. Combined optical and photoelectric study of the photocycle of 13-cis bacteriorhodopsin. Biophys J. 1994 Aug;67(2):855–861. doi: 10.1016/S0006-3495(94)80545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerwert K., Hess B., Soppa J., Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison G. S., Smith S. O., Pardoen J. A., Winkel C., Lugtenburg J., Herzfeld J., Mathies R., Griffin R. G. Dark-adapted bacteriorhodopsin contains 13-cis, 15-syn and all-trans, 15-anti retinal Schiff bases. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1706–1709. doi: 10.1073/pnas.81.6.1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R., Baldwin J. M., Ceska T. A., Zemlin F., Beckmann E., Downing K. H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990 Jun 20;213(4):899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- Hofrichter J., Henry E. R., Lozier R. H. Photocycles of bacteriorhodopsin in light- and dark-adapted purple membrane studied by time-resolved absorption spectroscopy. Biophys J. 1989 Oct;56(4):693–706. doi: 10.1016/S0006-3495(89)82716-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W., Logunov I., Schulten K., Sheves M. Molecular dynamics study of bacteriorhodopsin and artificial pigments. Biochemistry. 1994 Mar 29;33(12):3668–3678. doi: 10.1021/bi00178a025. [DOI] [PubMed] [Google Scholar]
- Kalisky O., Goldschmidt C. R., Ottolenghi M. On the photocycle and light adaptation of dark-adapted bacteriorhodopsin. Biophys J. 1977 Aug;19(2):185–189. doi: 10.1016/S0006-3495(77)85579-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorana H. G. Bacteriorhodopsin, a membrane protein that uses light to translocate protons. J Biol Chem. 1988 Jun 5;263(16):7439–7442. [PubMed] [Google Scholar]
- Korenstein R., Hess B. Hydration effects on cis--trans isomerization of bacteriorhodopsin. FEBS Lett. 1977 Oct 1;82(1):7–11. doi: 10.1016/0014-5793(77)80874-0. [DOI] [PubMed] [Google Scholar]
- Lanyi J. K. Proton transfer and energy coupling in the bacteriorhodopsin photocycle. J Bioenerg Biomembr. 1992 Apr;24(2):169–179. doi: 10.1007/BF00762675. [DOI] [PubMed] [Google Scholar]
- Mathies R. A., Lin S. W., Ames J. B., Pollard W. T. From femtoseconds to biology: mechanism of bacteriorhodopsin's light-driven proton pump. Annu Rev Biophys Biophys Chem. 1991;20:491–518. doi: 10.1146/annurev.bb.20.060191.002423. [DOI] [PubMed] [Google Scholar]
- Mukohata Y., Ihara K., Uegaki K., Miyashita Y., Sugiyama Y. Australian Halobacteria and their retinal-protein ion pumps. Photochem Photobiol. 1991 Dec;54(6):1039–1045. doi: 10.1111/j.1751-1097.1991.tb02127.x. [DOI] [PubMed] [Google Scholar]
- Oesterhelt D., Tittor J., Bamberg E. A unifying concept for ion translocation by retinal proteins. J Bioenerg Biomembr. 1992 Apr;24(2):181–191. doi: 10.1007/BF00762676. [DOI] [PubMed] [Google Scholar]
- Ohno K., Takeuchi Y., Yoshida M. Effect of light-adaptation on the photoreaction of bacteriorhodopsin from Halobacterium halobium. Biochim Biophys Acta. 1977 Dec 23;462(3):575–582. doi: 10.1016/0005-2728(77)90102-5. [DOI] [PubMed] [Google Scholar]
- Ottolenghi M., Sheves M. Synthetic retinals as probes for the binding site and photoreactions in rhodopsins. J Membr Biol. 1989 Dec;112(3):193–212. doi: 10.1007/BF01870951. [DOI] [PubMed] [Google Scholar]
- Rath P., Marti T., Sonar S., Khorana H. G., Rothschild K. J. Hydrogen bonding interactions with the Schiff base of bacteriorhodopsin. Resonance Raman spectroscopy of the mutants D85N and D85A. J Biol Chem. 1993 Aug 25;268(24):17742–17749. [PubMed] [Google Scholar]
- Roepe P. D., Ahl P. L., Herzfeld J., Lugtenburg J., Rothschild K. J. Tyrosine protonation changes in bacteriorhodopsin. A Fourier transform infrared study of BR548 and its primary photoproduct. J Biol Chem. 1988 Apr 15;263(11):5110–5117. [PubMed] [Google Scholar]
- Sawatzki J., Fishcer R., Scheer H., Siebert F. Fourier-transform Raman spectroscopy applied to photobiological systems. Proc Natl Acad Sci U S A. 1990 Aug;87(15):5903–5906. doi: 10.1073/pnas.87.15.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulten K., Tavan P. A mechanism for the light-driven proton pump of Halobacterium halobium. Nature. 1978 Mar 2;272(5648):85–86. doi: 10.1038/272085a0. [DOI] [PubMed] [Google Scholar]
- Smith S. O., Myers A. B., Pardoen J. A., Winkel C., Mulder P. P., Lugtenburg J., Mathies R. Determination of retinal Schiff base configuration in bacteriorhodopsin. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2055–2059. doi: 10.1073/pnas.81.7.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. O., de Groot H. J., Gebhard R., Courtin J. M., Lugtenburg J., Herzfeld J., Griffin R. G. Structure and protein environment of the retinal chromophore in light- and dark-adapted bacteriorhodopsin studied by solid-state NMR. Biochemistry. 1989 Oct 31;28(22):8897–8904. doi: 10.1021/bi00448a032. [DOI] [PubMed] [Google Scholar]
- Song L., El-Sayed M. A., Lanyi J. K. Protein catalysis of the retinal subpicosecond photoisomerization in the primary process of bacteriorhodopsin photosynthesis. Science. 1993 Aug 13;261(5123):891–894. doi: 10.1126/science.261.5123.891. [DOI] [PubMed] [Google Scholar]
- Sperling W., Carl P., Rafferty Ch, Dencher N. A. Photochemistry and dark equilibrium of retinal isomers and bacteriorhodopsin isomers. Biophys Struct Mech. 1977 Jun 29;3(2):79–94. doi: 10.1007/BF00535798. [DOI] [PubMed] [Google Scholar]
- Warshel A. Bicycle-pedal model for the first step in the vision process. Nature. 1976 Apr 22;260(5553):679–683. doi: 10.1038/260679a0. [DOI] [PubMed] [Google Scholar]
- Zhou F., Windemuth A., Schulten K. Molecular dynamics study of the proton pump cycle of bacteriorhodopsin. Biochemistry. 1993 Mar 9;32(9):2291–2306. doi: 10.1021/bi00060a022. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Liu R. S. Divergent pathways in photobleaching of 7,9-dicis-rhodopsin and 9,11-dicis-12-fluororhodopsin: one-photon-two-bond and one-photon-one-bond isomerization. Biochemistry. 1993 Sep 28;32(38):10233–10238. doi: 10.1021/bi00089a045. [DOI] [PubMed] [Google Scholar]




