Abstract
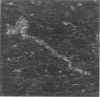
Atomic force microscopy (AFM) has been used to study the structure of rabbit skeletal muscle myosin deposited onto a mica substrate from glycerol solution. Images of the myosin molecule have been obtained using contact mode AFM with the sample immersed in propanol. The molecules have two heads at one end of a long tail and have an appearance similar to those prepared by glycerol deposition techniques for electron microscopy, except that the separation of the two heads is not so well defined. The average length of the tail (155 +/- 5 nm) agrees well with previous studies. Bends in the myosin tail have been observed at locations similar to those observed in the electron microscope. By raising the applied force, it has been possible locally to separate the two strands of the alpha-helical coiled-coil tail. We conclude that the glycerol-mica technique is a useful tool for the preparation of fibrous proteins for examination by scanning probe microscopy.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Binnig G, Quate CF, Gerber C. Atomic force microscope. Phys Rev Lett. 1986 Mar 3;56(9):930–933. doi: 10.1103/PhysRevLett.56.930. [DOI] [PubMed] [Google Scholar]
- Clemmer C. R., Beebe T. P., Jr Graphite: a mimic for DNA and other biomolecules in scanning tunneling microscope studies. Science. 1991 Feb 8;251(4994):640–642. doi: 10.1126/science.1992517. [DOI] [PubMed] [Google Scholar]
- Drake B., Prater C. B., Weisenhorn A. L., Gould S. A., Albrecht T. R., Quate C. F., Cannell D. S., Hansma H. G., Hansma P. K. Imaging crystals, polymers, and processes in water with the atomic force microscope. Science. 1989 Mar 24;243(4898):1586–1589. doi: 10.1126/science.2928794. [DOI] [PubMed] [Google Scholar]
- Elliott A., Offer G., Burridge K. Electron microscopy of myosin molecules from muscle and non-muscle sources. Proc R Soc Lond B Biol Sci. 1976 Mar 30;193(1110):45–53. doi: 10.1098/rspb.1976.0030. [DOI] [PubMed] [Google Scholar]
- Elliott A., Offer G. Shape and flexibility of the myosin molecule. J Mol Biol. 1978 Aug 25;123(4):505–519. doi: 10.1016/0022-2836(78)90204-8. [DOI] [PubMed] [Google Scholar]
- Faruqi A. R., Cross R. A., Kendrick-Jones J. Structural studies on the conformations of myosin. Adv Exp Med Biol. 1993;332:81–91. doi: 10.1007/978-1-4615-2872-2_8. [DOI] [PubMed] [Google Scholar]
- Hansma H. G., Vesenka J., Siegerist C., Kelderman G., Morrett H., Sinsheimer R. L., Elings V., Bustamante C., Hansma P. K. Reproducible imaging and dissection of plasmid DNA under liquid with the atomic force microscope. Science. 1992 May 22;256(5060):1180–1184. doi: 10.1126/science.256.5060.1180. [DOI] [PubMed] [Google Scholar]
- Häberle W., Hörber J. K., Ohnesorge F., Smith D. P., Binnig G. In situ investigations of single living cells infected by viruses. Ultramicroscopy. 1992 Jul;42-44(Pt B):1161–1167. doi: 10.1016/0304-3991(92)90418-j. [DOI] [PubMed] [Google Scholar]
- Mould A. P., Holmes D. F., Kadler K. E., Chapman J. A. Mica sandwich technique for preparing macromolecules for rotary shadowing. J Ultrastruct Res. 1985 Apr;91(1):66–76. doi: 10.1016/0889-1605(85)90077-1. [DOI] [PubMed] [Google Scholar]
- Murray M. N., Hansma H. G., Bezanilla M., Sano T., Ogletree D. F., Kolbe W., Smith C. L., Cantor C. R., Spengler S., Hansma P. K. Atomic force microscopy of biochemically tagged DNA. Proc Natl Acad Sci U S A. 1993 May 1;90(9):3811–3814. doi: 10.1073/pnas.90.9.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I., Rypniewski W. R., Schmidt-Bäse K., Smith R., Tomchick D. R., Benning M. M., Winkelmann D. A., Wesenberg G., Holden H. M. Three-dimensional structure of myosin subfragment-1: a molecular motor. Science. 1993 Jul 2;261(5117):50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- Slayter H. S., Lowey S. Substructure of the myosin molecule as visualized by electron microscopy. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1611–1618. doi: 10.1073/pnas.58.4.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler J. M., Branton D. Rotary shadowing of extended molecules dried from glycerol. J Ultrastruct Res. 1980 May;71(2):95–102. doi: 10.1016/s0022-5320(80)90098-2. [DOI] [PubMed] [Google Scholar]
- Vesenka J., Guthold M., Tang C. L., Keller D., Delaine E., Bustamante C. Substrate preparation for reliable imaging of DNA molecules with the scanning force microscope. Ultramicroscopy. 1992 Jul;42-44(Pt B):1243–1249. doi: 10.1016/0304-3991(92)90430-r. [DOI] [PubMed] [Google Scholar]
- Walker M., Knight P., Trinick J. Negative staining of myosin molecules. J Mol Biol. 1985 Aug 5;184(3):535–542. doi: 10.1016/0022-2836(85)90300-6. [DOI] [PubMed] [Google Scholar]
- Walzthöny D., Eppenberger H. M., Ueno H., Harrington W. F., Wallimann T. Melting of myosin rod as revealed by electron microscopy. II. Effects of temperature and pH on length and stability of myosin rod and its fragments. Eur J Cell Biol. 1986 Jun;41(1):38–43. [PubMed] [Google Scholar]
- Weisenhorn AL, Maivald P, Butt H, Hansma PK. Measuring adhesion, attraction, and repulsion between surfaces in liquids with an atomic-force microscope. Phys Rev B Condens Matter. 1992 May 15;45(19):11226–11232. doi: 10.1103/physrevb.45.11226. [DOI] [PubMed] [Google Scholar]