Abstract
Serotyped strains of group B streptococci can be divided into subtypes based on restriction endonuclease digestion patterns (RDP). Profiles of cell-bound proteins were compared among RDP types. Proteins that showed a remarkable difference in the degree of expression were found among strains of RDP Ia-3, which has been considered potentially virulent, as well as of RDP Ib-1. For RDP Ia-3 strains, the protein was predominant in strains from cerebrospinal fluid (CSF) but was mostly a minor component in other strains. For RDP Ib-1 strains, the protein was predominant in strains from CSF, showed diversity in strains from blood, and was mostly a minor component in other strains. By N-terminal sequencing analysis, the protein was identified as a C protein β antigen. The level of bound immunoglobulin A (IgA) or anti-β antigen monoclonal antibody correlated with the level of expressed β antigen, and invasive strains showed remarkably high levels of binding; the exception was a CSF-derived strain of RDP Ib-1 which produced a large amount of β antigen and showed a high level of binding of anti-β antigen monoclonal antibody but no IgA binding. PCR-based amplification revealed that the β antigen gene was detected in all RDP Ia-3 and Ib-1 strains but was not found in any strains of other RDP types. Competitive reverse transcriptase PCR demonstrated that the difference in the amount of protein produced was due to the difference in the level of expression of the β antigen mRNA. Our findings imply that differences in gene expression for a protein may contribute to the invasiveness of RDP Ia-3 and Ib-1 strains for the host.
Group B streptococci (GBS) are important pathogens causing serious infections in neonates. GBS are classified serologically on the basis of capsular polysaccharide antigens, and serotypes Ia, Ib, II, III, IV, and V (25), VI (13), and VII and VIII (13) have thus far been recognized. Serotypes III and Ia are predominant among strains isolated from neonates with invasive infections (8, 11).
A previous study demonstrated that serotyped strains of GBS can be further divided into subtypes on the basis of a numerical analysis of HindIII restriction endonuclease digestion patterns (RDP) with chromosomal DNA and showed that strains from neonates with invasive infections belong to particular RDP types, RDP Ia-3 and III-3 (21). These RDP types are found at a low frequency in vaginal strains (21, 29). It was also shown that, among serotype Ia and III strains, RDP Ia-3 and III-3 strains are more resistant to opsonophagocytosis, suggesting that potentially virulent strains of GBS with genetic homogeneity may exist (30). Evidence of the increased virulence of RDP III-3 strains was further accumulated in subsequent studies (29, 31).
The GBS capsular polysaccharide antigens are well recognized as potential virulence factors due to their ability to impart resistance to opsonophagocytosis. In addition, the importance of the role of several cell surface proteins, such as C protein α and β antigens, R proteins, and Rib protein, in pathogenicity has been suggested (5, 27). The C protein antigens are found in approximately 60% of GBS clinical isolates and are expressed mainly by strains of serotypes Ia, Ib, and II and rarely by serotype III strains (2, 10). It has been suggested that C protein antigens may contribute to resistance to opsonization (22) and resistance to intracellular killing by phagocytes (23). These antigens also elicit protective immunity (3, 17, 19). In addition, β antigen is known to bind specifically to human immunoglobulin A (IgA) via its Fc portion (15, 26). Rib protein has been discovered in most strains of serotype III (27), R proteins have been found predominantly in serotype II and III strains (6), and alpha-like protein has been found in many serotype V strains (14). These three types of proteins are phenotypically similar to α antigen, which shows a ladder-like pattern on sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) and is resistant to trypsin degradation (7, 14, 27). Several of these proteins also provide protective immunity in animal models (14, 16, 19, 27).
In this study, we compared the profiles of cell-bound proteins expressed by strains of each RDP type to determine whether or not particular proteins exist among certain RDP types. The results revealed proteins that were remarkably different in the degree of expression among strains of RDP Ia-3, which has been considered potentially virulent, as well as of RDP Ib-1. The level of expression of the genes coding for the proteins of interest reflected the amounts produced, and it is suggested that such differences in gene expression levels may contribute to the pathogenicity of RDP types.
MATERIALS AND METHODS
Bacterial strains.
A total of 37 GBS strains, including 16 from infected infants and 21 vaginal strains, selected on the basis of RDP types previously reported (21), are listed in Table 1. For RDP Ia-3 and Ib-1 strains, which showed remarkable differences in the degree of expression of particular proteins on SDS-PAGE, the numbers of strains used in the study were increased. GBS strains were grown overnight in Todd-Hewitt broth (THB) (BBL Microbiology Systems, Cockeysville, Md.) and then stored in 10% skim milk (Difco Laboratories, Detroit, Mich.) at −85°C until use.
TABLE 1.
Sources and characteristics of GBS strains
| Strain no. | Source | Serotype | RDP type | β antigen genea | Bindingb of:
|
β antigen mRNA transcripts (amol/ng of total RNA)c | |
|---|---|---|---|---|---|---|---|
| Anti-β monoclonal antibody | IgA | ||||||
| 1 | Vagina | Ia | Ia-1 | − | − | − | |
| 2 | Vagina | Ia | Ia-1 | − | − | − | |
| 3 | Vagina | Ia | Ia-2 | − | − | − | |
| 4 | Blood | Ia | Ia-2 | − | − | − | |
| 5 | Vagina | Ia | Ia-3 | + | ± | ± | 2.897 ± 0.155 |
| 6 | Vagina | Ia | Ia-3 | + | ± | ± | 3.343 ± 0.327 |
| 7 | Vagina | Ia | Ia-3 | + | − | − | 4.243 ± 0.224 |
| 8 | Vagina | Ia | Ia-3 | + | − | − | 0.127 ± 0.029 |
| 9 | Vagina | Ia | Ia-3 | + | − | − | 0.017 ± 0.009 |
| 10 | Blood | Ia | Ia-3 | + | − | − | 0.157 ± 0.005 |
| 11 | CSF | Ia | Ia-3 | + | +++ | +++ | 8.937 ± 0.279 |
| 12 | CSF | Ia | Ia-3 | + | +++ | +++ | 9.273 ± 0.303 |
| 13 | CSF | Ia | Ia-3 | + | +++ | +++ | 10.33 ± 1.036 |
| 14 | CSF | Ia | Ia-3 | + | +++ | +++ | 10.53 ± 0.66 |
| 15 | CSF | Ia | Ia-3 | + | +++ | +++ | 12.50 ± 0.938 |
| 16 | Vagina | Ib | Ib-1 | + | − | − | 0.053 ± 0.005 |
| 17 | Vagina | Ib | Ib-1 | + | − | − | 0.803 ± 0.176 |
| 18 | Vagina | Ib | Ib-1 | + | + | + | 3.143 ± 0.398 |
| 19 | Blood | Ib | Ib-1 | + | ± | ± | 2.657 ± 0.333 |
| 20 | Blood | Ib | Ib-1 | + | ++ | ++ | 11.23 ± 0.893 |
| 21 | Blood | Ib | Ib-1 | + | +++ | +++ | 13.73 ± 1.318 |
| 22 | CSF | Ib | Ib-1 | + | +++ | − | 12.96 ± 0.39 |
| 23 | CSF | Ib | Ib-1 | + | +++ | +++ | 16.25 ± 0.777 |
| 24 | Vagina | II | II-1 | − | − | − | |
| 25 | Vagina | II | II-2 | − | − | − | |
| 26 | Vagina | III | III-1 | − | − | − | |
| 27 | CSF | III | III-1 | − | − | − | |
| 28 | Vagina | III | III-2 | − | − | − | |
| 29 | Blood | III | III-2 | − | − | − | |
| 30 | Vagina | III | III-3 | − | − | − | |
| 31 | Blood | III | III-3 | − | − | − | |
| 32 | Vagina | V | V-1 | − | − | − | |
| 33 | Vagina | V | V-2 | − | − | − | |
| 34 | Vagina | VI | VI-1 | − | − | − | |
| 35 | CSF | VI | VI-1 | − | − | − | |
| 36 | Vagina | VIII | VIII-1 | − | − | − | |
| 37 | Vagina | VIII | VIII-1 | − | − | − | |
The β antigen gene was detected by PCR. +, present; −, absent.
The binding assay was performed by Western blotting.
Values are the means and standard deviations for three independent cRT-PCR determinations.
Protein extraction and SDS-PAGE.
Overnight cultures of GBS in 120 ml of THB were harvested by centrifugation and washed twice with 40 ml of 40 mM Tris-50 mM EDTA (pH 6.8). The pellets were resuspended in 2 ml of sample buffer (50 mM Tris-HCl [pH 6.8], 2% SDS, 1 mM dithiothreitol, 10% glycerol, 0.005% bromphenol blue), which was boiled for 5 min and then centrifuged at 12,000 × g at 4°C for 10 min. Protein determination by the Lowry method was performed with the supernatants by using the DC protein assay (Bio-Rad Laboratories, Richmond, Calif.).
Approximately 15-μg portions of the protein samples were subjected to SDS-PAGE with 5 to 20% gradient gels and were stained with SYPRO Red (BioWhittaker Molecular Applications, Rockland, Maine) under light-shielded conditions. Molecular masses were determined by using Perfect Protein Markers (10 to 225 kDa; Novagen Inc., Madison, Wis.) with Scanning Imager PDSI (Amersham Pharmacia Biotech, Piscataway, N.J.).
N-terminal sequence analysis.
N-terminal sequencing of 20 amino acid residues was performed on the dominant 144-kDa band obtained from strain no. 13 which, among the RDP Ia-3 strains analyzed by SDS-PAGE, had expressed a large amount of the protein of interest. The protein was eluted from the gel portion containing the 144-kDa fragment by adding 0.1% SDS-0.1 M NH4HCO3 and allowing the sample to stand overnight at 37°C. The protein was analyzed by the Edman degradation method with an automated sequencer (HP G1005A protein sequencing system; Hewlett-Packard, Palo Alto, Calif.).
Immunoblotting.
Protein samples for immunoblotting were prepared as described above. Approximately 5-μg portions of proteins separated by SDS-PAGE were electroblotted onto polyvinylidene difluoride membranes (Clearblot P membranes; Atto Corp., Tokyo, Japan). After two washes with phosphate-buffered saline (PBS) plus 0.1% Tween 20 (T-PBS) for 5 min, the membranes were blocked in T-PBS containing 3% skim milk for 30 min. After washing with T-PBS for 10 min, the membranes were incubated in T-PBS containing 10 μg of human IgA from colostrum (Sigma, St. Louis, Mo.)/ml overnight. The membranes were washed three times with T-PBS and then were incubated for 2 h in T-PBS containing a 1/1,000 dilution of horseradish peroxidase-conjugated goat anti-human IgA (Sigma). Finally, the membranes were washed three times with T-PBS for 10 min, and 4-chloro-1-naphthol was added for color development. Other samples from electroblotted membranes were processed in the same manner by using the combination of a 1/1,000 dilution of anti-β antigen (anti-β) monoclonal antibody (20) and a 1/1,000 dilution of peroxidase-labeled goat anti-mouse IgG (Sigma). The anti-β monoclonal antibody was kindly provided by Lars Bevanger, University of Trondheim, Trondheim, Norway. The amounts of IgA and anti-β monoclonal antibody bound were analyzed by densitometry (Scanning Imager PDSI and ImageQuaNT software [Amersham Pharmacia Biotech]).
PCR-based β antigen gene amplification.
For detection of the β antigen gene, we used 58 strains comprising the 37 strains listed in Table 1 and the 21 strains from infected infants and vaginal strains selected on the basis of RDP types. Oligonucleotide primers were synthesized on the basis of two 24-mer primers designed by Maeland et al. (18) by using the sequence data for the IgA-binding β antigen gene from Jerlström et al. (9). These primers, corresponding to nucleotides 1337 to 1360 (5′-AAGGCTATGAGTGAGAGCTTGGAG-3′) and 1917 to 1940 (5′-CTGCTCTGGTGTTTTAGGAACTTG-3′), amplified a 604-bp fragment. PCR was run for 30 cycles, each cycle consisting of denaturation at 94°C for 1 min, primer annealing at 45°C for 1 min, and extension at 72°C for 1 min.
Total RNA extraction.
Total RNA was extracted from 1 ml of GBS cells in the mid-exponential growth phase (108 CFU/ml) by using RNAprotect Bacteria Reagent and an RNeasy Mini Kit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's instructions. First, for RNA stabilization, 1 ml of cell culture in THB was added to 2 ml of RNAprotect Bacteria Reagent, and the mixture was incubated for 5 min at room temperature. The pelleted cells were resuspended in 100 μl of mutanolysin (Sigma) solution (100 U/ml of Tris-EDTA), incubated for 20 min at room temperature, and then treated with 350 μl of buffer RLT (Qiagen) for lysis. After the addition of 250 μl of ethanol to the lysate, RNase-free DNase I (Qiagen) treatment was performed for DNA removal. The RNA was purified in succeeding steps with spin columns, a process which yielded 50 μl of RNA in RNase-free water. The integrity of total RNA was checked by agarose gel electrophoresis. The RNA was quantified spectrophotometrically and stored in aliquots at −70°C.
Construction of control RNA.
Control RNA was designed to generate a 60-bp deletion product in the 540-bp target RNA by using the DNA sample from strain no. 13, with which the β antigen had been identified by N-terminal sequencing. Oligonucleotide 24-mer primers were synthesized on the basis of published sequence data (9) and corresponded to nucleotides 372 to 395 (5′-GTGTAGGAGTAGCTAGTGTAGCGG-3′; RT primer forward) and 888 to 911 (5′-GCTCTTTGTCAATATTGCTTAGAT-3′; RT primer reverse). PCR was run for 30 cycles consisting of 1 min of denaturation at 94°C, 1 min of annealing at 54°C, and 1 min of extension at 72°C per cycle; the last cycle was followed by a final extension at 72°C for 10 min.
A 5-μg portion of the 540-bp PCR amplification product was digested with 20 U of restriction enzyme MseI (New England Biolabs, Beverly, Mass.), which produced fragments of 352, 128, and 60 bp. A 60-bp MseI digest fragment was removed to create a 480-bp modified fragment by a ligation reaction. A phage T7 RNA polymerase promoter was added in the sense direction to the 480-bp fragment through a Lig'nScribe reaction (Ambion Inc., Austin, Tex.) according to the manufacturer's instructions. A 10-μl ligation reaction mixture including 1 μl (25 ng) of the 480-bp fragment DNA, 1 μl of T4 DNA ligase, 1 μl of T7 promoter adapter, and 1 μl of 10× ligation buffer was incubated for 15 min at room temperature, and a 2-μl portion of the ligated product was subjected to PCR. The PCR conditions were the same as those used for amplification of the 540-bp PCR product, except for the primer combination of PCR adapter primer (forward; Ambion) and our reverse primer (RT primer reverse). PCR-generated DNA thus obtained was purified and resuspended in 10 μl of sterile water.
For in vitro transcription, a 20-μl reaction mixture containing 1 μg (2 μl) of T7 promoter-containing template DNA, 1 μl of each deoxynucleoside triphosphate (10 mM each), 2 μl of T7 RNA polymerase, and 2 μl of 10× transcription buffer was prepared according to the manufacturer's instructions (MAXIscript; Ambion). After incubation for 1 h at 37°C, 1 μl (2 U) of RNase-free DNase I was added to the mixture, which was incubated for 15 min at 37°C. Free nucleotides were removed from the transcription reaction by ethanol precipitation and resuspended in 50 μl of RNase-free water. The transcripts were quantified spectrophotometrically and stored in aliquots at −70°C.
In every step of the control RNA construction process, the resulting products were confirmed by agarose gel electrophoresis.
cRT-PCR.
For the quantification of GBS mRNA, a competitive reverse transcriptase PCR (cRT-PCR) was performed by using OneStep RT-PCR (Qiagen) according to the manufacturer's instructions. Coamplification of a constant amount of total RNA with known amounts of twofold serial dilutions of control RNA was achieved with a 50-μl reaction mixture including 3 μl each of 0.6 μM RT primer forward and 0.6 μM RT primer reverse, 20 U of RNase inhibitor (Takara Shuzo Co., Kyoto, Japan), and 1 μl each of total RNA and control RNA. A reverse transcription reaction for 30 min at 50°C was followed by an initial heating step of PCR amplification at 95°C for 15 min to activate Taq DNA polymerase as well as to inactivate reverse transcriptase. After 30 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 54°C, and extension for 1 min at 72°C per cycle, a final extension for 10 min at 72°C was carried out. A control reaction, in which RNA was added during the initial heating step without prior reverse transcription, was included in each cRT-PCR to confirm no contamination of genomic DNA.
Five-microliter portions of the amplified products were subjected to agarose gel electrophoresis with 3% NuSieve 3:1 agarose, and then the gels were stained with SYBR Green I. The intensities of both 540- and 480-bp bands were analyzed by densitometry (Scanning Imager PDSI). The logarithm of the ratio of the intensity of the 480-bp band to that of the 540-bp band was plotted as a function of the logarithm of the amount of input control RNA. The initial amount of target RNA was given when the ratio was 1 (log1 = 0) in the linear regression analysis. Target RNA was expressed as attomoles of β antigen mRNA transcripts per nanogram of total RNA.
RESULTS
Protein profiles on SDS-PAGE.
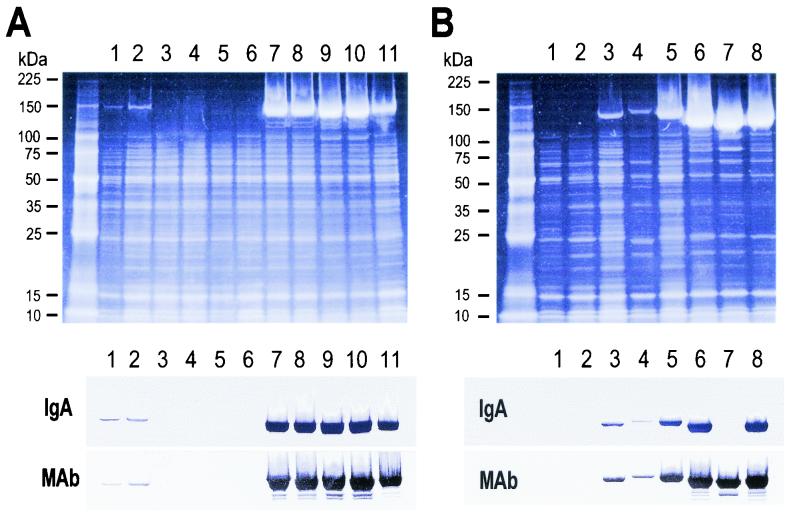
To determine whether or not particular proteins are expressed by certain RDP types, we compared the SDS-PAGE profiles of cell-bound proteins from 37 strains of known RDP types. A specific protein that showed a remarkable difference in the degree of expression was observed among strains of RDP Ia-3 and Ib-1. For RDP Ia-3 strains, such a protein was predominant in all five strains derived from the cerebrospinal fluid (CSF) of infants with meningitis (strain no. 11, 12, 13, and 14 [144 kDa] and 15 [147 kDa]). It was either found in trace amounts or was not recognized in a blood isolate from a case of septicemia and in vaginal isolates of RDP Ia-3 (Fig. 1A). For RDP Ib-1 strains, the corresponding protein was very predominant in strain no. 22 and 23 from the CSF of infants with meningitis (120 kDa). Among strains from the blood of infants with septicemia, strain no. 21 (124 kDa) was predominant, strain no. 20 (135 kDa) occurred at an intermediate level, and strain no. 19 (142 kDa) occurred at a low level. The specific protein was either found in trace amounts or was not observed in vaginal isolates of RDP Ib-1 (Fig. 1B). The specific protein was not recognized among strains of RDP types other than RDP Ia-3 and Ib-1.
FIG. 1.
SDS-PAGE profiles of cell-bound proteins of GBS of RDP Ia-3 and Ib-1. SDS-solubilized proteins from strain no. 5 to 15 (lanes 1 to 11) of RDP Ia-3 (A) and strain no. 16 to 23 (lanes 1 to 8) of RDP Ib-1 (B) were analyzed with 5 to 20% gradient gels. For each RDP, SYPRO Red staining and immunoblotting are shown. The proteins separated were blotted onto a polyvinylidene difluoride membrane and probed with human IgA (IgA) or with an anti-β monoclonal antibody (MAb). Numbers at left indicate molecular mass standards.
N-terminal amino acid sequencing.
N-terminal sequencing of 20 amino acids of the predominant 144-kDa protein from strain no. 13 (RDP Ia-3) revealed the sequence SELVKDDSVKTTEVAAKPYP. A database homology search with the Swiss Institute of Bioinformatics BLAST Network Service indicated that the sequence showed 100% homology to that of β antigen at the corresponding positions.
Immunoreactivity with IgA or anti-β monoclonal antibody.
For the specific protein showing a remarkable difference in the degree of expression, immunoreactivity with IgA or anti-β monoclonal antibody was investigated. Immunoblotting results for all 11 strains of RDP Ia-3 and all 8 strains of RDP Ib-1 are shown in Fig. 1, where all bands which were reactive with IgA or anti-β monoclonal antibody or both corresponded to bands for specific proteins on SDS-PAGE. In addition, the level of binding observed with IgA or anti-β monoclonal antibody when these 19 strains and strains of other RDP types were probed with IgA or anti-β monoclonal antibody is expressed as a percentage of the result obtained for the strain showing the highest density in each blotted membrane and then categorized as follows (Table 1): −, 0%; ±, <10%; +, 10 to 33%; ++, 34 to 66%; and +++, 67 to 100%. For RDP Ia-3 strains, the amounts of IgA and anti-β monoclonal antibody bound were remarkably high (+++) in five strains from the CSF of infants with meningitis (strain no. 11 to 15), but binding was not observed in strain no. 10, from the blood of an infant with septicemia. The binding was either in trace amounts or was not recognized in vaginal isolates of RDP Ia-3. For RDP Ib-1 strains, the amounts bound were remarkably high (+++) in two strains from blood and CSF (strain no. 21 and 23, respectively), intermediate (++) in strain no. 20 (from blood), and low (±) in strain no. 19 (from blood). The binding was either in trace amounts or was not recognized in vaginal isolates of RDP Ib-1. For strain no. 22, which was derived from CSF, IgA binding was not recognized, whereas a remarkable level of binding of anti-β monoclonal antibody (+++) was noted. No binding to IgA or anti-β monoclonal antibody was found among strains of other RDP types. These results demonstrated that the level of binding of IgA or anti-β monoclonal antibody correlated with the level of expressed β antigen on SDS-PAGE, except for strain no. 22.
Distribution of β antigen gene among RDP types.
For 58 strains selected on the basis of RDP types, the presence of the β antigen gene was analyzed by PCR. In all 11 strains of RDP Ia-3 and all 8 strains of RDP Ib-1, the β antigen gene was detected as a 604-bp PCR product with size homogeneity (100%). The β antigen gene was not found in any of the five strains each of RDP Ia-1, Ia-2, III-1, III-2, or III-3, in any of the three strains each of RDP VI-1 or VIII-1, and in either of the two strains each of RDP II-1, II-2, V-1, or V-2.
Quantification of β antigen transcripts.
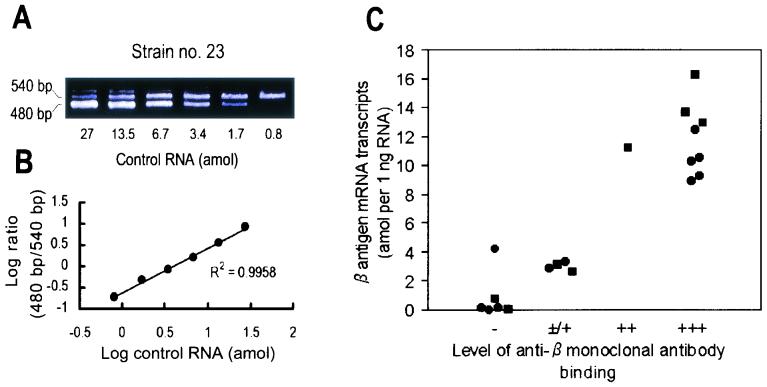
Agarose gel electrophoresis results showed bands with theoretical molecular sizes for each of the PCR products of target DNA, their MseI digests, ligation reaction products, T7 promoter addition products, and in vitro transcripts (data not shown), and each step of the control RNA construction process was confirmed to be properly achieved. The integrity of total RNA was confirmed as conserved (data not shown). The β antigen mRNA transcripts were quantified for 19 strains having the β antigen gene. For each strain, the appropriate combination of the range of six serial twofold dilutions for control RNA (27 to 0.00625 amol) and total RNA (1, 0.5, or 0.25 ng) in the reaction mixture was adopted based on preliminary experiments. Representative agarose gels and plotting of the data are shown in Fig. 2A and B.
FIG. 2.
Quantification of β antigen mRNA transcripts by cRT-PCR. (A) Agarose gel profile of β antigen transcripts of samples from a representative strain, strain no. 23 (RDP Ib-1), which shows the level of binding of anti-β monoclonal antibody to be +++. A constant amount of total RNA (0.25 ng) was reverse transcribed and amplified with twofold serial dilutions of control RNA (0.8 to 27 amol). Amplified products were visualized by staining with SYBER Green I after electrophoresis as described in Materials and Methods. The positions of the cRT-PCR products from 540-bp target RNA and 480-bp control RNA are indicated. (B) Plotting of the logarithm of the ratio of target to control intensities against the logarithm of the amount of control RNA added to the reaction mixture. The amounts of β antigen mRNA transcripts were determined from linear regression analysis of the plotted data. Scanning of the gels and quantitative analyses were done as described in Materials and Methods. (C) Quantification of β antigen mRNA transcripts and levels of anti-β monoclonal antibody binding. The quantification results are plotted as mean values. Symbols:•, RDP Ia-3 strains; ▪, RDP Ib-1 strains. A significant difference in the amount of transcripts was obtained for strains with anti-β monoclonal antibody binding levels of ++ and +++ compared to those with binding levels of − and ± or +.
An analysis of the gels after SYBER Green I staining showed cRT-PCR-amplified products of total RNA and control RNA, as expected, and two bands that differed in length by 60 bp were distinguishable (Fig. 2A). Plotting of the logarithm of the ratio of control RNA to total RNA band intensities as a function of the logarithm of the amount of input control RNA in the reaction mixture allowed calculation of the amounts of β antigen mRNA transcripts by linear regression analysis (Fig. 2B). The quantified results are shown in Table 1 and Fig. 2C. Among 19 strains comprising 11 strains of RDP Ia-3 and 8 strains of RDP Ib-1, the level of transcripts ranged from 16.25 to 0.017 amol per ng of total RNA. The detection limit of the cRT-PCR was estimated to be ≤0.00625 amol in our experimental system. The findings shown in Fig. 2C demonstrate a correlation between the amounts of anti-β monoclonal antibody bound and the amounts of β antigen mRNA transcripts. A significantly greater increase in the amounts of transcripts was obtained for samples from 9 strains (11.75 ± 2.346 amol) with a higher level of anti-β monoclonal antibody binding (++ and +++) than for 10 strains (1.744 ± 1.659 amol) with a lower level of anti-β monoclonal antibody binding (− and ± or +) (P < 0.0001, as determined by Student's t test).
DISCUSSION
In this study, we tried to detect differences in cell-bound protein profiles expressed by strains of each RDP type. Dissimilarity in the degree of expression of specific protein fragments was found among strains belonging to certain RDP types, Ia-3 and Ib-1. The specific protein was produced in remarkable amounts in RDP Ia-3 strains from the CSF of infants with meningitis, whereas it was either produced only in trace amounts or not observed in other RDP Ia-3 strains, including a strain from the blood of an infant with septicemia. For RDP Ib-1 strains, the specific protein was produced at remarkably high levels in strains from the CSF of infants with meningitis and showed remarkable differences in strains from the blood of infants with septicemia, being produced at extremely high, medium, and low levels. It was either present in trace amounts or not found in other RDP Ib-1 strains. Such a specific protein showing remarkable differences in the degree of expression was not observed among strains of RDP types other than RDP Ia-3 and Ib-1.
By N-terminal sequencing analysis of the first 20 amino acids, the specific protein was identified as β antigen with 100% homology; this result was also confirmed by anti-β monoclonal antibody binding.
The β antigen specifically binds to the Fc region of human IgA (15, 26). Human IgA is the predominant immunoglobulin class on mucosal surfaces. On mucosal surfaces, which are the major potential invasion sites, secretory IgA, as the main antibody in secretions, provides a first line of defense against pathogenic microorganisms (12). Our results showed that binding of human IgA was found among strains of limited RDP types, Ia-3 and Ib-1. Moreover, the level of IgA bound correlated with the level of β antigen expressed on SDS-PAGE, in which invasive strains showed a remarkably high level of binding.
A previous study based on PCR analysis showed that the β antigen gene is found only in serotype Ia, Ib, and II strains, with significant occurrences of 93% for type Ib, 57% for type II, and 6% for type Ia, whereas it is not found in serotype III strains (1). It was also reported that, through analysis with a β gene probe, hybridization is found for 21% of type Ia, 73% of type Ib, and 83% of type II strains but is not found for type III strains (28).
We performed a PCR-based analysis of the β antigen gene by using strains selected on the basis of RDP types; therefore, it might not be appropriate for determining the distribution of the gene among serotypes. Nevertheless, our findings, in which the β antigen gene was detected in serotypes Ia and Ib and was not detected in serotype III, are not inconsistent with those of previous studies. In addition, our RDP type-based analysis results showed that the β antigen gene was detectable specifically only in strains of RDP Ia-3, a subtype of serotype Ia, and RDP Ib-1, a subtype of serotype Ib. The RDP Ia-3 strains showed overall frequencies of 15.8 and 77.8% for vaginal strains and invasive strains, respectively, of serotype Ia (21; unpublished data) and were more resistant to opsonophagocytosis with serum containing type-specific antibodies (30). The data on β antigen gene distribution support our implication that RDP Ia-3 strains may be potentially virulent and also serve as important basic findings in analyzing the virulence of RDP Ib-1 strains. The present study did not detect the β antigen gene in any strains of RDP II-1 and II-2, which are subtypes of serotype II; this finding was not in agreement with the findings of other studies. Such differences may reflect geographical variations in the distribution of GBS strains. However, even when we took this possibility into consideration, our finding that the β antigen gene was limited to RDP Ia-3 and Ib-1 strains seems very interesting with regard to the evolutionary background of GBS strains.
The mRNA transcripts of the β antigen genes of RDP Ia-3 and Ib-1 strains were quantified through the cRT-PCR method. The results indicated that the amounts of the β antigen transcripts showed diversity among these strains, ranging from 16.25 to 0.017 amol per ng of total RNA, in spite of all of these strains having β antigen genes. The purpose of the quantification was to investigate whether the level of β antigen production is attributable to the level of the corresponding gene expression; our findings demonstrate a correlation between the amounts of the mRNA transcripts and the amounts of anti-β monoclonal antibody bound, which reflected the protein production level. There was a statistically significant difference in the mRNA transcript amounts between samples from strains with higher levels of anti-β monoclonal antibody binding, ++ and +++, and samples from strains with lower levels of anti-β monoclonal antibody binding, − and ± or +. Vaginal strain no. 7 showed an mRNA transcript level of 4.243 amol, and binding was not observed to either anti-β monoclonal antibody or IgA, whereas strain no. 5, 6, 18, and 19, with mRNA transcript levels ranging from 2.657 to 3.343 amol, showed ± or + levels of binding. By immunoblotting analysis with both anti-β monoclonal antibody and IgA, we confirmed that, in each strain, the amounts and the molecular masses of the major reactive bands in concentrated culture supernatants correlated with those in cell-bound protein extracts (data not shown). Therefore, in strain no. 7, the β antigen also could not be found in the concentrated culture supernatant.
The target sequence used in this study for the quantification of β antigen mRNA transcripts contains a signal sequence and a portion of IgA-binding domain A (9). Therefore, the difference in the amounts of the protein produced was due to the difference in the levels of expression of the β antigen mRNA derived from the target sequence. Furthermore, our findings that most invasive strains of RDP Ia-3 and Ib-1 achieved higher levels of expression of β antigen transcripts but that others showed, on average, only 15% the expression of those invasive strains suggest that such differences in gene expression may be involved in the virulence of strains of these RDP types. The proposed role of β antigen in virulence is that nonimmune binding of the Fc portion of IgA by β antigen may block the deposition of IgG or complement on the GBS surface, rendering them opsonically inactive. Alternatively, the ability of streptococcal IgA-binding proteins to inhibit the binding of IgA to human IgA receptor CD89, an important mediator of IgA effector function, has been shown (24). Perhaps the binding of the IgA Fc portion to β antigen allows GBS to evade the elimination processes that would normally be triggered by the binding of the IgA Fc portion to CD89 (32). Through these mechanisms or other mechanisms, RDP Ia-3 and Ib-1 strains with higher levels of expression of β antigen transcripts may become virulent. The genetic mechanism that increases the amounts of β antigen mRNA transcripts in these invasive strains needs to be further clarified.
Low-molecular-weight secreted forms of β antigen lacking IgA Fc-binding ability from some GBS strains have been reported (4). A CSF-derived strain, strain no. 22, which showed a higher level of expression of β antigen transcripts, produced a remarkable amount of the protein, but lacked IgA-binding ability, was found to belong to RDP Ib-1. The protein is considered to be a mature one, with a molecular mass of 120 kDa, and such a mature protein with no IgA-binding ability has not been reported. Further study is needed to define whether the lack of IgA binding is due to mutations in the coding sequence of the main IgA-binding domain or another mechanism as well as to define the possible role of a lack of IgA binding in virulence.
Acknowledgments
We are indebted to Shinji Takahashi, Joshi-Eiyoh University, for expert suggestions. We are grateful to Lars Bevanger, University of Trondheim, for providing the anti-β monoclonal antibody. We thank T. Uehara, Y. Ida, and the Medical Microbiology Laboratory staff of Funabashi Medical Center for their cooperation.
Editor: E. I. Tuomanen
REFERENCES
- 1.Berner, R., A. Bender, C. Rensing, J. Forster, and M. Brandis. 1999. Low prevalence of the immunoglobulin-A-binding β antigen of the c protein among. Streptococcus agalactiae isolates causing neonatal sepsis. Eur. J. Clin. Microbiol. Infect. Dis. 18:545-550. [DOI] [PubMed] [Google Scholar]
- 2.Bevanger, L. 1983. Ibc proteins as serotype markers of group B streptococci. Acta Pathol. Microbiol. Immunol. Scand. Sect. B 91:231-234. [DOI] [PubMed] [Google Scholar]
- 3.Bevanger, L., and A. I. Naess. 1985. Mouse protective antibodies against the Ibc proteins of group B streptococci. Acta Pathol. Microbiol. Immunol. Scand. Sect. B 93:121-124. [DOI] [PubMed] [Google Scholar]
- 4.Brady, L. J., and M. D. P. Boyle. 1989. Identification of non-immunoglobulin A-Fc-binding forms and low-molecular-weight secreted forms of the group B streptococcal β antigen. Infect. Immun. 57:1573-1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferrieri, P. 1988. Surface-localized protein antigens of group B streptococci. Rev. Infect. Dis. 10(Suppl. 2):S363-S366. [DOI] [PubMed] [Google Scholar]
- 6.Flores, A. E., and P. Ferrieri. 1989. Molecular species of R-protein antigens produced by clinical isolates of group B streptococci. J. Clin. Microbiol. 27:1050-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flores, A. E., and P. Ferrieri. 1996. Molecular diversity among the trypsin resistant surface proteins of group B streptococci. Zentbl. Bakteriol. 285:44-51. [DOI] [PubMed] [Google Scholar]
- 8.Harrison, L. H., J. A. Elliott, D. M. Dwyer, J. P. Libonati, P. Ferrieri, L. Billmann, and A. Schuchat. 1998. Serotype distribution of invasive group B streptococcal isolates in Maryland: implications for vaccine formulation. Maryland emerging infections program. J. Infect. Dis. 177:998-1002. [DOI] [PubMed] [Google Scholar]
- 9.Jerlström, P. G., G. S. Chhatwal, and K. N. Timmis. 1991. The IgA-binding antigen of the c protein complex of group B streptococci: sequence determination of its gene and detection of two binding regions. Mol. Microbiol. 5:843-849. [DOI] [PubMed] [Google Scholar]
- 10.Johnson, D. R., and P. Ferrieri. 1984. Group B streptococcal Ibc protein antigen: distribution of two determinants in wild-type strains of common serotypes. J. Clin. Microbiol. 19:506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kalliola, S., J. Vuopio-Varkila, A. K. Takala, and J. Eskola. 1999. Neonatal group B streptococcal disease in Finland: a ten-year nationwide study. Pediatr. Infect. Dis. J. 18:806-810. [DOI] [PubMed] [Google Scholar]
- 12.Kerr, M. A. 1990. The structure and function of human IgA. Biochem. J. 271:285-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachenauer, C. S., D. L. Kasper, J. Shimada, Y. Iciman, H. Ohtsuka, M. Kaku, L. C. Paoletti, and L. C. Madoff. 1999. Serotypes VI and VIII predominate among group B streptococci isolated from pregnant Japanese women. J. Infect. Dis. 179:1030-1033. [DOI] [PubMed] [Google Scholar]
- 14.Lachenauer, C. S., and L. C. Madoff. 1996. A protective surface protein from type V group B streptococci shares N-terminal sequence homology with the alpha C protein. Infect. Immun. 64:4255-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl, G., B. Åkerström, J. P. Vaerman, and L. Stenberg. 1990. Characterization of an IgA receptor from group B streptococci: specificity for serum IgA. Eur. J. Immunol. 20:2241-2247. [DOI] [PubMed] [Google Scholar]
- 16.Linden, V., K. K. Christensen, and P. Christensen. 1983. Correlation between low levels of maternal IgG antibodies to R protein and neonatal septicemia with group B streptococci carrying R protein. Int. Arch. Allergy Appl. Immunol. 71:168-172. [DOI] [PubMed] [Google Scholar]
- 17.Madoff, L. C., J. L. Michel, E. W. Gong, A. K. Rodewald, and D. L. Kasper. 1992. Protection of neonatal mice from group B streptococcal infection by maternal immunization with beta C protein. Infect. Immun. 60:4989-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maeland, J. A., O. G. Brakstad, L. Bevanger, and A. I. Kvam. 1997. Streptococcus agalactiae β gene and gene product variations. J. Med. Microbiol. 46:999-1005. [DOI] [PubMed] [Google Scholar]
- 19.Michel, J. L., L. C. Madoff, D. E. Kling, D. L. Kasper, and F. M. Ausubel. 1991. Cloned alpha and beta C-protein antigens of group B streptococci elicit protective immunity. Infect. Immun. 59:2023-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naess, A. I., L. Bevanger, O.-J. Iversen, and J. A. Maeland. 1991. Evaluation of monoclonal antibodies in serovar classification of group B streptococci (GBS). Acta Pathol. Microbiol. Immunol. Scand. 103:731-736. [DOI] [PubMed] [Google Scholar]
- 21.Nagano, Y., N. Nagano, S. Takahashi, K. Murono, K. Fujita, F. Taguchi, and Y. Okuwaki. 1991. Restriction endonuclease digest patterns of chromosomal DNA from group B β-haemolytic streptococci. J. Med. Microbiol. 35:297-303. [DOI] [PubMed] [Google Scholar]
- 22.Payne, N. R., and P. Ferrieri. 1985. The relation of the Ibc protein antigen to the opsonization differences between strains of type II group B streptococci. J. Infect. Dis. 151:672-681. [DOI] [PubMed] [Google Scholar]
- 23.Payne, N. R., Y. K. Kim, and P. Ferrieri. 1987. Effect of differences in antibody and complement requirements on phagocytic uptake and intracellular killing of “c” protein-positive and -negative strains of type II group B streptococci. Infect. Immun. 55:1243-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pleass, R. J., T. Areschoug, G. Lindahl, and J. M. Woof. 2001. Streptococcal IgA-binding proteins bind in the Cα2-Cα3 interdomain region and inhibit binding of IgA to human CD89. J. Biol. Chem. 276:8197-8204. [DOI] [PubMed] [Google Scholar]
- 25.Rench, M. A., and C. J. Baker. 1993. Neonatal sepsis caused by a new group B streptococcal serotype. J. Pediatr. 122:638-640. [DOI] [PubMed] [Google Scholar]
- 26.Russell-Jones, G. J., E. C. Gotschlich, and M. S. Blake. 1984. A surface receptor specific for human IgA on group B streptococci possessing the Ibc protein antigen. J. Exp. Med. 160:1467-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stalhammar-Carlemalm, M., L. Stenberg, and G. Lindahl. 1993. Protein Rib: a novel group B streptococcal cell surface protein that confers protective immunity and is expressed by most strains causing invasive infections. J. Exp. Med. 177:1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suvorov, A., A. Dmitriev, I. Ustinovitch, C. Schalen, and A. A. Totolian. 1997. Molecular analysis of clinical group B streptococcal strains by use of alpha and beta gene probes. FEMS Immunol. Med. Microbiol. 17:149-154. [DOI] [PubMed] [Google Scholar]
- 29.Takahashi, S., E. E. Adderson, Y. Nagano, N. Nagano, M. R. Briesacher, and J. F. Bohnsack. 1998. Identification of a highly encapsulated, genetically related group if invasive type III group B streptococci. J. Infect. Dis. 177:1116-1119. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi, S., Y. Nagano, N. Nagano, K. Fujita, F. Taguchi, and Y. Okuwaki. 1993. Opsonisation of group B streptococci and restriction endonuclease digestion patterns of their chromosomal DNA. J. Med. Microbiol. 38:191-196. [DOI] [PubMed] [Google Scholar]
- 31.Takahashi, S., Y. Nagano, N. Nagano, O. Hayashi, F. Taguchi, and Y. Okuwaki. 1995. Role of C5a-ase in group B streptococcal resistance to opsonophagocytic killing. Infect. Immun. 63:4764-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Egmond, M., E. van Garderen, A. B. van Spriel, C. A. Damen, E. S. van Amersfoort, G. van Zandbergen, J. van Hattum, J. Kuiper, and J. G. van de Winkel. 2000. FcalphaRI-positive liver Kupffer cells: reappraisal of the function of immunoglobulin A in immunity. Nat. Med. 6:680-685. [DOI] [PubMed] [Google Scholar]