Abstract
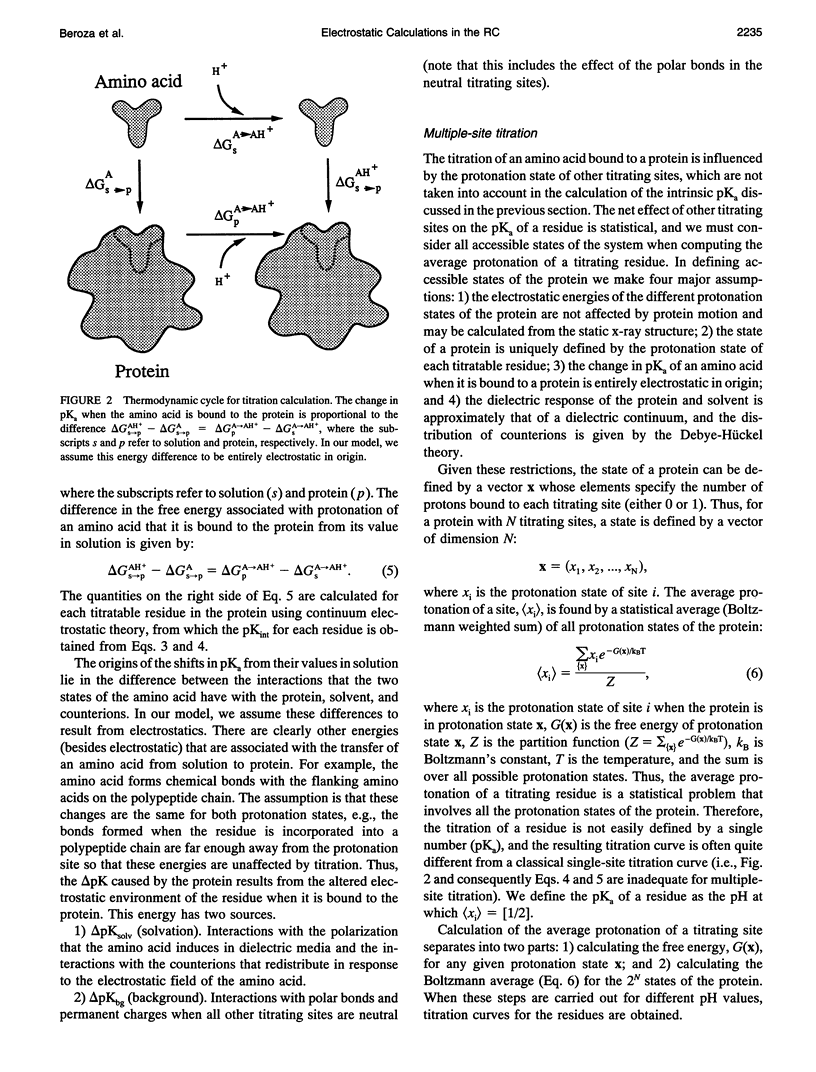
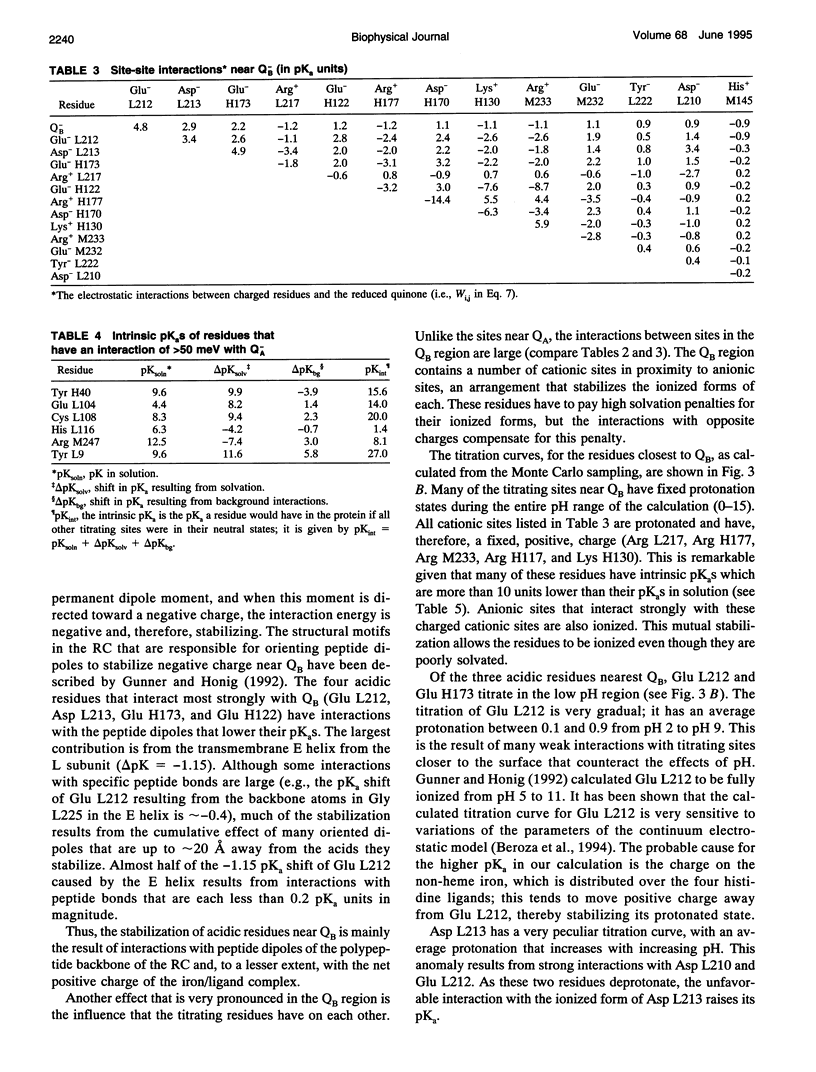
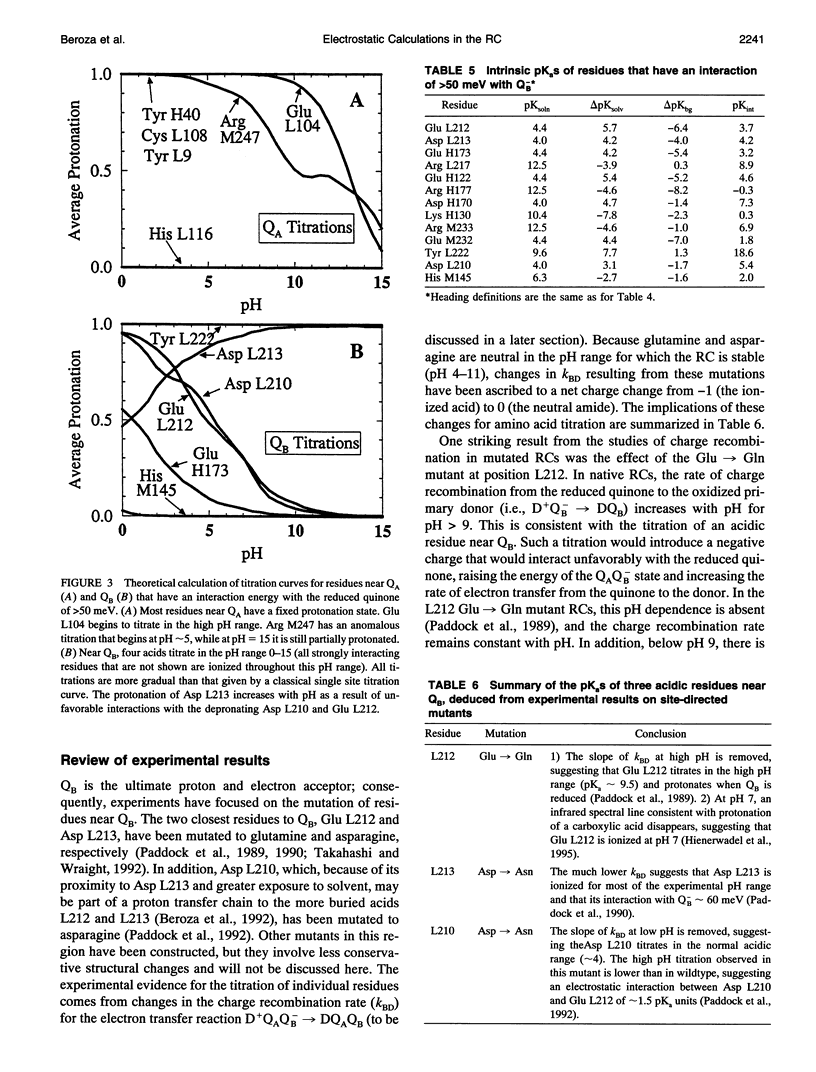
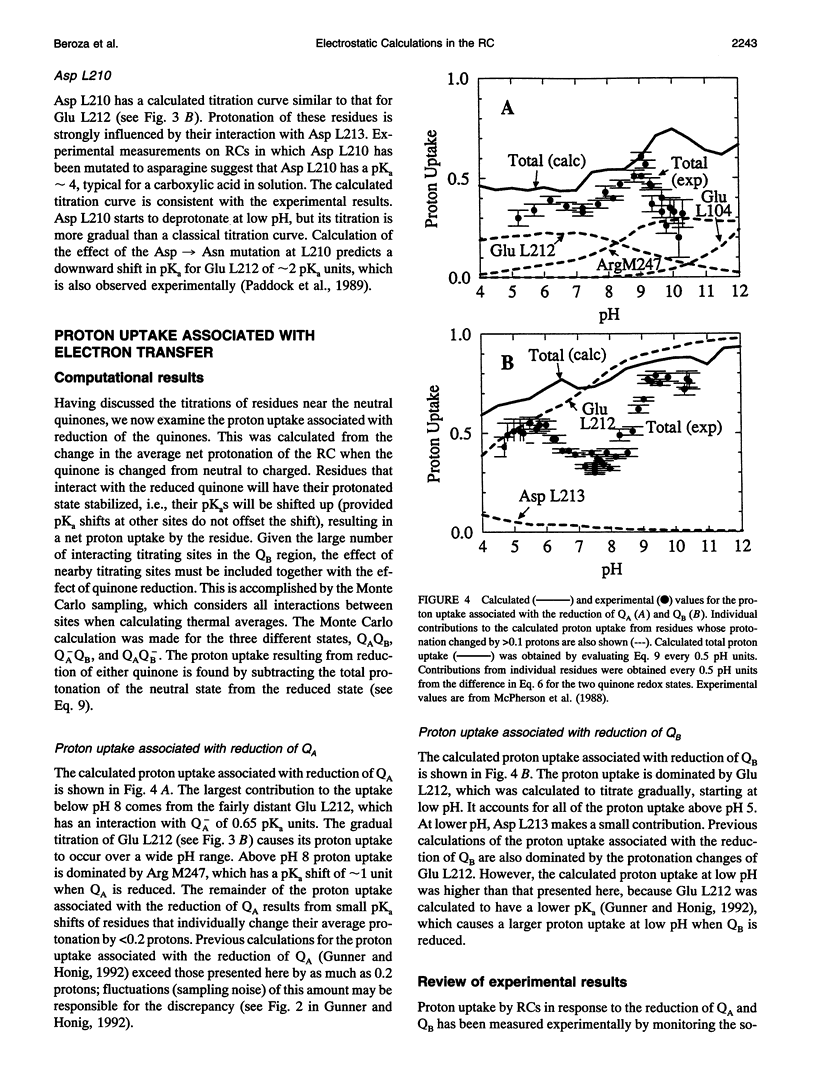
The titration of amino acids and the energetics of electron transfer from the primary electron acceptor (QA) to the secondary electron acceptor (QB) in the photosynthetic reaction center of Rhodobacter sphaeroides are calculated using a continuum electrostatic model. Strong electrostatic interactions between titrating sites give rise to complex titration curves. Glu L212 is calculated to have an anomalously broad titration curve, which explains the seemingly contradictory experimental results concerning its pKa. The electrostatic field following electron transfer shifts the average protonation of amino acids near the quinones. The pH dependence of the free energy between Q-AQB and QAQ-B calculated from these shifts is in good agreement with experiment. However, the calculated absolute free energy difference is in severe disagreement (by approximately 230 meV) with the observed experimental value, i.e., electron transfer from Q-A to QB is calculated to be unfavorable. The large stabilization energy of the Q-A state arises from the predominantly positively charged residues in the vicinity of QA in contrast to the predominantly negatively charged residues near QB. The discrepancy between calculated and experimental values for delta G(Q-AQB-->QAQ-B) points to limitations of the continuum electrostatic model. Inclusion of other contributions to the energetics (e.g., protein motion following quinone reduction) that may improve the agreement between theory and experiment are discussed.
Full text
PDF












Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the cofactors. Proc Natl Acad Sci U S A. 1987 Aug;84(16):5730–5734. doi: 10.1073/pnas.84.16.5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J. P., Feher G., Yeates T. O., Komiya H., Rees D. C. Structure of the reaction center from Rhodobacter sphaeroides R-26: the protein subunits. Proc Natl Acad Sci U S A. 1987 Sep;84(17):6162–6166. doi: 10.1073/pnas.84.17.6162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antosiewicz J., McCammon J. A., Gilson M. K. Prediction of pH-dependent properties of proteins. J Mol Biol. 1994 May 6;238(3):415–436. doi: 10.1006/jmbi.1994.1301. [DOI] [PubMed] [Google Scholar]
- Bashford D., Gerwert K. Electrostatic calculations of the pKa values of ionizable groups in bacteriorhodopsin. J Mol Biol. 1992 Mar 20;224(2):473–486. doi: 10.1016/0022-2836(92)91009-e. [DOI] [PubMed] [Google Scholar]
- Bashford D., Karplus M. pKa's of ionizable groups in proteins: atomic detail from a continuum electrostatic model. Biochemistry. 1990 Nov 6;29(44):10219–10225. doi: 10.1021/bi00496a010. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Beroza P., Fredkin D. R., Okamura M. Y., Feher G. Protonation of interacting residues in a protein by a Monte Carlo method: application to lysozyme and the photosynthetic reaction center of Rhodobacter sphaeroides. Proc Natl Acad Sci U S A. 1991 Jul 1;88(13):5804–5808. doi: 10.1073/pnas.88.13.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. H. Energetics of charge-charge interactions in proteins. Proteins. 1988;3(1):32–52. doi: 10.1002/prot.340030104. [DOI] [PubMed] [Google Scholar]
- Gilson M. K., Honig B. H. The dielectric constant of a folded protein. Biopolymers. 1986 Nov;25(11):2097–2119. doi: 10.1002/bip.360251106. [DOI] [PubMed] [Google Scholar]
- Gilson M. K. Multiple-site titration and molecular modeling: two rapid methods for computing energies and forces for ionizable groups in proteins. Proteins. 1993 Mar;15(3):266–282. doi: 10.1002/prot.340150305. [DOI] [PubMed] [Google Scholar]
- Hagler A. T., Huler E., Lifson S. Energy functions for peptides and proteins. I. Derivation of a consistent force field including the hydrogen bond from amide crystals. J Am Chem Soc. 1974 Aug 21;96(17):5319–5327. doi: 10.1021/ja00824a004. [DOI] [PubMed] [Google Scholar]
- Harvey S. C. Treatment of electrostatic effects in macromolecular modeling. Proteins. 1989;5(1):78–92. doi: 10.1002/prot.340050109. [DOI] [PubMed] [Google Scholar]
- Hienerwadel R., Grzybek S., Fogel C., Kreutz W., Okamura M. Y., Paddock M. L., Breton J., Nabedryk E., Mäntele W. Protonation of Glu L212 following QB- formation in the photosynthetic reaction center of Rhodobacter sphaeroides: evidence from time-resolved infrared spectroscopy. Biochemistry. 1995 Mar 7;34(9):2832–2843. doi: 10.1021/bi00009a013. [DOI] [PubMed] [Google Scholar]
- Kleinfeld D., Okamura M. Y., Feher G. Electron transfer in reaction centers of Rhodopseudomonas sphaeroides. I. Determination of the charge recombination pathway of D+QAQ(-)B and free energy and kinetic relations between Q(-)AQB and QAQ(-)B. Biochim Biophys Acta. 1984 Jul 27;766(1):126–140. doi: 10.1016/0005-2728(84)90224-x. [DOI] [PubMed] [Google Scholar]
- Oberoi H., Allewell N. M. Multigrid solution of the nonlinear Poisson-Boltzmann equation and calculation of titration curves. Biophys J. 1993 Jul;65(1):48–55. doi: 10.1016/S0006-3495(93)81032-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura M. Y., Feher G. Proton transfer in reaction centers from photosynthetic bacteria. Annu Rev Biochem. 1992;61:861–896. doi: 10.1146/annurev.bi.61.070192.004241. [DOI] [PubMed] [Google Scholar]
- Paddock M. L., Rongey S. H., Feher G., Okamura M. Y. Pathway of proton transfer in bacterial reaction centers: replacement of glutamic acid 212 in the L subunit by glutamine inhibits quinone (secondary acceptor) turnover. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6602–6606. doi: 10.1073/pnas.86.17.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards F. M. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 1977;6:151–176. doi: 10.1146/annurev.bb.06.060177.001055. [DOI] [PubMed] [Google Scholar]
- Sampogna R. V., Honig B. Environmental effects on the protonation states of active site residues in bacteriorhodopsin. Biophys J. 1994 May;66(5):1341–1352. doi: 10.1016/S0006-3495(94)80925-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp K. A., Honig B. Electrostatic interactions in macromolecules: theory and applications. Annu Rev Biophys Biophys Chem. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- Takahashi E., Wraight C. A. Proton and electron transfer in the acceptor quinone complex of Rhodobacter sphaeroides reaction centers: characterization of site-directed mutants of the two ionizable residues, GluL212 and AspL213, in the QB binding site. Biochemistry. 1992 Jan 28;31(3):855–866. doi: 10.1021/bi00118a031. [DOI] [PubMed] [Google Scholar]
- Tanford C., Roxby R. Interpretation of protein titration curves. Application to lysozyme. Biochemistry. 1972 May 23;11(11):2192–2198. doi: 10.1021/bi00761a029. [DOI] [PubMed] [Google Scholar]
- Treutlein H., Schulten K., Brünger A. T., Karplus M., Deisenhofer J., Michel H. Chromophore-protein interactions and the function of the photosynthetic reaction center: a molecular dynamics study. Proc Natl Acad Sci U S A. 1992 Jan 1;89(1):75–79. doi: 10.1073/pnas.89.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshel A., Russell S. T. Calculations of electrostatic interactions in biological systems and in solutions. Q Rev Biophys. 1984 Aug;17(3):283–422. doi: 10.1017/s0033583500005333. [DOI] [PubMed] [Google Scholar]
- Warwicker J., Watson H. C. Calculation of the electric potential in the active site cleft due to alpha-helix dipoles. J Mol Biol. 1982 Jun 5;157(4):671–679. doi: 10.1016/0022-2836(82)90505-8. [DOI] [PubMed] [Google Scholar]
- Yang A. S., Gunner M. R., Sampogna R., Sharp K., Honig B. On the calculation of pKas in proteins. Proteins. 1993 Mar;15(3):252–265. doi: 10.1002/prot.340150304. [DOI] [PubMed] [Google Scholar]


