Abstract
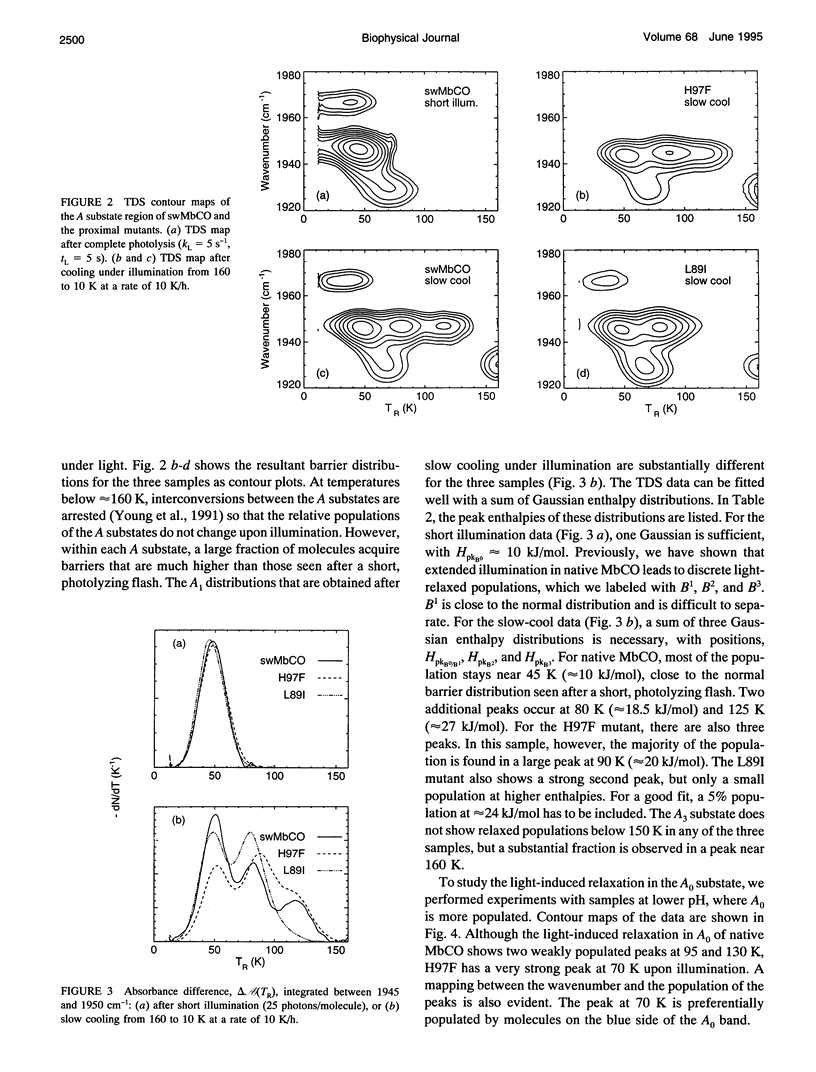
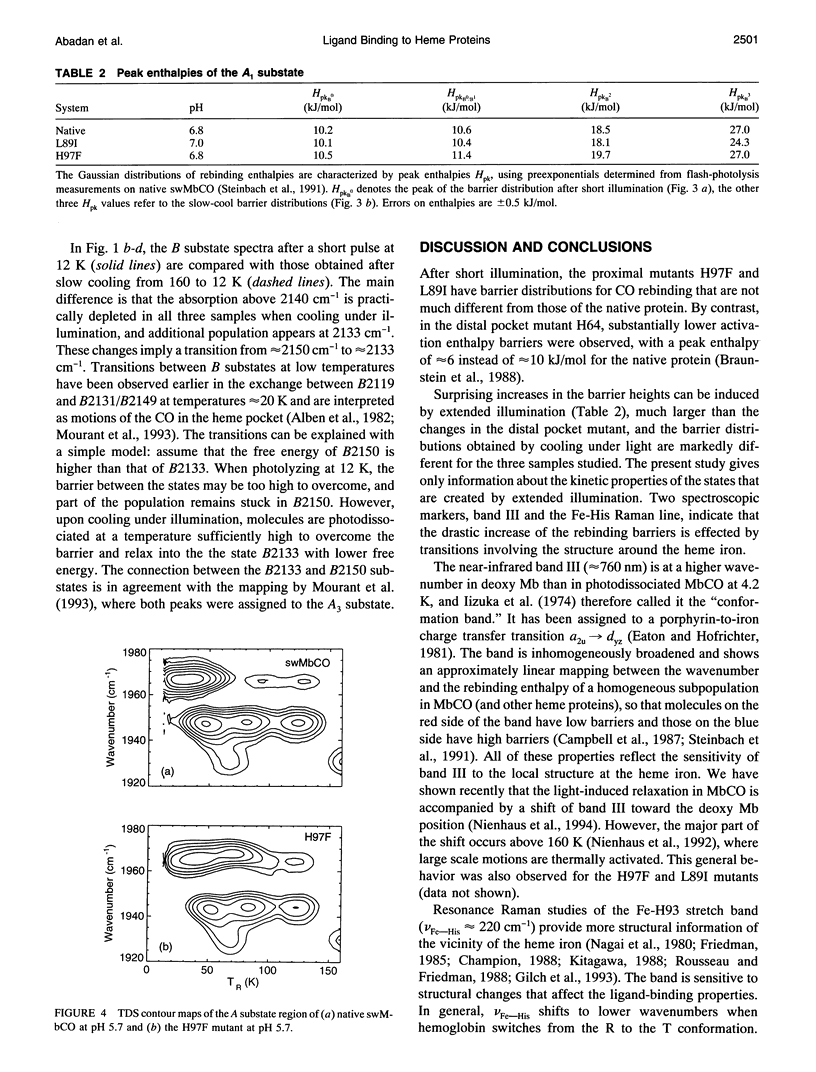
We have studied the proximal mutants L89I and H97F of MbCO with FTIR and temperature-derivative spectroscopy at temperatures between 10 and 160 K. The mutations give rise only to minor alterations of the stretch spectra of the bound and photodissociated CO ligand. The most pronounced difference is a larger population in the A3 substate at approximately 1930 cm-1 in the mutants. The barrier distributions, as determined by temperature-derivative spectroscopy, are very similar to native MbCO after short illumination. Extended illumination leads to substantial increases of the rebinding barriers in native MbCO and the proximal mutants. A larger fraction of light-relaxed states is found in the proximal mutants, implying that the conformational energy landscape has been modified to more easily allow light-induced transitions. These and other spectroscopic data imply that the large changes in the binding properties are brought about by a light-induced conformational relaxation involving the structure at the heme iron. Similarities with spectral hole-burning studies and physical models are discussed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adachi S., Nagano S., Ishimori K., Watanabe Y., Morishima I., Egawa T., Kitagawa T., Makino R. Roles of proximal ligand in heme proteins: replacement of proximal histidine of human myoglobin with cysteine and tyrosine by site-directed mutagenesis as models for P-450, chloroperoxidase, and catalase. Biochemistry. 1993 Jan 12;32(1):241–252. doi: 10.1021/bi00052a031. [DOI] [PubMed] [Google Scholar]
- Ansari A., Berendzen J., Braunstein D., Cowen B. R., Frauenfelder H., Hong M. K., Iben I. E., Johnson J. B., Ormos P., Sauke T. B. Rebinding and relaxation in the myoglobin pocket. Biophys Chem. 1987 May 9;26(2-3):337–355. doi: 10.1016/0301-4622(87)80034-0. [DOI] [PubMed] [Google Scholar]
- Ansari A., Jones C. M., Henry E. R., Hofrichter J., Eaton W. A. Conformational relaxation and ligand binding in myoglobin. Biochemistry. 1994 May 3;33(17):5128–5145. doi: 10.1021/bi00183a017. [DOI] [PubMed] [Google Scholar]
- Austin R. H., Beeson K. W., Eisenstein L., Frauenfelder H., Gunsalus I. C. Dynamics of ligand binding to myoglobin. Biochemistry. 1975 Dec 2;14(24):5355–5373. doi: 10.1021/bi00695a021. [DOI] [PubMed] [Google Scholar]
- Barrick D. Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93-->Gly. Biochemistry. 1994 May 31;33(21):6546–6554. doi: 10.1021/bi00187a023. [DOI] [PubMed] [Google Scholar]
- Berendzen J., Braunstein D. Temperature-derivative spectroscopy: a tool for protein dynamics. Proc Natl Acad Sci U S A. 1990 Jan;87(1):1–5. doi: 10.1073/pnas.87.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein D., Ansari A., Berendzen J., Cowen B. R., Egeberg K. D., Frauenfelder H., Hong M. K., Ormos P., Sauke T. B., Scholl R. Ligand binding to synthetic mutant myoglobin (His-E7----Gly): role of the distal histidine. Proc Natl Acad Sci U S A. 1988 Nov;85(22):8497–8501. doi: 10.1073/pnas.85.22.8497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell B. F., Chance M. R., Friedman J. M. Linkage of functional and structural heterogeneity in proteins: dynamic hole burning in carboxymyoglobin. Science. 1987 Oct 16;238(4825):373–376. doi: 10.1126/science.3659921. [DOI] [PubMed] [Google Scholar]
- Carver T. E., Olson J. S., Smerdon S. J., Krzywda S., Wilkinson A. J., Gibson Q. H., Blackmore R. S., Ropp J. D., Sligar S. G. Contributions of residue 45(CD3) and heme-6-propionate to the biomolecular and geminate recombination reactions of myoglobin. Biochemistry. 1991 May 14;30(19):4697–4705. doi: 10.1021/bi00233a009. [DOI] [PubMed] [Google Scholar]
- Cheng X. D., Schoenborn B. P. Neutron diffraction study of carbonmonoxymyoglobin. J Mol Biol. 1991 Jul 20;220(2):381–399. doi: 10.1016/0022-2836(91)90020-7. [DOI] [PubMed] [Google Scholar]
- Chu K, Ernst RM, Frauenfelder H, Mourant JR, Nienhaus GU, Philipp R. Light-induced and thermal relaxation in a protein. Phys Rev Lett. 1995 Mar 27;74(13):2607–2610. doi: 10.1103/PhysRevLett.74.2607. [DOI] [PubMed] [Google Scholar]
- Eaton W. A., Hofrichter J. Polarized absorption and linear dichroism spectroscopy of hemoglobin. Methods Enzymol. 1981;76:175–261. doi: 10.1016/0076-6879(81)76126-3. [DOI] [PubMed] [Google Scholar]
- Egeberg K. D., Springer B. A., Martinis S. A., Sligar S. G., Morikis D., Champion P. M. Alteration of sperm whale myoglobin heme axial ligation by site-directed mutagenesis. Biochemistry. 1990 Oct 23;29(42):9783–9791. doi: 10.1021/bi00494a004. [DOI] [PubMed] [Google Scholar]
- Egeberg K. D., Springer B. A., Sligar S. G., Carver T. E., Rohlfs R. J., Olson J. S. The role of Val68(E11) in ligand binding to sperm whale myoglobin. Site-directed mutagenesis of a synthetic gene. J Biol Chem. 1990 Jul 15;265(20):11788–11795. [PubMed] [Google Scholar]
- Friedman J. M., Campbell B. F., Noble R. W. A possible new control mechanism suggested by resonance Raman spectra from a deep ocean fish hemoglobin. Biophys Chem. 1990 Aug 31;37(1-3):43–59. doi: 10.1016/0301-4622(90)88006-e. [DOI] [PubMed] [Google Scholar]
- Friedman J. M. Structure, dynamics, and reactivity in hemoglobin. Science. 1985 Jun 14;228(4705):1273–1280. doi: 10.1126/science.4001941. [DOI] [PubMed] [Google Scholar]
- Gilch H., Schweitzer-Stenner R., Dreybrodt W. Structural heterogeneity of the Fe(2+)-N epsilon (HisF8) bond in various hemoglobin and myoglobin derivatives probed by the Raman-active iron histidine stretching mode. Biophys J. 1993 Oct;65(4):1470–1485. doi: 10.1016/S0006-3495(93)81216-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry E. R., Eaton W. A., Hochstrasser R. M. Molecular dynamics simulations of cooling in laser-excited heme proteins. Proc Natl Acad Sci U S A. 1986 Dec;83(23):8982–8986. doi: 10.1073/pnas.83.23.8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iben IE, Braunstein D, Doster W, Frauenfelder H, Hong MK, Johnson JB, Luck S, Ormos P, Schulte A, Steinbach PJ. Glassy behavior of a protein. Phys Rev Lett. 1989 Apr 17;62(16):1916–1919. doi: 10.1103/PhysRevLett.62.1916. [DOI] [PubMed] [Google Scholar]
- Lambright D. G., Balasubramanian S., Decatur S. M., Boxer S. G. Anatomy and dynamics of a ligand-binding pathway in myoglobin: the roles of residues 45, 60, 64, and 68. Biochemistry. 1994 May 10;33(18):5518–5525. doi: 10.1021/bi00184a021. [DOI] [PubMed] [Google Scholar]
- Li P., Champion P. M. Investigations of the thermal response of laser-excited biomolecules. Biophys J. 1994 Feb;66(2 Pt 1):430–436. doi: 10.1016/s0006-3495(94)80793-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews A. J., Rohlfs R. J., Olson J. S., Tame J., Renaud J. P., Nagai K. The effects of E7 and E11 mutations on the kinetics of ligand binding to R state human hemoglobin. J Biol Chem. 1989 Oct 5;264(28):16573–16583. [PubMed] [Google Scholar]
- Maxwell J. C., Caughey W. S. An infrared study of NO bonding to heme B and hemoglobin A. Evidence for inositol hexaphosphate induced cleavage of proximal histidine to iron bonds. Biochemistry. 1976 Jan 27;15(2):388–396. doi: 10.1021/bi00647a023. [DOI] [PubMed] [Google Scholar]
- Mourant J. R., Braunstein D. P., Chu K., Frauenfelder H., Nienhaus G. U., Ormos P., Young R. D. Ligand binding to heme proteins: II. Transitions in the heme pocket of myoglobin. Biophys J. 1993 Oct;65(4):1496–1507. doi: 10.1016/S0006-3495(93)81218-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K., Kitagawa T., Morimoto H. Quaternary structures and low frequency molecular vibrations of haems of deoxy and oxyhaemoglobin studied by resonance raman scattering. J Mol Biol. 1980 Jan 25;136(3):271–289. doi: 10.1016/0022-2836(80)90374-5. [DOI] [PubMed] [Google Scholar]
- Nagai K., Luisi B., Shih D., Miyazaki G., Imai K., Poyart C., De Young A., Kwiatkowsky L., Noble R. W., Lin S. H. Distal residues in the oxygen binding site of haemoglobin studied by protein engineering. 1987 Oct 29-Nov 4Nature. 329(6142):858–860. doi: 10.1038/329858a0. [DOI] [PubMed] [Google Scholar]
- Nienhaus G. U., Mourant J. R., Chu K., Frauenfelder H. Ligand binding to heme proteins: the effect of light on ligand binding in myoglobin. Biochemistry. 1994 Nov 15;33(45):13413–13430. doi: 10.1021/bi00249a030. [DOI] [PubMed] [Google Scholar]
- Nienhaus G. U., Mourant J. R., Frauenfelder H. Spectroscopic evidence for conformational relaxation in myoglobin. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2902–2906. doi: 10.1073/pnas.89.7.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson J. S., Mathews A. J., Rohlfs R. J., Springer B. A., Egeberg K. D., Sligar S. G., Tame J., Renaud J. P., Nagai K. The role of the distal histidine in myoglobin and haemoglobin. Nature. 1988 Nov 17;336(6196):265–266. doi: 10.1038/336265a0. [DOI] [PubMed] [Google Scholar]
- Park K. D., Guo K. M., Adebodun F., Chiu M. L., Sligar S. G., Oldfield E. Distal and proximal ligand interactions in heme proteins: correlations between C-O and Fe-C vibrational frequencies, oxygen-17 and carbon-13 nuclear magnetic resonance chemical shifts, and oxygen-17 nuclear quadrupole coupling constants in C17O- and 13CO-labeled species. Biochemistry. 1991 Mar 5;30(9):2333–2347. doi: 10.1021/bi00223a007. [DOI] [PubMed] [Google Scholar]
- Phillips G. N., Jr, Arduini R. M., Springer B. A., Sligar S. G. Crystal structure of myoglobin from a synthetic gene. Proteins. 1990;7(4):358–365. doi: 10.1002/prot.340070407. [DOI] [PubMed] [Google Scholar]
- Post F., Doster W., Karvounis G., Settles M. Structural relaxation and nonexponential kinetics of CO-binding to horse myoglobin. Multiple flash photolysis experiments. Biophys J. 1993 Jun;64(6):1833–1842. doi: 10.1016/S0006-3495(93)81554-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers L., Chance B., Chance M., Campbell B., Friedman J., Khalid S., Kumar C., Naqui A., Reddy K. S., Zhou Y. Kinetic, structural, and spectroscopic identification of geminate states of myoglobin: a ligand binding site on the reaction pathway. Biochemistry. 1987 Jul 28;26(15):4785–4796. doi: 10.1021/bi00389a028. [DOI] [PubMed] [Google Scholar]
- Schlichting I., Berendzen J., Phillips G. N., Jr, Sweet R. M. Crystal structure of photolysed carbonmonoxy-myoglobin. Nature. 1994 Oct 27;371(6500):808–812. doi: 10.1038/371808a0. [DOI] [PubMed] [Google Scholar]
- Springer B. A., Egeberg K. D., Sligar S. G., Rohlfs R. J., Mathews A. J., Olson J. S. Discrimination between oxygen and carbon monoxide and inhibition of autooxidation by myoglobin. Site-directed mutagenesis of the distal histidine. J Biol Chem. 1989 Feb 25;264(6):3057–3060. [PubMed] [Google Scholar]
- Springer B. A., Sligar S. G. High-level expression of sperm whale myoglobin in Escherichia coli. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8961–8965. doi: 10.1073/pnas.84.24.8961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srajer V., Reinisch L., Champion P. M. Investigation of laser-induced long-lived states of photolyzed MbCO. Biochemistry. 1991 May 21;30(20):4886–4895. doi: 10.1021/bi00234a008. [DOI] [PubMed] [Google Scholar]
- Steinbach P. J., Ansari A., Berendzen J., Braunstein D., Chu K., Cowen B. R., Ehrenstein D., Frauenfelder H., Johnson J. B., Lamb D. C. Ligand binding to heme proteins: connection between dynamics and function. Biochemistry. 1991 Apr 23;30(16):3988–4001. doi: 10.1021/bi00230a026. [DOI] [PubMed] [Google Scholar]
- Zollfrank J., Friedrich J., Parak F. Spectral hole burning study of protoporphyrin IX substituted myoglobin. Biophys J. 1992 Mar;61(3):716–724. doi: 10.1016/S0006-3495(92)81876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]


