Abstract
Holoprosencephaly (HPE), a human developmental brain defect, usually is also associated with varying degrees of midline facial dysmorphism. Heterozygous mutations in the Sonic hedgehog (SHH) gene are the most common genetic lesions associated with HPE, and loss of Shh function in the mouse produces cyclopia and alobar forebrain development. The N-terminal domain (ShhNp) of Sonic hedgehog protein, generated by cholesterol-dependent autoprocessing and modification at the C terminus and by palmitate addition at the N terminus, is the active ligand in the Shh signal transduction pathway. Here, we analyze seven reported missense mutations (G31R, D88V, Q100H, N115K, W117G, W117R, and E188Q) that alter the N-terminal signaling domain of Shh protein, and show that two of these mutations (Q100H and E188Q), which are questionably linked to HPE, produce no detectable effects on function. The remaining five alterations affect normal processing, Ptc binding, and signaling to varying degrees. These effects include introduction of a recognition site for furin-like proteases by the G31R alteration, resulting in cleavage of 11 amino acid residues from the N terminus of ShhNp and consequent reduced signaling potency. Two other alterations, W117G and W117R, cause temperature-dependent misfolding and retention in the sterol-poor endoplasmic reticulum, thus disrupting cholesterol-dependent autoprocessing.
Keywords: autoprocessing, development, protein misfolding, endoplasmic reticulum retention
Holoprosencephaly (HPE) occurs with a prevalence of ≈1 in 250 during early embryogenesis, decreasing throughout gestation to a frequency of ≈1 in 16,000 live births (1). In its most severe form, HPE is associated with alobar brain development, cyclopia, and other facial abnormalities. However, the disease is highly heterogeneous and can also be associated with milder brain malformations and midfacial dysmorphisms, as well as abnormalities such as microcephaly, mental retardation, and epilepsy.
A condition similar to severe human HPE is associated with homozygous mutation of the Sonic hedgehog (Shh) gene in the mouse (2). The Shh member of the Hedgehog protein family plays critical roles in the patterning of the developing neural tube, the limbs, the axial skeleton, and other derivatives of the somites and in induction of ventral forebrain structures (2, 3). This latter activity is relevant to HPE, and homozygous mutation of the mouse Shh gene provided the first association of cyclopia with a single gene mutation. About a dozen human chromosomal loci are associated with HPE (1), and mutations in HPE3 affect the human Sonic hedgehog (SHH) gene (4, 5). Human gene function appears to be haploinsufficient, but the HPE phenotype associated with heterozygous mutations is less severe and the cosegregation of phenotype with genotype in some cases is unclear. In addition, more recent studies have begun to implicate environmental influences (6) and cosegregating mutations at other loci (7) in the etiology of HPE.
To clarify the effects of SHH mutations in human HPE, we have undertaken a molecular analysis of seven reported HPE missense mutations that affect specific amino acid residues within the mature SHH protein signal. The Shh protein is synthesized as a ≈45-kDa precursor that undergoes an autocatalytic processing event that produces a ≈19-kDa N-terminal product, responsible for all signaling activities, and a ≈25-kDa C-terminal fragment (8). The N-terminal product triggers Hh pathway activation by binding to Patched (Ptc), the primary receptor for Hh signaling, which suppresses pathway activity in the absence of Hh stimulation (9). The role of the Hh C-terminal domain is to mediate autoprocessing of the precursor, leading to covalent modification of the N-terminal fragment by cholesterol (10). A second lipid modification subsequently results from attachment of palmitate at the N terminus (11) in a reaction requiring activity of a gene encoding an apparent acyltransferase, skinny hedgehog (12). This N-terminal modification is required for many in vivo activities of the Sonic hedgehog protein (reviewed in ref. 8), although neural plate explants appear to respond well in vitro in the absence of this modification (13, 14).
Some HPE mutations truncate protein synthesis, thus directly disrupting production of the N-terminal signaling domain or disrupting autoprocessing, which requires the C-terminal domain. All seven of the HPE mutations selected for this study alter individual residues within the N-terminal signaling domain, and our expectation was that they might affect the signaling potency of the processed N-terminal signaling domain. Although some of these mutations indeed appear to affect receptor affinity and signaling potency, we also found that N-terminal domain mutations can affect processing, and one such mutation creates a processing site for a furin-like protease. Two of the SHH mutations produced no detectable effect on processing, receptor binding, or signaling.
Materials and Methods
Ptch-Binding and Neural Plate Signaling Assays of Recombinant ShhN Variants. Alterations corresponding to human HPE missense mutations were introduced into a construct for Escherichia coli expression of mouse ShhN and purified as described (15). Human and mouse ShhN amino acid sequences are identical except for a Thr in mouse and Ser in human at position 67 in the human sequence (Fig. 1). The numbering of corresponding human and mouse residues differs by one because of differences in the human and mouse signal sequences (Fig. 1): here, the human numbering is used throughout for consistency. 32P-labeled ShhN protein was prepared essentially as described, by using the catalytic subunit of protein kinase A (Sigma), [γ-32P]ATP, and recombinant ShhN protein containing a protein kinase A site (RRASV) at the C terminus (15).
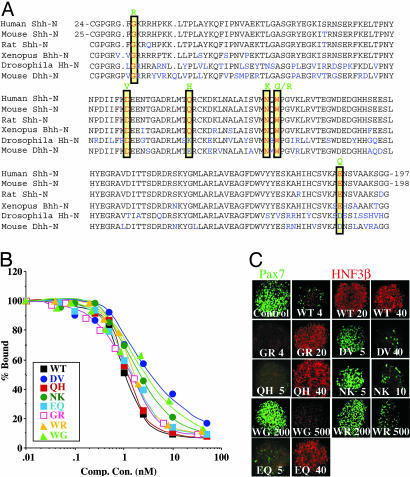
Fig. 1.
Ptch-binding and signaling activities of HPE mutant ShhN proteins. (A) Sequences of human ShhN, mouse ShhN, rat ShhN, Xenopus Banded hedgehog (Bhh), Drosophila Hh, and mouse Desert hedgehog (Dhh) are aligned with nonidentical residues in blue. Human and mouse sequences are identical except for a Ser in place of Thr at position 67 in the human sequence; the numbering of corresponding human and mouse residues differs by one because of different signal sequence lengths. Residues altered in human HPE are boxed in yellow, and the altered residues are shown above in green. (B) Binding of altered ShhN mutant proteins to the Ptch receptor. Binding of 32P-ShhN to EcR-293 cells stably expressing Ptch-CTD (15) was measured in the presence of unlabelled recombinant proteins; binding was normalized to the amount (100%) bound in absence of any competitor. (C) Signaling activities of altered ShhN proteins in intermediate neural plate explants from chick embryos. After incubation in presence of recombinant proteins (concentrations in nM are indicated by the numbers in each panel), explants were double stained with antibodies against Pax-7 (green) and the floor plate marker HNF3-β (red).
Assays of 32P-ShhN binding to cells expressing Ptc-CTD with competition by ShhN variants were essentially as described (15).
ShhN signaling assays in neural plate explants were as described (15).
Expression of Shh Variants in Mammalian Cells and Glycosidase Treatment. For expression of Shh protein variants containing HPE mutations, mouse Shh constructs containing the mutations were subcloned into the pRK5 expression vector. Recombinant plasmids were then transiently transfected into HEK-293 cells by using FuGENE 6 (Roche Diagnostics). Two days later, conditioned medium was collected and cells were washed with cold PBS. Cells were lysed in RIPA buffer (150 mM NaCl/50 mM Tris·HCl, pH 8.0/1% Nonidet P-40/0.5% Deoxycholate/0.1% SDS) containing 0.5 mM PMSF, 2 μg/ml pepstatin A, and 10 μg/ml leupeptin. After lysis, cell lysates and conditioned media were cleared by centrifugation and separated in SDS/15% PAGE and immunoblotted, with detection using anti-ShhN or anti ShhC antibodies (15).
For glycosidase treatment, HEK-293 cells grown and transiently transfected in six-well plates were lysed in 125 μl of RIPA lysis buffer. Conditioned medium was obtained by replacing FBS with Opti-MEM (Invitrogen) and N2 supplement 1 day after transfection. After growth for another 30 h, conditioned medium was collected and cleared at 2,000 rpm (400 × g) in an Eppendorf centrifuge. For glycosidase treatment, 20 μl of cell extracts or conditioned medium was mixed with 2.2 μl of 10× denaturing buffer (NEB) and incubated at 100°C for 10 min. A total of 2.5 μl of Endo H or PNGase F reaction buffer (10×) and 20 units of Endo H or PNGase F were added, and the reaction mixture was incubated at 37°C for 1 h before addition of 12.5 μl of 3× gel loading buffer. Treated proteins were subjected to SDS/10% or 15% PAGE, and immunoblotted with either anti-ShhN or anti-ShhC antibodies.
For stable expression of Shh protein containing the GR mutation in EcR-293 cells (Invitrogen), a Shh construct containing the mutation subcloned into pIND vector (Invitrogen) (pIND-GR-Shh) was transfected into EcR-293 cells by using FuGENE 6 (Roche Diagnostics). Stable clones were generated and amplified in a medium containing DMEM, 10% FBS, 1% penicillin/streptomycin, Zeocin (360 μg/ml), and G418 (400 μg/ml) as selection antibiotic, and generation of processed N-terminal domain (GR-ShhNp) was measured by immunoblotting of the cell-free extract using anti-ShhN antibodies.
Shh Signaling Assay in C3H10T1/2 Cells. Induction of alkaline phosphatase in C3H10T1/2 cells (American Type Culture Collection) by ShhN signaling was measured by growing cells to full confluency in 24-well plates in growth medium containing DMEM, 10% FBS, 2% penicillin/streptomycin, 100 μM 2-mercaptoethanol, and 0.5 μg/ml ZnSO4. Growth medium was then replaced with medium in which 10% FBS was substituted with N2 supplement (Invitrogen). WT or mutant ShhN was added, and after 4-5 days of further incubation at 37°C, cells were washed in cold PBS and lysed in a buffer containing 10 mM dimethylethanol amine and 1 mM MgCl2 at 37°C for 1 h. The lysate was then mixed with chromogenic substrate p-nitrophenyl phosphate (Sigma) dissolved in lysis buffer and incubated at room temperature for 15 min, and absorbance at 405 nm was measured.
Purification and Sequencing of ShhN*. For purification and sequencing of ShhN*, a shorter form generated from GR-Shh in vivo, conditioned medium from HEK-293 cells transfected with GR-ShhN-Myc construct was passed through heparin agarose, which retains GR-ShhN (minor amount) but not ShhN* (the major form). The flow-through containing ShhN* was then purified by using monoclonal antibody 5E1 linked to Affigel essentially as described (16). A Coomassie-stained band corresponding to ShhN* (16-17 kDa) was excised from SDS/15% PAGE and processed for N-terminal sequence analysis by Edman degradation.
Processing of GR-Shh Mutant Protein in Vivo in Presence of Furin Inhibitor. EcR-293 cells stably integrating Shh or GR-Shh full-length constructs were grown to confluency in growth medium containing DMEM, 10% FBS, 1% penicillin/streptomycin, Zeocin (360 μg/ml), and G418 (400 μg/ml). The medium was then changed to DMEM, 1% penicillin/streptomycin, and 1% N2 supplement-containing medium. 2.5 μM of ponasterone A (to induce expression of Shh protein) and varying concentrations of Furin inhibitor I (Calbiochem) were then added to the above medium. The cells were then grown for 2 days at 37°C. Conditioned media was collected, and cells were lysed in 100 μl of RIPA lysis buffer. Lysates (20 μl) were mixed with 10 μl of 3× gel loading buffer and run in SDS/15% PAGE, followed by immunoblotting with anti-ShhN antibodies.
Results
Patched Binding of Altered ShhN Proteins. To determine how human HPE mutations might affect SHH function, sequence alterations corresponding to the seven human missense mutations associated with HPE (G31R, D88V, Q100H, N115K, W117G, W117R, and E188Q; refs. 4, 5, and 17) were introduced into an expression construct for the N-terminal fragment (ShhN) of mouse Sonic hedgehog protein (Fig. 1 A). Human and mouse ShhN amino acid sequences are identical except for a Thr in mouse and Ser in human at position 67 in the human sequence. The altered proteins were expressed in E. coli and purified to homogeneity to test their affinity for binding to the primary Hh receptor, Ptc. The assay used cells expressing mPtc and 32P-labeled ShhN as ligand, with the altered recombinant proteins as unlabelled competitors (see Materials and Methods) (15). The Ptc-binding affinities measured for the altered proteins in these competition assays (Fig. 1B and Table 1) were substantially similar to WT for most of the proteins, differing by ≈3-fold in the most affected protein (D88V).
Table 1. Properties of variant ShhN proteins.
| Protein | Mutation (human) | Autoprocessing in vivo | Pax-7 repression, nM* | HNF3-β induction, nM* | Dissociation constant (KD) for Ptc-CTD, nM | Relative activity at 50 nM in C3H10T1/2 assay |
|---|---|---|---|---|---|---|
| WT | ShhN (aa 24–197) | + | ≈4 | ≈20 | 0.48 | 1.0 |
| GR | G31R | + | ≈4 | ≈20 | 0.8 | 1.7 |
| DV | D88V | +/– | 40–50 | ≈80 | 1.58 | 0.07 |
| QH | Q100H | + | ≈5 | <40 | 0.5 | 1.4 |
| NK | N115K | + | >10 | ND | 0.86 | 0.2 |
| WG | W117G | – | 400–500 | ND | 1.5 | 0.03 |
| WR | W117R | – | >500 | ND | 0.7 | 0.13 |
| EQ | E188Q | + | ≈5 | <40 | 0.62 | 1.9 |
| N-11 | (aa 35–197) | ND | ≈9 | 45–90 | 1.1 | 0.06 |
ND, not determined, aa, amino acids
Minimum concentration required
Neural Plate Signaling of Altered ShhN Proteins. The similar Ptc-binding affinity of purified recombinant WT and altered proteins prompted us to examine their signaling potency in neural plate explants. Various concentrations of each mutant protein were applied to cultures of intermediate neural plate explants from chicken embryos, and suppression of dorsal marker Pax-7 (green) and induction of floor plate marker HNF3-β (red) were monitored (see Materials and Methods and ref. 15). WT ShhN protein applied to neural plate explant culture repressed Pax-7 expression almost completely at 4 nM, and at higher concentrations (20 and 40 nM) was able to induce expression of HNF3-β in all of the cells of the explants. The G31R, Q100H, and E188Q altered proteins also repressed Pax-7 expression at 4 nM, and induced HNF3-β expression at 20 nM protein concentrations (Fig. 1C and Table 1). The D88V and N115K proteins required 2- to 3-fold higher concentrations for suppression of Pax-7 (Fig. 1C and Table 1). However, the most striking change in signaling potency was observed for the W117G and W117R proteins, which failed to suppress Pax-7 at concentrations a 100-fold higher than WT and appeared incapable of inducing HNF3-β expression.
W117G and W117R Proteins Are Temperature Sensitive. The extreme loss of signaling potency for W117G and W117R proteins was surprising in light of their relatively modest reductions in Ptc-binding affinities (15). As signaling potencies were tested at 37°C versus 4°C for Ptc-binding, we tested for temperature-dependent effects on signaling potencies by culturing neural plate explants at 32°C. Survival of the explants and suppression of Pax-7 expression by WT ShhN at 4 nM (Fig. 2A) were not compromised at this temperature; suppression of Pax-7 expression by both W117G and W117R was significantly enhanced, and these altered proteins also were able to induce HNF3-β at 200 and 400 nM concentrations (Fig. 2 A).
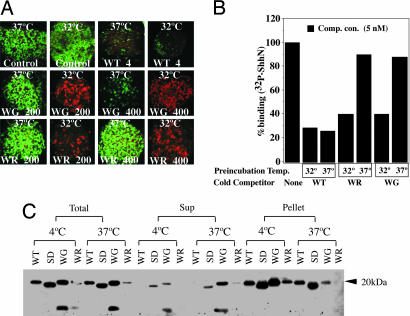
Fig. 2.
Temperature dependence of WG and WR protein activities. (A) Neural plate signaling of WG and WR variants. Neural plate explants were incubated at 37°C and 32°C at the indicated concentrations of ShhN (in nM) and stained to monitor expression of Pax-7 (green; control panels) and HNF3β (red). Stimulation with WT protein (4 nM) suppressed Pax-7 expression at both temperatures, whereas variant proteins at 37°C failed to suppress Pax-7 at 200 nM protein concentrations (WG and WR) or at 400 nM protein concentrations (WR). However, at 32°C, WG and WR proteins at 200 and 400 nM suppressed Pax-7 (data not shown) and induced HNF3-b expression (red). (B) Ptch-binding of WG and WR variants. Proteins were preincubated at 37°C or 32°C for 1 h, then added (5 nM) to EcR-293 cells stably expressing Ptc-CTD in the presence of 32P-ShhN. WG and WR variants failed to compete with 32P-ShhN when preincubated at 37°C, but showed some activity when preincubated at 32°C. (C) Immunoprecipitation of variant proteins by 5E1 monoclonal antibody. WT ShhN and surface D (SD) (15), WG, and WR variants were all immunoprecipitated by 5E1-monoclonal antibody when preincubated at 4°C (bulk of proteins found in pellet, not supernatant), but after preincubation at 37°C, WG and WR were predominantly found in the supernatant. Proteins were detected by immunoblotting with anti-ShhN polyclonal antibodies.
We further tested whether the temperature sensitivity of these proteins in signaling was due to Ptc-binding affinity by preincubating WT, W117G, and W117R proteins at 32°C and 37°C for 1 h (Fig. 2B), and then by using them as competitors for 32P-ShhN binding to Ptch. We observed equivalent inhibition of 32P-ShhN binding by 5 nM WT ShhN at both temperatures. Preincubation with the W117G and W117R proteins also inhibited 32P-ShhN binding at 32°C, albeit to a lesser extent than WT. However, at 37°C, neither W117G nor W117R proteins was able to compete for binding of 32P-ShhN.
To further characterize the basis of this temperature-sensitive Ptc-binding behavior, we used the 5E1 monoclonal antibody, which recognizes a discontinuous epitope that coincides with the Ptc binding surface of the ShhN protein (15). We found that 5E1 immunoprecipitated most or all of ShhN at 4°C and 37°C. In contrast, although both W117G and W117R proteins were immunoprecipitated at 4°C, at 37°C the great bulk of these proteins was not immunoprecipitated, indicating a dramatic reduction in binding at the higher temperature. Because W117 is not within the surface recognized by 5E1, we surmise that the W117G and W117R mutations induce a temperature-dependent conformational change that indirectly affects the Ptc-binding surface.
We also tested the D88V and N115K proteins for temperature sensitivity in a Ptc-binding competition assay, and these proteins showed no change in activities at different temperatures (data not shown).
Processing Defects of Altered HPE-Associated Sonic Hedgehog Proteins. Having found no apparent defect in Ptc-binding or neural plate signaling for some of the bacterially produced proteins, we examined the biogenesis of these proteins by using electrophoretic mobility of glycosidase-treated proteins to monitor autoprocessing and glycosylation. Constructs for expression of altered Shh proteins were transiently transfected into HEK 293 cells, and cell extracts and conditioned medium were immunoblotted by using antibodies against the N- and C-terminal domains of the Shh proteins. The bulk of the overexpressed protein under these conditions is processed to yield ShhNp and ShhC, although a fraction remains unprocessed and is associated with the cell extract (Fig. 3 A and B). This residual unprocessed precursor is sensitive to digestion with EndoH (Fig. 3C), indicating the presence of immature glycosyl adducts and suggesting retention within the endoplasmic reticulum (ER). The autoprocessing reaction appears to occur after initial glycosylation but before Golgi-mediated maturation of the glycosyl chains, as indicated by EndoH sensitivity of cell-associated ShhC (Fig. 3D). All ShhC in the medium is sensitive to PNGaseF, but not EndoH (Fig. 3E), indicative of glycosyl adduct maturation in Golgi before secretion from the cell.
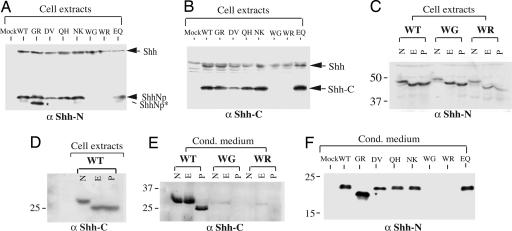
Fig. 3.
Effect of HPE mutations on Shh protein processing. (A and B) Western blot analysis of cell extracts from HEK-293 cells transiently transfected with full-length WT and variant Shh constructs. Anti-ShhN polyclonal antibodies detected the full-length Shh protein (45 kDa, indicated by arrow), the processed and lipid-modified N-terminal fragment (ShhNp, indicated by arrowhead), and an abnormal processed N-terminal fragment (ShhNp*, indicated by star) generated from the GR variant (A), whereas anti-ShhC antibodies detected full-length Shh and the ShhC fragment (B). The WG and WR variants produced neither ShhNp nor ShhC fragments. (C-E) Glycosidase digestion. Extracts (C and D) or conditioned medium (E) from HEK-293 cells transfected with Shh constructs were immunoblotted after denaturation and no further treatment (N) or treatment with Endo H (E) or PNGase F (P). Proteins were separated in 10% (C) or 15% (D and E) SDS/PAGE and immunoblotted with anti-ShhN (C) or anti-ShhC (D and E) antibodies. (F) Conditioned medium from transfected HEK-293 cells was immunoblotted and analyzed with anti-ShhN antibodies. Note the greater abundance of the abnormally processed ShhN GR variant in conditioned medium.
No apparent autoprocessing defect is caused by D88V, Q100H, N115K, and E188Q alterations, as precursors of these proteins are processed normally to give rise to ShhNp and ShhC (Fig. 3 A and B). In contrast, the W117G and W117R alterations produce no detectable levels of ShhNp and ShhC either within cells or within medium, (Fig. 3 A, B, E, and F), despite the presence of an intact C-terminal processing domain. The precursor of these proteins remains entirely within cells and with an immature glycosylation pattern, suggestive of retention within the ER.
One additional alteration, G31R, produced a mixture of ShhNp of apparently normal mobility, and a second form (ShhNp*) that migrated faster than WT ShhNp (Fig. 3A). Furthermore, although the majority of ShhNp is retained by producing cells (ref. 13 and data not shown), the majority of the ShhNp* protein (apparent molecular mass, 16-17 kDa) was released into the medium (compare Fig. 3 F with A).
Proteolysis of 11 N-Terminal Residues in the G31R-ShhN Protein. The additional processing reaction in the G31R-Shh protein (Fig. 3 A and F) appears not to depend on autoprocessing, because expression of a G31R-ShhN-Myc N-terminal domain protein like full-length G31R-Shh, still produced two forms of N-terminal protein, in contrast to the single N-terminal form produced by WT-Shh or WT-ShhN (Fig. 4A). The alteration produced by this additional processing event appears to occur at the N terminus, as both forms of the G31R-ShhN-Myc altered protein retained the C-terminal Myc epitope (data not shown).
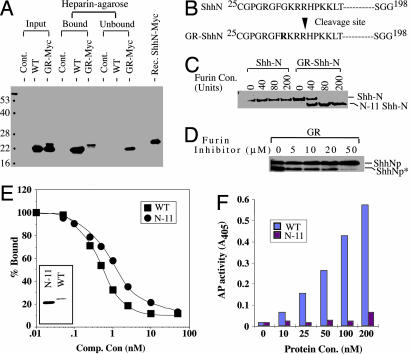
Fig. 4.
Abnormal processing and functional characterization of ShhN from the GR variant. (A) Heparin-agarose binding. Conditioned media from HEK-293 cells transfected with ShhN or GR-ShhN-Myc were separated with heparin-agarose and immunoblotted with anti-ShhN polyclonal antibodies. The C-terminal Myc epitope can be detected in both the slower- and faster-migrating forms of GR-ShhN-Myc (data not shown), indicating that the abnormal truncation occurs at the N terminus. WT ShhN and the longer form of GR-ShhN-Myc are both retained by heparin agarose, but the truncated form of the GR variant is not. Note the comigration of slower-migrating GR-ShhN-Myc and recombinant ShhN-Myc protein purified from E. coli.(B) Determination of cleavage site. The abnormally truncated ShhN*-Myc protein was purified by lack of binding to heparin-agarose and with the monoclonal antibody 5E1 and subjected to N-terminal sequence analysis by Edman degradation. This sequence indicated that cleavage occurred between Arg (35) and His (36) residues. (C and D) Furin cleavage of recombinant GR-ShhN protein in vitro and in vivo. (C) Purified, recombinant ShhN (1.5 μg) and GR-ShhN (2.0 μg) proteins treated with 0, 40, 80, and 200 units of furin protease (Sigma) were analyzed by SDS/PAGE and Coomassie blue staining. (D) EcR-293 cells with stably integrated construct for expression of GR-Shh full-length protein were induced with ponasterone in the presence of 0, 5, 10, 20, and 50 μM of furin inhibitor I, and cell extracts were analyzed by immunoblotting with anti-ShhN antibodies. (E and F) Ptc-binding and Shh signaling activities of recombinant ShhN*. (E) Recombinant WT ShhN and protein lacking 11 N-terminal residues (N-11) proteins were purified (Inset) and used as competitor for 32P-ShhN binding to EcR-293 cells expressing Ptc-CTD. (F) Signaling activities of ShhN WT and N-11 in C3H10T1/2 cells.
To further analyze potential N-terminal differences between these two forms of G31R-ShhN protein, we separated them by using heparin agarose beads, which bind to the slower, but not the faster, migrating form, consistent with the binding of WT ShhN protein to heparin through its N terminus (Fig. 4A) (15, 18). These two forms were further immunoaffinity purified with 5E1 monoclonal antibody immobilized on Affigel, and bound proteins were eluted with low pH buffer (16). Edman degradation of the lower mass protein revealed the N-terminal residues to be H-P-K-K-L-T. This form, ShhN*, thus appears to have been generated by cleavage of the G31R-ShhN mutant protein between Arg-35 and His-36 amino acid residues, thus removing 11 amino acid residues (Fig. 4B).
Because the G31R mutation adds an additional basic amino acid residue adjacent to three preexisting basic residues, this sequence alteration may create a cleavage site for furin or a furin-like protease. We found that furin (Sigma) was indeed able to cleave G31R-ShhN but not WT protein in vitro (Fig. 4C). We further noted that treatment of cells expressing G31R-ShhN protein with furin inhibitor 1 (Calbiochem) reduced the relative level of ShhN* (Fig. 4D), indicating that a furin-like activity is responsible for this cleavage in vivo.
Reduced Activity of ShhN Protein Lacking 11 N-Terminal Residues. To determine whether loss of 11 N-terminal residues affects Ptc-binding and signaling potency, we prepared ShhN* recombinant protein lacking 11 N-terminal residues (N-11) as compared to the WT protein (Fig. 4E Inset). Competitive Ptc-binding analysis in presence of N-11 protein indicated that affinity of N-11 mutant protein for Ptc is reduced 2.3-fold (Fig. 4E), and that signaling potency in neural plate explant assays was reduced by a similar factor (Table 1).
ShhN signaling potency in vivo is largely dependent on N-terminal modification (reviewed in ref. 8). In this respect, cultured cell lines appear to more faithfully reflect determinants of ShhN potency than neural plate explants, which appear to be insensitive to N-terminal modification of the ShhN protein (14). We therefore tested response of C3H10T1/2 cells to N-11 protein by measuring induction of alkaline phosphatase (AP). As previously noted, 50-100 nM WT recombinant ShhN protein was required for significant induction of AP expression, a concentration many-fold higher than that required for the modified protein (14, 16). The N-11 protein displays an even more dramatically reduced activity in this assay (Fig. 4F), with little induction of AP even at 200 nM. However, this protein did not display the dominant negative effect previously reported for recombinant ShhN proteins lacking 10 N-terminal residues (ref. 14 and data not shown).
Q100H and E188Q Alterations Do Not Affect Shh Activity. The Q100H and E188Q alterations did not affect signaling activity in any of our Shh signaling assays (Fig. 1C and Table 1) and caused no apparent defects in cholesterol-mediated autoprocessing reactions (Fig. 3 A and B).
Discussion
Multiple examples of cosegregation with heterozygous SHH gene deletions or truncations have demonstrated haploinsufficiency of SHH gene function in HPE. More subtle SHH mutations have also been reported to cause HPE. Here, we have characterized the effects of seven such missense mutations affecting the N-terminal signaling domain of SHH, to both examine causality and learn more about SHH function. Our assays have included measurements of binding to the Ptch receptor, examination of SHH protein processing in cultured cells, and assessment of the ability of altered recombinant proteins to elicit responses in neural plate explants and in C3H 10T1/2 cultured cells, which consistently is the most sensitive assay in revealing differences in signaling activity of altered proteins as compared to WT.
Weak SHH Mutations Show Poor HPE Cosegregation in Pedigree Analysis. Five of the seven altered proteins showed decrements in function. The three of these five that clearly cosegregate with an HPE phenotype (G31R, W117G, and W117R) showed the strongest functional impairments (see below). A fourth, D88V, although not associated with HPE in the heterozygous mother of the proband, shows a pattern of cosegregation in second-degree relatives that could be consistent with incomplete penetrance of the mutation (19). The fifth alteration, N115K, is not associated with HPE in the mother of the proband, and no other evidence of cosegregation with phenotype exists. This protein appears to be processed normally and only differs from WT protein in Ptch-binding and neural tube signaling by ≈2-fold. Nevertheless, induction of alkaline phosphatase in C3H 10T1/2 cells by recombinant ShhN carrying the N115K alteration is reduced 5-fold; if indicative of function in vivo, this decrement in signaling potency would reduce SHH signaling activity by 40%, only slightly less than the 50% that is causally associated with HPE in the case of null function mutations. Thus, it seems reasonable, although unproven, that the N115K mutation might be causally linked to the phenotype of the proband, but that its effect is incompletely penetrant, and is possibly influenced by other genetic or environmental factors.
For the Q100H and E188Q altered proteins, no defect in processing and little, if any, difference in Ptch binding or in signaling potency in neural plate and cultured cell assays were observed. Although these mutations were inherited from the mothers of the probands, they were not associated with HPE in any family member other than the probands themselves. The HPE phenotype in these probands thus could be due to mutant effects of variable penetrance on some aspect of Shh function that we cannot measure, or alternatively could be due to other genetic or environmental causes.
A Furin-Like Cleavage Site Created by the G31R HPE Mutation. The G31R mutation generates a tetrabasic furin cleavage site by substituting Arg for Gly at a position adjacent to three preexisting basic residues. This cleavage site is efficiently used in vivo, because most of the ShhN protein in cells and all of that released into the medium is cleaved. The resulting protein lacks 11 residues (N-11), including the N-terminal site of attachment for palmitate modification of ShhN. Despite loss of the site for palmitate attachment, which is critical for signaling potency in vivo (12, 20, 21), a recombinant N-11 protein was only moderately impaired (≈2- to 3-fold) in binding to Ptch and in neural plate signaling assays, consistent with similar previous observations (14, 15). However, this protein is 15- to 20-fold reduced in signaling potency in C3H 10T1/2 cells. The greater decrement in signaling potency of the N-11 protein in the C3H 10T1/2 assay could be due to absence of basic residues involved in mediating ShhN binding to heparin (see above; refs. 14 and 18). Several previous reports have demonstrated a dominant interfering effect for proteins lacking 9 or 10 N-terminal residues (N-9 and N-10), but we observed no such effect for N-11 (data not shown). The difference in properties of these proteins could be due to the presence of more basic residues in N-9 and N-10, which might render them better able to interact with sulfated sugars and thus interfere with signaling.
Lack of Shh Processing Correlates with Misfolding and ER Retention. The W117G and W117R ShhN protein variants prepared from bacteria are inactive at 37°C, but display some activity in neural plate signaling assays at 32°C. Binding to the 5E1 monoclonal antibody also is temperature dependent. The nonlinear ShhN determinants recognized by this antibody coincide with those required for binding to Ptch and do not include W117 (15), suggesting that the effects of W117 alterations on signaling are indirect, possibly because of temperature-dependent changes in conformation. Consistent with this notion, the W117G and W117R proteins also display dramatic temperature-dependent changes in their circular dichroism spectra (data not shown).
The immature glycosyl adducts (EndoH sensitive) of these proteins at their nonnative temperature suggests that they are trapped in the ER, a common fate for misfolded proteins. The absence of processed products at this temperature further suggests that ER localization is incompatible with autoprocessing, despite the presence of a WT ShhC processing domain. Maturation of glycosyl adducts is not a factor in function of the ShhC processing domain, as processed ShhC with EndoH-sensitive adducts from WT Shh can be found within cells. Taken together, these observations suggest that Shh cleavage and processing likely occur in the cis-Golgi, post-ER but before arrival in the medial Golgi, where glycosyl adducts mature (22). One possible mechanism governing intracellular localization of the processing event is the availability of cholesterol, found at only low levels in the ER and at higher levels in the Golgi and other more distal compartments of the secretory pathway (23). Consistent with availability of cholesterol as a critical determinant of processing, extreme depletion of cholesterol from cells using β-methyl cyclodextrin can inhibit Shh processing (24). A lack of signaling activity was also reported for W117G and W117R proteins overexpressed in chick embryos (25), and recently, a failure of these proteins expressed in HEK-293 cells to undergo autoprocessing was independently reported (26). These findings are consistent with our observations and support our hypothesis that temperature-dependent misfolding disrupts cholesterol-dependent autoprocessing by causing retention in the sterol-poor ER.
The temperature-sensitive folding and signaling behavior of the W117G and W117R proteins also suggests the possibility of using amino acid substitutions at the W117 position to design proteins that make temperature-regulated functional transitions useful for biological studies of Shh signaling in nonthermogenic experimental models such as fish and frogs, where growth temperatures can be externally manipulated.
In conclusion, our analysis of ShhN missense alterations in HPE patients suggests that mutations that are questionably linked to HPE are less likely to detectably affect Shh biogenesis and signaling potency. Most missense mutations that clearly cosegregate with HPE appear to cause defects in production of the mature ShhN signal by destabilizing ShhN protein or altering its processing. The G31R variant thus creates a processing site for a furin-like enzyme, producing an abnormally processed protein of reduced activity. The W117G and W117R alterations, on the other hand, produce folding defects and consequent ER retention, consistent with a requirement for exit from the sterol-poor environment of the ER for cholesterol-dependent autoprocessing.
Author contributions: T.M., N.F., and P.A.B. designed research; T.M. and N.F. performed research; T.M., N.F., and P.A.B. analyzed data; and T.M. and P.A.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: HPE, holoprosencephaly; ER, endoplasmic reticulum.
References
- 1.Muenke, M. & Beachy, P. A. (2001) in The Metabolic and Molecular Bases of Inherited Disease, eds. Scriver, C., Beaudet, A., Sly, W. & Valle, D. (McGraw-Hill, New York), pp. 6203-6230.
- 2.Chiang, C., Litingtung, Y., Lee, E., Young, K., Corden, J. L., Westphal, H. & Beachy, P. (1996) Nature 383, 407-413. [DOI] [PubMed] [Google Scholar]
- 3.Ingham, P. W. & McMahon, A. P. (2001) Genes Dev. 15, 3059-3087. [DOI] [PubMed] [Google Scholar]
- 4.Roessler, E., Belloni, E., Gaudenz, K., Jay, P., Berta, P., Scherer, S., Tsui, L. & Muenke, M. (1996) Nat. Genet. 14, 357-360. [DOI] [PubMed] [Google Scholar]
- 5.Belloni, E., Muenke, M., Roessler, E., Traverso, G., Siegel-Bartelt, J., Frumkin, A., Mitchell, H., Donis-Keller, H., Helms, C., Hing, A., et al. (1996) Nat. Genet. 14, 353-356. [DOI] [PubMed] [Google Scholar]
- 6.Cooper, M. K., Wassif, C. A., Krakowiak, P. A., Taipale, J., Gong, R., Kelley, R. I., Porter, F. D. & Beachy, P. A. (2003) Nat. Genet. 33, 508-513. [DOI] [PubMed] [Google Scholar]
- 7.Ming, J. E. & Muenke, M. (2002) Am. J. Hum. Genet. 71, 1017-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann, R. K. & Beachy, P. A. (2004) Annu. Rev. Biochem. 73, 891-923. [DOI] [PubMed] [Google Scholar]
- 9.Lum, L. & Beachy, P. A. (2004) Science 304, 1755-1759. [DOI] [PubMed] [Google Scholar]
- 10.Porter, J., Young, K. & Beachy, P. (1996) Science 274, 255-259. [DOI] [PubMed] [Google Scholar]
- 11.Pepinsky, R., Zeng, C., Wen, D., Rayhorn, P., Baker, D., Williams, K., Bixler, S., Ambrose, C., Garber, E., Miatkowski, K., et al. (1998) J. Biol. Chem. 273, 14037-14045. [DOI] [PubMed] [Google Scholar]
- 12.Chamoun, Z., Mann, R., Nellen, D., von Kessler, D., Bellotto, M., Beachy, P. & Basler, K. (2001) Science 293, 2080-2084. [DOI] [PubMed] [Google Scholar]
- 13.Roelink, H., Porter, J., Chiang, C., Tanabe, Y., Chang, D., Beachy, P. & Jessell, T. (1995) Cell 81, 445-455. [DOI] [PubMed] [Google Scholar]
- 14.Williams, K., Rayhorn, P., Chi-Rosso, G., Garber, E., Strauch, K., Horan, G., Reilly, J., Baker, D., Taylor, F., Koteliansky, V. & Pepinsky, R. (1999) J. Cell Sci. 112, 4405-4414. [DOI] [PubMed] [Google Scholar]
- 15.Fuse, N., Maiti, T., Wang, B., Porter, J., Tanaka Hall, T., Leahy, D. & Beachy, P. (1999) Proc. Natl. Acad. Sci. USA 96, 10992-10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taipale, J., Chen, J., Cooper, M., Wang, B., Mann, R., Milenkovic, L., Scott, M. & Beachy, P. (2000) Nature 406, 1005-1009. [DOI] [PubMed] [Google Scholar]
- 17.Odent, S., Attié-Bitach, T., Blayau, M., Mathieu, M., Jugé, J., Delezoïde, A. L., Le Gall, J. Y., Le Marec, B., Munnich, A., David, V. & Vekemans, M. (1999) Hum. Mol. Genet. 8, 1683-1689. [DOI] [PubMed] [Google Scholar]
- 18.Rubin, J., Choi, Y. & Segal, R. (2002) Development (Cambridge, U.K.) 129, 2223-2232. [DOI] [PubMed] [Google Scholar]
- 19.Nanni, L., Ming, J., Bocian, M., Steinhaus, K., Bianchi, D., Die-Smulders, C., Giannotti, A., Imaizumi, K., Jones, K., Campo, M., et al. (1999) Hum. Mol. Genet. 8, 2479-2488. [DOI] [PubMed] [Google Scholar]
- 20.Lee, J., Kraus, P., Gaiano, N., Nery, S., Kohtz, J., Fishell, G., Loomis, C. & Treisman, J. (2001) Dev. Biol. 233, 122-136. [DOI] [PubMed] [Google Scholar]
- 21.Kohtz, J., Lee, H., Gaiano, N., Segal, J., Ng, E., Larson, T., Baker, D., Garber, E., Williams, K. & Fishell, G. (2001) Development (Cambridge, U.K.) 128, 2351-2363. [DOI] [PubMed] [Google Scholar]
- 22.Lowe, J. B. & Marth, J. D. (2003) Annu. Rev. Biochem. 72, 643-691. [DOI] [PubMed] [Google Scholar]
- 23.Mitra, K., Ubarretxena-Belandia, I., Taguchi, T., Warren, G. & Engelman, D. M. (2004) Proc. Natl. Acad. Sci. USA 101, 4083-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guy, R. (2000) Proc. Natl. Acad. Sci. USA 97, 7307-7312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schell-Apacik, C., Rivero, M., Knepper, J. L., Roessler, E., Muenke, M. & Ming, J. E. (2003) Hum. Genet. 113, 170-177. [DOI] [PubMed] [Google Scholar]
- 26.Traiffort, E., Dubourg, C., Faure, H., Rognan, D., Odent, S., Durou, M. R., David, V. & Ruat, M. (2004) J. Biol. Chem. 279, 42889-42897. [DOI] [PubMed] [Google Scholar]