Abstract
We recently described the use of mini-Tn5 to generate complement-sensitive mutants derived from a complement-resistant Klebsiella pneumoniae clinical isolate deficient in the lipopolysaccharide O side chain. One mutant with a reduced capacity to survive in nonimmune human sera carried the transposon inserted in the htrA gene. We cloned and sequenced the gene and predicted from the deduced amino acid sequence that the putative HtrA homolog contains structural features similar to those of previously described HtrA proteins. To investigate the biological functions and the role of the htrA gene in the virulence of K. pneumoniae, we constructed an isogenic mutant by insertion-duplication mutagenesis. Characterization of the mutant showed that it had greater sensitivity to temperature (50°C) and oxidative stress (H2O2) than the parent strain. Furthermore, the htrA mutant produced less capsule, bound more molecules of complement component C3, and was more sensitive to complement and whole-blood killing than was the parent strain. Finally, disruption of the htrA gene in a virulent K. pneumoniae strain caused a reduction of its virulence in a mice model. Our results indicate that the htrA gene plays an important role in the virulence of K. pneumoniae.
Klebsiella pneumoniae is a frequent pathogen of the human urinary tract that occasionally causes bacteremia and pneumonia, particularly in immunocompromised patients (5, 11). Two virulence factors are essential for the microorganism to be able to spread through the blood and to cause sepsis: lipopolysaccharide (LPS) and the capsular polysaccharide (CPS) (28). LPS is required to resist complement-mediated killing (20), while CPS is mainly involved in resistance to phagocytosis, acting as a physical barrier (28), although it may be also involved in resistance to the complement system (3, 20). Almost all K. pneumoniae clinical isolates express both virulence factors; however, resistance to the bactericidal effect of the complement varies among them (23). This experimental observation suggests that resistance to complement is probably multifactorial and that, although CPS and LPS are key components in resistance to complement-mediated killing, other virulence factors may contribute to bacterial survival in serum.
Recently, we investigated the prevalence of LPS O types among 638 K. pneumoniae clinical isolates. In that study, 17.4% of the strains were nontypeable and almost half of the nontypeable clinical isolates (8.3% of the total) lacked the O side chain of LPS (O− strains) independently of the isolation source (urine, blood, or other sites) (13). Since the LPS O side chain is the major complement resistance factor described in K. pneumoniae (20), we decided to characterize these O− clinical isolates and their mechanisms of resistance to complement-mediated killing. To investigate these mechanisms, we generated a set of mutants with mini-Tn5. A transposon mutant that showed a reduction in its capacity to survive in human serum was selected for further studies. Genomic analysis of the mutant chromosome identified the presence of the transposon in the htrA gene.
htrA is involved in the virulence in other species such as Salmonella enterica serovar Typhimurium (6), Brucella abortus (10), and Yersinia enterocolitica (18). htrA mutants from these species exhibited a reduction in their capacity to survive in macrophages and in mice: their 50% lethal doses (LD50) decreased between 2 and 5 log units, probably because their increased sensitivity to oxidative stress makes them more susceptible to host's oxygen-dependent killing mechanisms. However, the mechanisms by which the virulence of htrA mutants is reduced are not completely understood.
In this report, we describe the role of the htrA gene in the virulence of K. pneumoniae and describe a new mechanism to explain why the K. pneumoniae htrA mutants are attenuated.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
K. pneumoniae strains included in this study were clinical isolates USA0352/78 (serotypes O− and K47) and 52145 (serotypes O1 and K2). Spontaneous mutants resistant to rifampin derived from these strains were selected for the conjugation experiments. Escherichia coli strains used in the cloning experiments were DH5α and S17-1 λpir (14), in which gene pir encodes protein π, which is essential for replication of plasmid pFS100.
Plasmid pUC19 (New England Biolabs) was used in the cloning experiments, and plasmid pFS100 was used to create insertion-duplication mutations by homologous recombination (24). Bacterial cells were grown in Luria-Bertani broth at 37°C with shaking or were solidified with 1.5% agar. When necessary, media were supplemented with ampicillin at 50 μg/ml, kanamycin at 50 μg/ml, and rifampin at 40 μg/ml.
DNA procedures.
Plasmid DNA was isolated using the Wizard Miniprep kit (Promega) according to the manufacturer's instructions. Isolation of genomic DNA, transformation, and electroporation were carried out by standard techniques (4). T4 DNA ligase and restriction endonucleases were used in accordance with the manufacturer's recommendations (Pharmacia). DNA fragments prepared by restriction enzyme digestion were separated by agarose gel electrophoresis and visualized by ethidium bromide staining. For Southern blot analysis and probe labeling and detection we used the ECL kit (Amersham) according to the manufacturer's protocol. DNA sequencing was performed with an automated sequencing apparatus (Applied Biosystems).
Blood, sera, and antisera.
Fresh blood collected from healthy volunteers was treated with heparin or clotted and centrifuged to obtain normal nonimmune human sera (NHS). NHS was pooled, aliquoted, and frozen at −70°C until its use or incubated at 56°C for 30 min to obtain heat-inactivated human sera (HI-NHS), which was also stored at −70°C.
Polyclonal antisera against human complement component C3 were obtained as described by Albertí et al. (1).
Bacterial survival experiments.
To determine the effect of high-temperature stress on K. pneumoniae, stationary-phase cells grown at 37°C were transferred to prewarmed 50°C tubes and incubated for 2 min. For oxidative-stress experiments, stationary-phase cells were incubated at 37°C in the presence of H2O2 (10 and 40 mM) for 30 min. To determine the number of viable bacteria after exposure to the appropriate stresses, an aliquot of each bacterial suspension was diluted and plated. Results were expressed as percentages of viable bacteria after treatment with respect to the viable bacteria before treatment (100% survival).
Analysis of complement C3 deposition.
A bacterial suspension (2 × 108 CFU/ml) was opsonized with NHS or HI-NHS diluted in phosphate-buffered saline (PBS; 25% final concentration) at 37°C for 15 min. Cells were washed three times with PBS-1% sodium dodecyl sulfate (SDS) by centrifugation. Pellets were resuspended in 50 mM carbonate-bicarbonate buffer (pH 9.0) containing 1 M NH4OH to disrupt ester bonds between C3 fragments and the bacterial surface. After 2 h at 37°C, the C3 fragment suspension was reduced and alkylated as described previously (12). Aliquots of C3 fragment suspension were diluted 1:1 in SDS-polyacrylamide gel electrophoresis (PAGE) sample buffer and subjected to SDS-PAGE and Western blotting. Filters were blocked with PBS-1% bovine serum albumin (BSA) and incubated sequentially with anti-human C3 (diluted 1:1,000) and alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G and developed with BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium. Finally, the filters were analyzed by densitometry using a Bio Image densitometer and Whole Band, version 3.1, software (Millipore).
Virulence studies of K. pneumoniae strains.
To determine the susceptibility of K. pneumoniae to complement-mediated killing, bacterial cells in the logarithmic phase were incubated in 25% NHS or HI-NHS, as a control, for 3 h at 37°C. Determination of the number of viable bacteria was made by dilution and plating on Luria-Bertani agar plates.
Whole-blood killing assays were performed by incubation of logarithmic-phase cells in fresh blood. Aliquots were withdrawn for quantitative culture immediately and after 3 h of end-over-end rotation at 37°C. In serum and whole-blood killing assays results were expressed as the percentages of viable bacteria after treatment with respect to the viable bacteria before treatment (100% survival).
Mouse lethality studies were performed with male CD1 mice, each weighing 20 to 30 g. Briefly, 10-fold dilutions of logarithmic-phase organisms were administered by intraperitoneal injection to groups of 10 mice. Mortality was determined 7 days after challenge. The LD50 was calculated for each strain from results of three independent experiments including a total of 30 animals for each strain.
Determination of CPS production.
CPS production was quantified by a competitive enzyme-linked immunosorbent assay (ELISA). CPS-containing extracts from 4 × 109 bacterial cells were obtained by a phenol-water minipreparation method (2). Phenol was eliminated by chloroform extraction and ethanol precipitation. Precipitates containing CPS extracts were resuspended in distilled water and used in the inhibition step in the competitive ELISA.
For this purpose, plates were coated with 1 μg of purified CPS type 47 per well. After a blocking step with PBS-1% BSA, plates were incubated with serial dilutions of CPS extracts and antisera against CPS type 47. Antibodies bound to the CPS-coated plates were detected with alkaline phosphatase-labeled goat anti-rabbit immunoglobulin G and developed with p-nitrophenyl phosphate. Incubations with antisera diluted in PBS-1% BSA were carried out at 37°C for 1 h and were always followed by PBS washes. Known amounts of CPS purified by the method of Wilkinson and Sutherland (27) were used to construct a standard curve.
Homology searches.
DNA and protein sequence analyses were performed by using BLASTN and BLASTP.
Nucleotide sequence accession number.
The K. pneumoniae htrA gene sequence has been submitted to the EMBL database under accession no. AJ430233.
RESULTS
K. pneumoniae HtrA.
To look for genes involved in complement resistance in K. pneumoniae, we generated a set of mutants derived from K. pneumoniae strain USA0352/78 using the mini-Tn5 transposon (3). We cloned in pUC19 the EcoRI chromosomal fragment containing the complete transposon from all mutants that showed a reduction in their capacity to survive in human serum. Sequence analysis of plasmid pLOKIM9, derived from the serum-sensitive mutant designated USA0352-M9, identified the transposon 360 bp upstream from the start codon of the putative K. pneumoniae htrA gene. Further sequencing of plasmid pLOKIM9 confirmed the presence of the complete htrA gene. The deduced amino acid sequence encoded by the complete K. pneumoniae htrA gene was that of a protein of 48,824 kDa with an extensive amino acid identity to the HtrA homologues from S. enterica serovar Typhimurium (88%), E. coli (87%), and Y. enterocolitica (73%).
Construction of a K. pneumoniae htrA mutant.
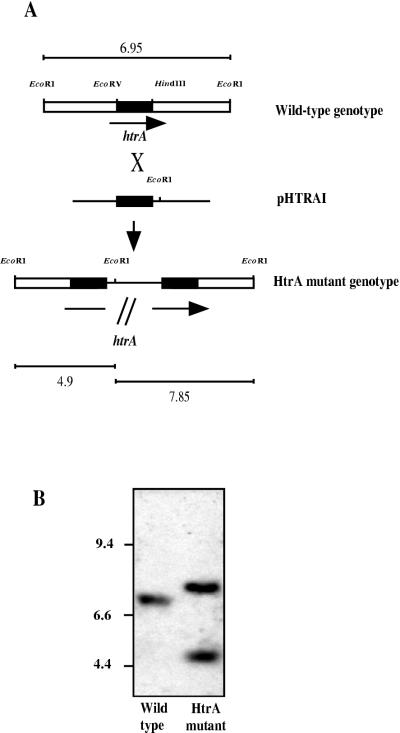
To study directly the role of the htrA gene in K. pneumoniae virulence, we constructed an htrA mutant by insertion-duplication mutagenesis as summarized in Fig. 1A. An internal EcoRV-HindII htrA fragment of 688 bp obtained from pLOKIM9 was cloned in the pFS100 EcoRV site to give pHTRAI, which was introduced into K. pneumoniae USA0352/78 by conjugation. To select for integrants of plasmid pHTRAI in the K. pneumoniae chromosome, thereby disrupting expression of htrA, an aliquot of the conjugate was spread on kanamycin-containing agar. Southern blot analysis of the chromosome of one integrant, designated K. pneumoniae USA0352-htrA−, using as the probe the EcoRV-HindII 688-bp htrA fragment mentioned above, confirmed that two incomplete copies of the htrA gene were generated by the integration of the plasmid (Fig. 1B).
FIG. 1.
Insertion-duplication mutagenesis of htrA in strain USA0352/78. (A) Diagram of insertion-duplication mutagenesis showing homologous recombination between the K. pneumoniae chromosome of wild-type strain USA0352/78 and the pHTRAI disruption construct to generate isogenic htrA mutant USA0352-htrA−. DNA fragment size in the diagram is not to scale. The line in the mutant genome represents the plasmid DNA integrated into the chromosome. Black boxes, EcoRV-HindII htrA internal fragment cloned in pHTRAI and used as probe in the Southern blot analysis. Expected sizes (kilobases) of the fragments that hybridize with the probe described above are indicated. (B) Southern blot analysis of K. pneumoniae wild-type and htrA mutant chromosomes digested with EcoRI. Molecular size markers (kilobases) are shown on the left.
Phenotypic and virulence characterization of a K. pneumoniae htrA mutant.
htrA mutant USA0352-htrA− was more sensitive to H2O2 than the parent strain (Table 1). We observed significant reductions of almost 4-fold and 15-fold in the viability of the htrA mutant compared to that of the parent strain after exposure to 10 and 40 mM H2O2, respectively. When parent strain USA0352/78 and its derived htrA mutant were grown at 37°C, they exhibited similar growth rates (data not shown). However, when both strains were incubated at 50°C, we observed a pronounced reduction in the viability of the htrA mutant compared to that of the parent strain (Table 1). Altogether, these results indicate that in K. pneumoniae the mutation of the htrA gene confers an increased susceptibility to high temperature and oxidative stress.
TABLE 1.
Percent survival of K. pneumoniae USA0352/78 and its derived mutant strain USA0352-htrA− after exposure to oxidative and temperature stresses
| Condition | % Survivala of:
|
|
|---|---|---|
| Wild-type strain | htrA mutant | |
| 10 mM H2O2 | 100 ± 0 | 24.07 ± 6.11 |
| 40 mM H2O2 | 63 ± 2.10 | 4.26 ± 1.03 |
| 50°C | 55.21 ± 3.84 | 2 ± 0.71 |
Data are means ± standard deviations of three independent experiments. P < 0.001 for all comparisons between the wild-type strain and the htrA mutant (two-tailed t test).
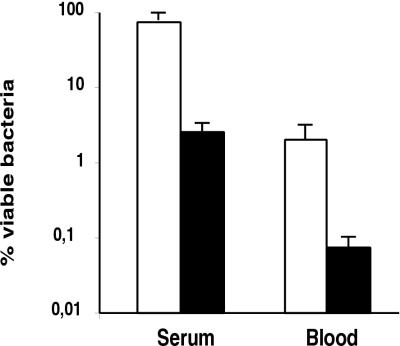
The HtrA mutant was more sensitive to complement-mediated killing than the parent strain. We observed a reduction of almost 2 log units in the viability of the htrA mutant cells exposed to NHS, a significant reduction compared with that for the parent strain, which exhibited a reduction of only approximately 1/2 log unit in its viability (Fig. 2).
FIG. 2.
Resistance to complement and to whole-blood killing of K. pneumoniae strains. Bars indicate percentages of viable bacteria after incubation for 3 h in NHS or in fresh human blood. The results represent the means ± standard errors of three independent experiments for wild-type strain USA0352/78 (open bars) and htrA mutant strain USA0352-htrA− (solid bars). A comparison of the htrA mutant and wild-type strains by a two-tailed t test yielded P values <0.01.
USA0352/78 and its derived htrA mutant failed to grow in human blood. The number of cells of the htrA mutant was reduced more than 1,500-fold after 3 h of incubation, compared with a minor reduction (100-fold) for wild-type strain USA0352/78 (Fig. 2).
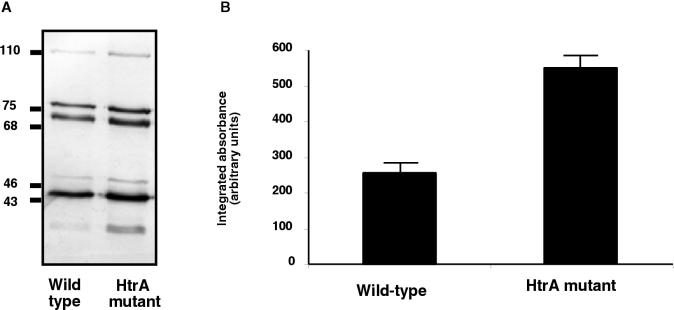
To investigate the mechanisms of the increased susceptibility to the complement activity showed by the htrA mutant, we studied by Western blotting analysis the deposition and degradation of the human C3 complement component on the cell surfaces of USA0352/78 and its derived mutant USA0352-htrA− (Fig. 3A). The patterns of complement C3 deposition on both strains were similar; C3b deposition and cleavage to iC3b occurred rapidly (<5 min; data not shown). Both strains bound the α′ chain of C3b (105 kDa), the β chain common to C3b and iC3b (75 kDa), and the 68-kDa α′1 chain of iC3b. Therefore, deposition of C3b, the active form of C3, and its breakdown to iC3b were detected on both strains. Densitometric analysis of filters indicated that the major difference was that USA0352-htrA− bound more C3b and its degradation fragments that did USA0352/78.
FIG. 3.
Analysis of complement C3 deposition on K. pneumoniae cell surfaces. (A) Cells of wild-type strain USA0352/78 and its derived isogenic mutant USA0352-htrA− were incubated in NHS. C3 fragments deposited on the bacterial surface were released, separated by SDS-PAGE, and identified by Western blotting with anti-human C3 serum and by comparison with purified C3 fragments. (B) Quantification of the 75-kDa band of the β chain common to C3b and iC3b was carried out by densitometric analysis. Data are means ± standard deviations of three independent experiments. Control values for each strain, i.e., C3 fragment depositions with heat-inactived serum, were subtracted from experimental results. A comparison of the htrA mutant and wild-type strains by a two-tailed t test yielded P values <0.01.
We showed previously that the LPS O side chain and CPS are the most important surface components involved in the modulation of complement deposition on K. pneumoniae (3, 20). Since strain USA0352/78 and its derived htrA mutant do not express the LPS O side chain (data not shown), we measured the amounts of CPS produced by the htrA mutant and the parent strain. For this purpose, we performed a competitive ELISA with CPS extracts from the wild-type strain and its derived htrA mutant. Parent strain USA0352/78 produced almost twofold more CPS (1,15.5 ± 14 fg/CFU) than the htrA mutant (59.8 ± 7.9 fg/CFU) (P < 0.01 for comparison between the parent strain and the htrA mutant strain; two-tailed t test).
To further characterize the role of htrA in K. pneumoniae virulence, we performed LD50 experiments with mice. Strain USA0352/78 was not virulent in this model (LD50 > 108 CFU; data not shown). We thus challenged mice with highly virulent K. pneumoniae strain 52145 and its isogenic htrA mutant derived in accordance with the strategy used for Fig. 1. In this study, the LD50 for 52145 was 10 CFU, compared with 5 × 103 CFU for the htrA mutant. The magnitude of the difference between the LD50 for the htrA mutant and that for the wild-type strain in these experiments was 500-fold. These results demonstrated that mutation of the htrA gene attenuated K. pneumoniae virulence.
DISCUSSION
HtrA is a heat shock-induced serine protease that has been described in diverse bacteria, as well as in some eukaryotes (21). First associated with thermosensitivity and survival in the face of environmental stress stimuli, the htrA gene is now of interest because htrA mutant versions of several gram-negative bacteria have been shown to be attenuated in animal models (8, 10, 18) and can also be used as vaccines (21).
In the present study, the K. pneumoniae htrA gene was cloned and sequenced. Alignment of the deduced amino acid sequence of K. pneumoniae HtrA revealed extensive amino acid identity with HtrA sequences from other bacteria. The conserved consensus sequence GDSGGPK surrounding the active serine residues of many serine proteases (21) and the PDZ domains involved in protein-protein interactions (21) are also present in the K. pneumoniae HtrA protein, suggesting that it may be involved in processes similar to those described above for other bacterial species.
To determine the contribution of the htrA gene to K. pneumoniae virulence, we derived an isogenic htrA mutant by an insertion-duplication method that, in contrast with transposon mutagenesis, avoids potential polar effects on other genes. Disruption of the htrA gene increased the susceptibility of K. pneumoniae to temperature and oxidative stresses, as has been shown to occur in other species such as B. abortus. However, there is a wide variation in the phenotype of the htrA mutants in different bacteria. For example, E. coli htrA mutants, in contrast with S. enterica serovar Typhimurium htrA mutants, are sensitive to temperature stress but insensitive to oxidative stress (15, 19). Furthermore, Elzer et al. reported that B. abortus htrA mutants were sensitive to both temperature and oxidative stress (10), but Tatum et al. observed that B. abortus htrA mutants were insensitive to both stimuli (26). It is likely that the contribution of HtrA to susceptibility to temperature and oxidative stress is crucial in some species but may depend on other genes in some species.
Resistance to complement and resistance to phagocytosis are two of the most important mechanisms associated with K. pneumoniae virulence (23). Our results demonstrated that the ability of the isogenic htrA mutant derived from a K. pneumoniae O side chain-deficient strain to grow in human sera was impaired. We used an encapsulated K. pneumoniae strain deficient in the O side chain of LPS, a component that plays a key role in complement resistance in K. pneumoniae. CPS of K. pneumoniae has also been shown to be important in resistance to complement-mediated killing (20), particularly in O side chain-deficient isolates where capsule is a major complement resistance factor (3) because it masks target molecules for complement C3 deposition. We found that synthesis of CPS, measured by quantitative competition ELISA, was reduced in a K. pneumoniae htrA mutant compared to that in the parent strain. In contrast, SDS-PAGE analysis of the outer membrane proteins, particularly porins, and LPS, the most important complement C3 ligands of K. pneumoniae (1), did not reveal any significant difference between the wild-type strain and the htrA mutant that could account for the increased C3 deposition observed in the htrA mutant (data not shown). Only CPS expression, not LPS expression, both quantified by ELISA, was altered in the htrA mutant. This reduction contributed to the increased efficiency of complement C3 binding, as we previously reported (9), and might explain the increased sensitivity to serum-mediated killing showed by the K. pneumoniae htrA mutants.
It has been speculated that HtrA's putative chaperone and protein-processing functions are needed for the folding of secreted proteins or that HtrA might be involved in the oligomerization and export of virulence factors (21). The periplasmic localization of HtrA allows and influences these functions (25). Our results suggest that dysfunction of these activities would interfere with the export of the CPS. Further studies will help to elucidate the interactions between HtrA and the biosynthesis and transport systems of CPS in K. pneumoniae.
We demonstrated that the K. pneumoniae htrA mutant was more sensitive to whole-blood killing than the parent strain, suggesting a reduced ability to resist phagocytosis. The reduction of the ability to resist phagocytosis showed by the htrA mutants has been attributed to a decreased capacity to resist oxidative stress that probably makes the bacteria more susceptible to host's oxygen-dependent killing (15, 18, 21, 22). K. pneumoniae CPS plays an important role in resistance to phagocytosis (9, 16, 28). Thus, in addition to the mechanism described above, the reduction of CPS expression observed in the K. pneumoniae htrA mutant might also affect the resistance to phagocytosis.
The importance of htrA gene in an animal model of bacterial pathogenesis was also studied. The K. pneumoniae htrA mutant derived from a highly virulent strain showed a 500-fold reduction in its virulence compared to the parent strain. Similar results for different microorganisms have been reported by other authors (10, 15, 18). Furthermore, these results led several authors to use htrA mutants as live attenuated vaccine strains to confer resistance against lethal challenge with virulent organisms (7, 8, 15, 17). A practical application of K. pneumoniae htrA mutants would be their use as live carrier vaccines, particularly of protective antigens from respiratory pathogens.
In summary, our results indicate that mutation of the htrA gene in K. pneumoniae affects the ability of K. pneumoniae to produce CPS and increases the deposition of complement and the sensitivity to complement and whole-blood killing, with subsequent virulence attenuation.
Acknowledgments
We thank Fernando Vivanco (Fundación Jiménez Diaz, Madrid) for providing antisera against human C3.
This work was supported by grants from Fondo de Investigaciones Sanitarias and Ministerio de Ciencia y Tecnología. G.C. and B.D.A. are the recipients of predoctoral fellowships from Universitat de les Illes Balears.
Editor: E. I. Tuomanen
REFERENCES
- 1.Albertí, S., D. Álvarez, S. Merino, M. T. Casado, F. Vivanco, J. M. Tomás, and V. J. Benedí. 1996. Analysis of complement C3 deposition and degradation on Klebsiella pneumoniae. Infect. Immun. 64:4726-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albertí, S., J. Imperial, J. M. Tomás, and V. J. Benedí. 1991. Bacterial lipopolysaccharide extraction in silica gel-containing tubes. J. Microbiol. Methods 14:63-69. [Google Scholar]
- 3.Álvarez, D., S. Merino, J. M. Tomás, V. J. Benedí, and S. Albertí. 2000. Capsular polysaccharide is a major complement resistance factor in lipopolysaccharide O side chain-deficient Klebsiella pneumoniae clinical isolates. Infect. Immun. 68:953-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1997. Current protocols in molecular biology. Greene Publishing and Wiley Interscience, New York, N.Y.
- 5.Bartlett, J. G., P. O'Keefe, F. P. Tally, T. J. Louie, and S. L. Gorbach. 1986. Bacteriology of hospital-acquired pneumonia. Arch. Intern. Med. 146:868-871. [PubMed] [Google Scholar]
- 6.Baumler, A. J., J. G. Kusters, I. Stojiljkovic, and F. Heffron. 1994. Salmonella typhimurium loci involved in survival within macrophages. Infect. Immun. 62:1623-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chabalgoity, J. A., C. M. Khan, A. A. Nash, and C. E. Hormaeche. 1996. A Salmonella typhimurium htrA live vaccine expressing multiple copies of a peptide comprising amino acids 8-23 of Herpes simplex virus glycoprotein D as a genetic fusion to tetanus toxin fragment C protects mice from Herpes simplex virus infection. Mol. Microbiol. 19:791-801. [DOI] [PubMed] [Google Scholar]
- 8.Chatfield, S. N., H. Strahan, D. Pickard, I. G. Charles, C. E. Hormaeche, and G. Dougan. 1992. Evaluation of Salmonella typhimurium strains harbouring defined mutations in htrA and aroA in the murine salmonellosis model. Microb. Pathog. 12:145-151. [DOI] [PubMed] [Google Scholar]
- 9.Cortés, G., N. Borrell, B. de Astorza, C. Gómez, J. Sauleda, and S. Albertí. 2002. Molecular analysis of the contribution of the capsular polysaccharide and the lipopolysaccharide O side chain to the virulence of Klebsiella pneumoniae in a murine model of pneumonia. Infect. Immun. 70:2583-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elzer, P. H., R. W. Phillips, G. T. Robertson, and R. M. Roop II. 1996. The Htra stress response protease contributes to resistance of Brucella abortus to killing by murine phagocytes. Infect. Immun. 64:4838-4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.García de la Torre, M., J. Romero-Vivas, J. Martínez-Beltrán, A. Guerrero, M. Messeguer, and E. Bouza. 1985. Klebsiella bacteremia: an analysis of 100 episodes. Rev. Infect. Dis. 7:143-150. [DOI] [PubMed] [Google Scholar]
- 12.Gordon, D. L., J. Rice, J. J. Finlay-Jones, P. J. MacDonald, and M. K. Hostetter. 1988. Analysis of C3 deposition and degradation on bacterial surfaces after opsonization. J. Infect. Dis. 157:697-704. [DOI] [PubMed] [Google Scholar]
- 13.Hansen, D. S., F. Mestre, S. Albertí, S. Hernández-Allés, D. Álvarez, A. Doménech, J. Gil, S. Merino, J. M. Tomás, and V. J. Benedí. 1999. Klebsiella pneumoniae lipopolysaccharide O-typing: revision of prototype strains and O-type distribution among clinical isolates of different sources and countries. J. Clin. Microbiol. 37:56-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrero, M., V. de Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, K., I. Charles, G. Dougan, D. Pickard, P. O'Gaora, G. Costa, T. Ali, I. Miller, and C. Hormaeche. 1991. The role of a stress-response protein in Salmonella typhimurium virulence. Mol. Microbiol. 5:401-407. [DOI] [PubMed] [Google Scholar]
- 16.Kabha, K., L. Nissimov, A. Athamma, Y. Keisari, H. Parolis, L. A. S. Parolis, R. M. Grue, J. Schlepper-Schafer, A. R. B. Ezekowitz, D. E. Ohma, and I. Ofek. 1995. Relationships among capsular structure, phagocytosis, and mouse virulence in Klebsiella pneumoniae. Infect. Immun. 63:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine, M. M., J. Galen, E. Barry, F. Noriega, S. Chatfield, M. Sztein, G. Dougan, and C. Tacket. 1996. Attenuated Salmonella as live oral vaccines against typhoid fever and as live vectors. J. Biotechnol. 44:193-196. [DOI] [PubMed] [Google Scholar]
- 18.Li, S. R., N. Dorrell, P. H. Everest, G. Dougan, and B. W. Wren. 1996. Construction and characterization of Yersinia enterocolitica O:8 high-temperature requirement (htrA) isogenic mutant. Infect. Immun. 64:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lipinska, B., O. Fayet, L. Baird, and C. Georgopoulus. 1989. Identification, characterization, and mapping of the Escherichia coli htrA gene, whose product is essential for bacterial growth only at elevated temperatures. J. Bacteriol. 171:1574-1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merino, S., S. Camprubí, S. Albertí, V. J. Benedí, and J. M. Tomás. 1992. Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 60:2529-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pallen, M. J., and B. W. Wren. 1997. The HtrA family of serine proteases. Mol. Microbiol. 26:209-221. [DOI] [PubMed] [Google Scholar]
- 22.Pedersen, L. L., M. Radulic, M. Doric, and Y. Abu Kwaik. 2001. HtrA homologue of Legionella pneumophila: an indispensable element for intracellular infection of mammalian but not protozoan cells. Infect. Immun. 69:2569-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Podschun, R., S. A. Fischer, and U. Ullmann. 1993. Serotypes, hemagglutinins, siderophore synthesis, and serum resistance of Klebsiella isolates causing human urinary tract infections. J. Infect. Dis. 168:1415-1421. [DOI] [PubMed] [Google Scholar]
- 24.Rubirés, X., F. Saigi, N. Puqué, N. Climent, S. Merino, S. Albertí, J. M. Tomás, and M. Regué. 1997. A gene (wbbL) from Serratia marcescens N28b (O4) complements the rfb-50 mutation of Escherichia coli K-12 derivates. J. Bacteriol. 179:7581-7586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skórko-Glonek, J., B. Lipínska, K. Krzenwski, G. Zolese, E. Bertoli, and F. Tanfani. 1997. HtrA heat shock protease interacts with phospholipid membranes and undergoes conformational changes. J. Biol. Chem. 272:8974-8982. [DOI] [PubMed] [Google Scholar]
- 26.Tatum, F. M., N. F. Cheville, and D. Morfitt. 1994. Cloning, characterization and construction of htrA and htrA-like mutants of Brucella abortus and their survival in BALB/c mice. Microb. Pathog. 17:23-26. [DOI] [PubMed] [Google Scholar]
- 27.Wilkinson, J. F., and I. W. Sutherland. 1971. Chemical extraction methods of microbial cells. Methods Microbiol. 5B:345-383. [Google Scholar]
- 28.Williams, P., and J. M. Tomás. 1990. The pathogenicity of Klebsiella pneumoniae. Rev. Med. Microbiol. 1:196-204. [Google Scholar]