Abstract
Shiga toxin-producing Escherichia coli (STEC) comprises a broad group of bacteria, some of which cause attaching and effacing (AE) lesions and enteritis in animals and humans. Non-O157 STEC serotypes contain a gene (efa1) that mediates attachment to cultured epithelial cells. An almost-identical gene in enteropathogenic E. coli (lifA) encodes lymphostatin, which inhibits the proliferation of mitogen-activated lymphocytes and the synthesis of proinflammatory cytokines. We have investigated the role of the efa1 gene in colonization of 4- and 11-day-old conventional calves by STEC serotypes O5 and O111. Our findings show that Efa1 is required for efficient colonization of the bovine intestinal tract by STEC, since efa1 deletion and insertion mutants were shed in the feces in significantly lower numbers. In addition, efa1 mutations dramatically reduced the number of bacteria associated with the intestinal epithelium. Expression and secretion of locus for enterocyte effacement-encoded type III secreted proteins that are required for adhesion and AE-lesion formation were impaired by mutation of efa1 in STEC but not by mutation of lifA in enteropathogenic E. coli. However, STEC efa1 mutants retain the ability to nucleate filamentous actin under sites of bacterial attachment to cultured eukaryotic cells. Efa1 is only the second STEC factor shown to influence carriage of the bacteria in the bovine intestine. Our data may have implications for strategies to reduce the prevalence of STEC in cattle.
Shiga toxin-producing Escherichia coli (STEC) comprises a group of zoonotic enteric pathogens (43). In humans, infections with some STEC serotypes result in bloody or nonbloody diarrhea, which may be complicated by hemorrhagic colitis and severe renal and neurological sequelae, including hemolytic-uremic syndrome (45). STEC is closely related to enteropathogenic E. coli (EPEC), which is a leading cause of infantile diarrhea in developing countries, and share many of the EPEC genes implicated in virulence (15, 49).
Cattle are an important reservoir of STEC (17), and human infections frequently result from direct or indirect contact with ruminant feces (19, 53). Strategies to lower the prevalence of STEC in cattle therefore offer the possibility of reducing the incidence of human infections. Natural and experimental infection of calves with STEC results in efficient colonization of the intestinal tract with large numbers of bacteria being shed in the feces for several weeks (3, 6, 7, 8, 9, 10, 21, 46). Clinical signs of STEC infection in calves may vary from subclinical to dysentery depending on the serotype (35). The molecular basis of intestinal colonization and STEC serotype host specificity is poorly understood.
The bacterial outer membrane protein intimin is required for intestinal colonization in colostrum-deprived neonatal calves by E. coli O157:H7 and the induction of colonic edema and diarrhea (9). Intimin is required for the formation of intestinal attaching and effacing (AE) lesions, which are characterized by intimate bacterial attachment to the apical surface of enterocytes and the localized destruction of microvilli (15). This histopathology is determined by the chromosomal locus for enterocyte effacement (LEE), and also requires the LEE-encoded Tir protein which is translocated into host cells via a type III protein secretion system, where it acts as a receptor for intimin (11). Intimin can also bind to β1-integrins (16) and cell-surface localized nucleolin (52), though the consequence of this is unknown. Intimin-null mutants still colonize some compartments of the bovine intestinal tract (9), indicating that other colonization factors and/or survival of the bacteria in the intestinal lumen may be required.
Recently, Nicholls et al. identified a gene (efa1 [for E. coli factor for adherence]) that mediates attachment of a clinical O111:H− STEC strain to cultured Chinese hamster ovary cells (44). A TnphoA insertion in the STEC O111:H− efa1 gene gave rise to an enzymatically active alkaline phosphatase-Efa1 fusion protein, indicating that the gene is functionally expressed and has an extracytoplasmic domain (44). The efa1 gene is identical in size and 99.9% identical in nucleotide sequence to the lifA gene in enteropathogenic E. coli (Table 1). The lifA gene confers upon EPEC the ability to inhibit the proliferation of human peripheral blood lymphocytes and the mitogen-stimulated synthesis of interleukin-2 (IL-2), IL-4, IL-5, and gamma interferon (30). The predicted product of lifA (lymphostatin) also inhibits the proliferation of human and murine gastrointestinal lymphocytes, indicating that it may modulate mucosal immunity in the gut (28, 31, 37). Lymphostatin specifically targets lymphoid cells and does not affect the proliferation of epithelial cells, exert direct cytotoxic effects or increase apoptosis (30). The EPEC lifA and STEC efa1 genes encode predicted proteins of 366 kDa that are 97.4% identical at the amino acid level.
TABLE 1.
LCT homologues in EPEC nd STECa
Nucleotide sequence accession numbers for the following are given parenthetically: EPEC O127:H6 lifA (AJ133705), STEC O111:H− efa1 (AF159462), STEC O157:H7 17095 (toxB) (AF074613), STEC O157:H7 efa1′ (AE005174).
REPEC, rabbit enteropathogenic E. coli.
ORF, open reading frame.
IFN-γ, gamma interferon.
The efa1/lifA gene is present not only in STEC O111 and EPEC but also in all non-O157 STEC serotypes tested and related enteropathogens such as Citrobacter rodentium, Hafnia alvei, and rabbit EPEC (30, 44). While E. coli O157:H7 lacks the full-length efa1 gene a truncated version of efa1 exists in the chromosome (Table 1) (24, 47). Screening of a bank of E. coli O157:H7 mini-Tn5Km2 mutants has recently shown that a transposon insertion upstream of the truncated efa1 gene reduces bacterial adherence to human colon carcinoma cells (57), indicating that the truncated version of the gene may also influence bacterial adhesion and intestinal colonization.
A large gene (toxB/l7095) with significant homology to efa1 also exists on the E. coli O157:H7 pO157 virulence plasmid (Table 1) (4, 30, 36, 44). E. coli O157:H7 strains containing derivatives of pO157 that lack toxB exhibit reduced adherence to cultured epithelial cells (58). The authors reported that toxB indirectly influences adherence by modulating the production and secretion of LEE-encoded type III secreted proteins that are required for the formation of AE lesions (58). The toxB gene has also been suggested to be a functional homologue of lymphostatin, since an E. coli O157:H7 strain cured of pO157 lacked the ability to inhibit IL-2 and IL-4 synthesis in mitogen-activated human peripheral blood mononuclear cells (PBMCs) (30).
STEC O111 Efa1, EPEC LifA, and the E. coli O157:H7 ToxB predicted protein share 40% amino acid homology over the N-terminal 470 amino acids (aa) with the N-terminal domains of Clostridium difficile toxins A and B (Table 1) (4, 30, 36, 44). Large clostridial toxins (LCTs) act by glucosylating small GTPases that regulate the actin cytoskeleton (29). This activity is determined by the N-terminal 546 aa of toxin B and is dependent on a DXD motif that is shared by eukaryotic glycosyltransferases (5, 26). Efa1, LifA, and ToxB share homology with LCTs and eukaryotic glycosyltransferases in this region and all three proteins contain the conserved DXD motif. Several open reading frames with homology to LCTs have also been identified in the genome of Chlamydia trachomatis (54) and were recently shown to be required for cytotoxicity (2), indicating that LCT-homologues may influence pathogenesis in diverse pathogens.
Since Efa1 influences adhesion of STEC to eukaryotic cells in vitro and may act as an inhibitor of lymphocyte proliferation, we proposed that Efa1 may influence colonization of the bovine intestine by non-O157 STEC. We have inoculated 4- and 11-day-old conventional calves orally with STEC O5 and O111 strains containing efa1 with deletion or insertion mutations, respectively. Our results suggest a pivotal role for Efa1 in intestinal colonization and enteropathogenesis in calves.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media. STEC strain S102-9 (O5:H−) was isolated from an outbreak of calf dysentery in the United Kingdom (7). S102-9 Nalr is a spontaneous nalidixic acid-resistant derivative of S102-9 and exhibits normal growth and adhesion characteristics in vitro. E. coli E45035N is a nalidixic acid-resistant derivative of STEC O111:H− isolated from the feces of a patient with hemolytic-uremic syndrome (44). EBK4 is derived from E45035N and contains an insertion of a kanamycin resistance cassette in efa1 (44). Both E45035N and EBK4 were kindly supplied by E. Hartland, Monash University, Melbourne, Australia. The prototypic EPEC strain E2348/69 (O127:H6) was isolated by Levine et al. (34). E. coli K-12 strain HB101 [F− Δ(gpt-proA)62 leuB6 supE44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20 xyl-5 mtl-1 recA13] was used as a control in some assays. Plasmid pCVD442 is a positive-selection suicide vector containing blaM, the Bacillus subtilis sacBR genes, the mob region from plasmid RP4 and a pir-dependent R6K origin of replication (13). pCVD442 and derivatives were maintained in E. coli DH5α λpir (48) and in E. coli S17-1λpir for conjugation (51). A cosmid containing lifA (pIV-8-A) (32) was kindly supplied by M. Donnenberg, University of Maryland. Bacteria were isolated on Luria-Bertani (LB) agar and cultivated in LB broth with the following antibiotics at the indicated concentrations: ampicillin, 100 μg/ml; kanamycin, 25 μg/ml; nalidixic acid, 25 μg/ml. For oral inoculation studies bacteria were amplified in brain heart infusion broth for 18 h at 37°C, and the absorbance at 600 nm was adjusted to within 0.1 optical density units.
Cell lines.
HeLa cells (ATCC CCL2) were cultivated in RPMI 1640 buffered with sodium bicarbonate (2 g/liter) and supplemented with 10% (vol/vol) fetal calf serum (PAA Laboratories GmbH, Linz, Austria) and l-glutamine (0.3 g/liter). For adhesion assays, cells were seeded at 2 × 105 cells/35-mm-diameter dish on glass coverslips and grown for 18 h at 37°C in a humidified 5% CO2 atmosphere.
Routine DNA manipulation.
Standard procedures were used for DNA extraction, cloning, PCR, and the verification of mutants by Southern hybridization (50).
Construction of STEC O5:H− efa1 and EPEC O127:H6 lifA mutants.
The STEC Δefa1 and EPEC ΔlifA mutants contain deletions of the entire gene and were constructed as follows. Sequences flanking the S102-9 efa1 gene were separately amplified by PCR with Vent proofreading DNA polymerase (New England Biolabs, Beverly, Mass.) using the primer pairs efa3 (5′ ATATATGAGCTCGTGTAGCGGTATTCGGC 3′) plus efa7 (5′ CCCGTTGCTGCAATTAATTACATTTCCGCTTAA 3′) and efa8 (5′ CGGAAATGTAATTAATTGCAGCAACGGGTA 3′) plus efa9 (5′ ATATATGAGCTCTTAGTTAAAAAGGTTGTC 3′) (based on the sequence of STEC O111 efa1). The primary PCR products were gel purified and combined in an overlapping PCR (27) using the flanking primers efa3 and efa9. The secondary PCR product was then cloned into pCVD442 via SacI sites incorporated into the primers. The resulting plasmid, pCVDΔefa1 was introduced into S102-9 Nalr and E2348/69 by conjugation from E. coli S17-1λpir and merodiploids isolated on LB agar containing ampicillin and nalidixic acid. Double recombinants were selected by growing merodiploids to late-logarithmic phase in LB broth lacking ampicillin and plating onto LB agar (minus NaCl) containing 5% (wt/vol) sucrose at 30°C. Sucrose-resistant colonies were screened for deletions by colony PCR and recombinants verified by Southern hybridization. The deletions result in juxtaposition of the predicted start and stop codons.
Adhesion assay.
Adhesion of STEC strains to HeLa cells was quantified essentially as described previously (39). Cells were stained with Hemacolor rapid staining solutions (Merck, Darmstadt, Germany) and multiple images captured at a magnification of ×400 using a Leica DMLS microscope with a Polaroid digital microscope camera. Fluorescent actin staining for the detection of F-actin under sites of bacteria adhesion to HeLa cells was performed as described previously (33), except that bacteria were incubated on the cells for a total of 8 h.
Lymphocyte proliferation assay.
Bacterial lysates were prepared from 50-ml overnight LB cultures. Cells were collected by centrifugation, washed, and resuspended in 5 ml of phosphate-buffered saline (PBS) and then disrupted by sonication for 2 min total at 2-s intervals on ice. The lysate was centrifuged (1,000 × g for 10 min at 4°C) to remove cell debris, and the protein concentration was determined by a colorimetric assay (Bio-Rad Laboratories, Hercules, Calif.). Bovine PBMCs were isolated from fresh venous blood of ca. 6 month old healthy calves by centrifugation at 2000 × g on Histopaque 1083 gradients (Sigma Chemical Company, St Louis, Mo.) for 40 min at room temperature. PBMCs were aspirated, washed in PBS and collected by centrifugation at 2,000 × g. The cells were resuspended in RPMI 1640 buffered with sodium bicarbonate (2 g/liter) and supplemented with 10% (vol/vol) fetal calf serum, l-glutamine (0.3 g/liter), penicillin (100 U/ml), and streptomycin (100 μg/ml). Cells were counted on a hemocytometer and dispensed into 96-well microtiter plates. Cells (5 × 105) were stimulated with concanavalin A (5 μg/ml) in the presence of 50 μg of bacterial protein. Cultures, set up in quadruplicate, were incubated for 3 days at 37°C in 5% CO2 in a humidified atmosphere. Sixteen hours before harvest, cells were pulsed with 1 μC of [3H]thymidine (Amersham Pharmacia Biotech, Little Chalfont, Buckinghamshire, United Kingdom) per well. Radioactivity incorporated into DNA was measured using a Beckman β-scintillation counter and is expressed as counts per minute.
Oral inoculation of calves and quantification of shedding.
All animal experiments were performed in accordance with the Animals (Scientific Procedures) Act 1986 and were approved by the local Ethical Review Committee. Conventional Friesian bull calves were fed on milk replacer twice daily with free access to water. In the week prior to infection calves were screened for the absence of STEC and Salmonella enterica by enrichment culture of rectal swabs on Sorbitol MacConkey agar containing tellurite, or on brilliant green agar (Oxoid, Basingstoke, United Kingdom), respectively. Calves with diarrhea or excreting STEC and/or Salmonella were excluded from the analysis. Total levels of immunoglobulin (Ig) in serum were measured 1 or 2 days prior to infection by zinc sulfate turbidity (ZST) assay. Only calves with a zinc sulfate turbidity measurement over 10 were used.
Two days before infection calves were transferred to high-security biocontainment accommodation and housed in tanks on tenderfoot mats. The calves were orally challenged with ca. 1010 CFU of STEC in 20 ml of antacid (1 g of MgO, 1 g of Mg trisilicate, and 1 g of NaHCO3 in H2O) just before their morning feed. The bacterial strains STEC O5, STEC O5 Δefa1, STEC O111, and STEC O111 efa1::kanr were each administered to two 4-day-old and two 11-day-old calves (16 animals in total). During the next 7 days, the calves were observed twice daily for signs of diarrhea, and fecal samples were taken. Viable STEC per gram of feces were enumerated by plating triplicate 10-fold serial dilutions onto sorbitol MacConkey agar medium containing nalidixic acid (20 μg/ml) and tellurite (2.5 μg/ml). The fecal shedding data were statistically analyzed after a 10log transformation for the effect of serotype, mutation, age of calf, and their interactions by means of an F test, with the data taken as repeated measurements (Proc Mixed, Statistical Analysis System; SAS Institute, Cary, N.C.).
Tissue sampling and microscopy.
To examine the effect of mutation of efa1 on bacterial association with the bovine intestinal mucosa, single 11-day-old calves were separately inoculated with STEC O111 or STEC O111 efa1::kanr as described above. Three days after inoculation samples of mucosa from the distal ileum, cecum, spiral colon, and rectum were excised ante mortem under terminal anesthesia and placed either in ice-cold mucosal medium for enumeration of adherent bacteria or fixed in 4% (wt/vol) paraformaldehyde in PBS. For confocal microscopy, 50-μm-thick unembedded sections were cut on a Leica vibrating microtome and permeabilized with 0.5% (vol/vol) Triton-X-100 in PBS for 30 min, and nonspecific binding sites were blocked with 0.5% (wt/vol) bovine serum albumin in PBS as described previously (42, 55). Bacteria were detected with 1:100 rabbit anti-O111 typing serum (Veterinary Laboratories Agency, Weybridge, United Kingdom) and 1:100 anti-rabbit Ig-Alexa568 (Molecular Probes, Leiden, The Netherlands). Filamentous actin was stained with 1:10 Oregon green 514-phalloidin (Molecular Probes). Stained sections were washed extensively with PBS, mounted with Vectashield (Vector Laboratories, Burlingame, Calif.), and analyzed at magnifications of ×630 to ×1,000 using a Leica TCS NT confocal laser scanning microscope with version 1.6.587 software. At least three 1-cm-long sections were examined.
For electron microscopy, paraformaldehyde-fixed colonic mucosa was transferred to 2% (vol/vol) glutaraldehyde in 50 mM phosphate buffer overnight at 4°C. The mucosa was then postfixed for 8 h at room temperature with 1% (wt/vol) osmium tetroxide in 50 mM phosphate buffer with the osmotic pressure adjusted to 350 mosmol by the addition of sucrose. Specimens were dehydrated in a graded series of ethanol solutions and embedded in Agar 100 resin (Agar Scientific Ltd., Stansted, United Kingdom). Sections (ca. 1 μm thick) were cut on a Leica UCT microtome and contrasted with uranyl acetate and lead citrate using a Leica EMStain machine (Leica Microsystems, Milton Keynes, United Kingdom). Sections were imaged using an FEI Technai 12 transmission electron microscope at 100 kV.
Detection of EspA and Tir by Western blotting.
To measure expression and secretion of LEE-encoded type III secreted proteins, bacteria were grown to mid-logarithmic phase (A600, ca. 0.6) in Dulbecco's modified Eagle medium. Cells from 1 ml of each culture were collected by centrifugation and resuspended in 50 μl of sample buffer. Secreted proteins were precipitated from 45 ml of filtered (pore size, 0.22 μm) culture supernatants with 10% (vol/vol) trichloroacetic acid on ice for 1 h, resuspended in 1% sodium dodecyl sulfate (SDS), and reprecipitated with acetone. Pellets were resuspended in 50 μl sample buffer. Proteins (ca. 25 μg) were resolved by SDS-10 polyacrylamide gel electrophoresis (SDS-10% PAGE) or -15% PAGE and transferred to nitrocellulose membranes using a Bio-Rad Semi-Dry Trans-Blot apparatus at 15 V for 1 h. Membranes were blocked with 5% (wt/vol) skim milk in Tris-buffered saline containing 0.1% (vol/vol) Tween 20. To detect EspA we used a combination of four anti-EspA monoclonal antibodies raised against purified EPEC EspA (23) and anti-mouse Ig-horseradish peroxidase conjugate (Amersham). Tir was detected using a rabbit polyclonal antiserum against EPEC Tir (22) and anti-rabbit Ig-horseradish peroxidase using an Amersham ECL chemiluminescence detection kit.
RESULTS
Efa1 influences adhesion of STEC O5 to cultured HeLa cells.
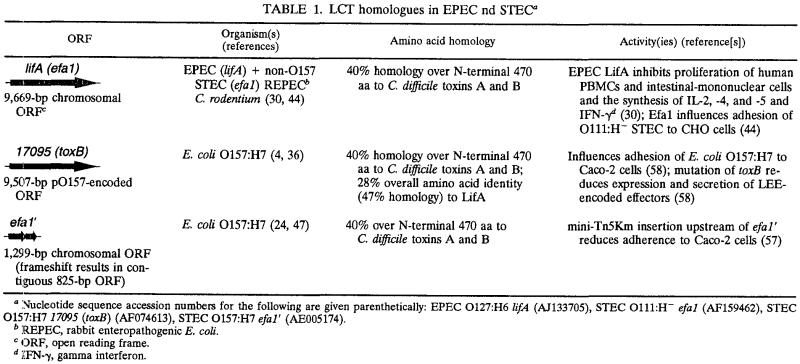
Consistent with the observations of Nicholls et al. (44), we observed a 19-fold reduction in adherence of STEC O5 Δefa1 compared to the wild-type strain in a standard HeLa cell adhesion assay (Fig. 1). Filamentous actin was nucleated under the few adherent efa1 mutant bacteria that could be detected (Fig. 1D), indicating that LEE-mediated attachment and rearrangement of cytoskeletal actin can still occur. No significant reduction in adherence of the EPEC ΔlifA mutant was observed (data not shown), as reported by Klapproth et al. (30). Mutation of the STEC O5 efa1 and EPEC lifA genes did not alter the in vitro growth rates of the mutants compared to the parent strains (data not shown).
FIG. 1.
Interaction of STEC O5 wild-type and Δefa1 mutant strains with cultured HeLa cells. Hemacolor-stained cells were examined for adherent bacteria at a magnification of ×630. (A) STEC O5; (B) STEC O5 Δefa1. (A and B) While the efa1 mutant strain adhered to HeLa cells in significantly lower numbers, no difference was observed in the ability of the wild-type and mutant strains to form microcolonies and to nucleate filamentous actin under the sites of bacterial attachment. (C) STEC O5; (D) STEC O5 Δefa1. (C and D) Bacteria were detected with rabbit anti-O5 lipopolysaccharide typing serum and anti-rabbit Ig-Alexa568 (red). Filamentous actin was stained with Oregon green 514-phalloidin (green). Magnification, ×1,000.
Lymphostatin inhibits the mitogen-activated proliferation of bovine lymphocytes.
We attempted to determine if lymphostatin is active against bovine peripheral blood lymphocytes and if STEC Efa1 also confers lymphostatin activity. Bovine PBMCs were stimulated with concanavalin A in the presence of lysates from EPEC, EPEC ΔlifA, STEC O5, STEC O5 Δefa1 or a laboratory strain of E. coli (HB101). Lysates from the EPEC wild-type strain, but not HB101, inhibited mitogen-activated proliferation (Fig. 2). Deletion of the EPEC lifA gene abrogated this inhibitory effect indicating that lymphostatin is active against bovine PBMCs. Lysates of the STEC O5 strains also inhibited the mitogen-activated proliferation of bovine PBMCs, however other cytotoxins such as Shiga toxin 1 and/or enterohemolysin may contribute to this effect since deletion of efa1 did not relieve the inhibitory effect.
FIG. 2.
LifA inhibits the mitogen-activated proliferation of bovine peripheral blood lymphocytes. Bovine PBMCs were stimulated with concanavalin A (5 μg/ml) in the presence of 50 μg of protein from E. coli HB101, EPEC, EPEC ΔlifA, STEC O5, or STEC O5 Δefa1, and proliferation was measured by incorporation of tritiated thymidine as described in Materials and Methods. The results are the means of quadruplicate measurements from two independent experiments ± standard errors of the means (error bars).
Efa1 influences STEC colonization and pathogenesis in calves.
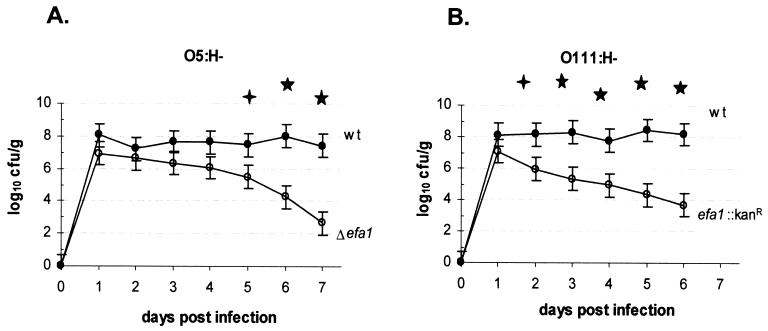
To investigate the role of Efa1 in colonization of the bovine intestine by STEC, we inoculated two 11-day-old conventional Friesian bull calves each with STEC O5, STEC O5 Δefa1, STEC O111 or STEC O111 efa1::kanr by the oral route. To investigate the induction of diarrhea, we also inoculated duplicate 4-day-old calves with the STEC O5 and O111 wild-type and efa1 mutant strains. No significant effect of age on the course of fecal excretion of STEC was detected (P = 0.08); therefore, we analyzed the data from all calves given the wild-type and efa1 mutant strain for the effect of the mutation. In all cases the STEC O5 and O111 efa1 mutant strains were shed in the feces in significantly lower numbers than the corresponding wild-type strains (P values < 0.05 at 6 days postinoculation) (Fig. 3).
FIG. 3.
Efa1 influences the course of fecal excretion of STEC serotypes O5 H− and O111:H− in calves. Mean fecal shedding data from four calves inoculated with STEC O5 and STEC O5 Δefa1 (A) and STEC O111 and STEC O111 efa1::kanr (B) are shown. No significant effect of age on fecal excretion was detected; therefore, the data obtained from 4- and 11-day-old calves inoculated with the same strain was pooled. Bacterial recoveries from the feces were measured twice each day. The mean daily fecal count ± standard error of the mean(error bars) is shown. Symbols: ✦, tendency towards significance (P < 0.1); ★, significant difference (P < 0.05).
Consistent with previous observations (6, 21), the STEC O5 strain induced dysentery in 4-day-old calves lasting 3 or 4 days from 3 days postinoculation, but did not induce dysentery in 11-day-old animals. The STEC O5 Δefa1 mutant did not induce dysentery in 4- or 11-day-old animals, and the calves remained clinically normal throughout with no overt pathology visible at postmortem examination.
The STEC O111 efa1 mutant exhibits reduced adherence to bovine colonic mucosa.
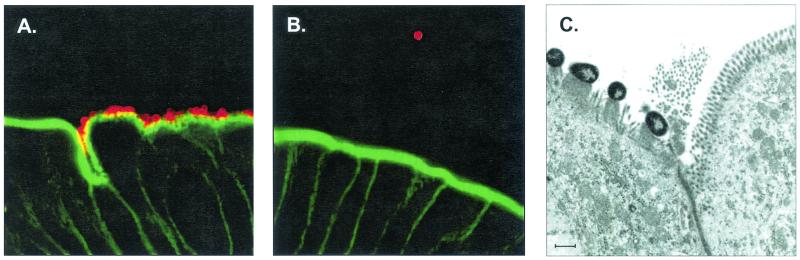
To determine if mutations affecting the efa1 gene influence adhesion to bovine intestinal epithelia we inoculated single 11 day calves with STEC O111 wild-type or STEC O111 efa1::kanr. Three days postinoculation samples of intestinal mucosa were removed into either fixative for microscopic analysis or mucosal medium for enumeration of bacteria. We detected large numbers of adherent wild-type STEC O111 on the epithelium of spiral colon in dense microcolonies by confocal microscopy, yet virtually no adherent bacteria at the same site in calves infected with the efa1::kanr mutant (Fig. 4A and B). Analysis of colonic mucosa from the STEC O111-infected calf by transmission electron microscopy showed the bacteria to have formed classical AE lesions (Fig. 4C). Consistent with the microscopic observations, we recovered a mean of 9.2 ± 0.06 log10 CFU/g STEC O111 wild-type from triplicate full-thickness biopsies of spiral colon but only < 2 log10 CFU/g by enrichment from the same site in the calf infected with the efa1::kanr mutant. Significantly fewer efa1 mutant bacteria were also recovered from ileal, cecal, and rectal mucosa compared to the wild-type strain (data not shown). Thus, mutation of efa1 significantly reduces association of STEC with the bovine intestinal epithelium.
FIG. 4.
Association of STEC O111 wild-type and efa1 mutant strains with bovine colonic epithelium. Confocal microscopy of 50-μm-thick vibrating microtome sections of spiral colon from 11-day-old calves 3 days after inoculation with STEC O111 wild-type (A) and STEC O111 efa1::kanr (B). Bacteria were detected with rabbit anti-O111 lipopolysaccharide typing serum and anti-rabbit Ig-Alexa568 (red). Filamentous actin was stained with Oregon green 514-phalloidin (green). Magnification, ×550. (C) Transmission electron micrograph showing AE lesions induced by STEC O111 on the same biopsy of colonic epithelium. Scale bar = 1 μm.
Deletion of efa1 influences expression and secretion of LEE-encoded type III secreted proteins in STEC.
To determine if efa1 mutations also influence bacterial adhesion and colonization by an indirect mechanism we analyzed expression and secretion of EspA and Tir by EPEC and the STEC O5 wild-type and mutant strains by Western blotting using specific antisera. Our analysis shows that expression and secretion of EspA and Tir by EPEC is not significantly affected by deletion of lifA (Fig. 5). In, the STEC O5 Δefa1 mutant however, we observed a reduction in the production and secretion of EspA and Tir. However, we and others have reported that efa1 mutations do not affect the ability of STEC to form microcolonies and to nucleate filamentous actin under the sites of bacterial attachment to cultured cells (Table 1) (44). This indicates that expression and secretion of LEE-encoded type III secreted proteins required for AE-lesion formation cannot be severely affected by mutation of efa1.
FIG. 5.
Effect of lifA and efa1 mutations on expression and secretion of LEE-encoded type III secreted proteins in EPEC and STEC O5, respectively. Whole-cell and secreted protein fractions of EPEC, EPEC ΔlifA, STEC O5, and STEC O5 Δefa1 were resolved by SDS-10% PAGE for detection of Tir and by SDS-15% PAGE for detection of EspA. Proteins (25 μg) were transferred to nitrocellulose membrane and probed using specific antisera. Equivalent loading of proteins was confirmed by Coomassie blue staining of gels (data not shown).
DISCUSSION
Colonization of the bovine intestinal tract is influenced by the bacterial outer membrane protein intimin (9). The LEE-encoded Tir, EspA, and EspB secreted proteins are also required for intestinal colonization by REPEC in a rabbit model following oral inoculation (1, 38). Intimin mutants do still colonize some compartments of the bovine intestinal tract, indicating that other accessory colonization factors may be required. Several accessory adhesins have been identified in STEC that mediate attachment to cultured cells, including EspA (14), Iha (56), Efa1 (44) and several other genes identified by transposon mutagenesis of E. coli O157:H7 (57). However, the role of these factors in colonization of the bovine intestine has so far not been tested.
Persistent colonization of the bovine intestine by STEC may also require bacterial interference with mucosal immune responses. Subversion of host cell functions and the modulation of mucosal immunity by enteric pathogens are emerging themes (12). Indeed it has been shown that EPEC uses the LEE-encoded type III protein secretion system to inhibit phagocytosis (18) and can also modulate apoptotic pathways (25), presumably to their own benefit.
We report here the effect of mutations in the efa1 gene of STEC serotypes O5 and O111 on intestinal colonization in 4- and 11-day-old calves. Deletion of the efa1 gene in an STEC O5 strain isolated from a case of calf dysentery dramatically reduced adherence of the bacteria to cultured cells, consistent with previous observations (44). Further, we found that lifA expressed by EPEC is active against bovine peripheral blood lymphocytes, as well as human and murine PBMCs (30, 32), indicating that the almost identical efa1 gene may modulate mucosal immune responses in the bovine host. We were unable to determine if Efa1 expressed by STEC confers lymphostatin activity against bovine PBMCs, probably owing to the secretion of other potent cytotoxins by our strains. Indeed it has been reported that Stx1 is a potent inhibitor of the proliferation of bovine lymphocytes (41). Until STEC Efa1 has been shown to act as a lymphotoxin in the same way as EPEC LifA we suggest that the efa1 nomenclature is more appropriate for the STEC gene despite the high levels of nucleotide and amino acid identity.
Deletion of the efa1 gene in STEC O5 strain significantly reduced fecal shedding of the organism following oral inoculation of 4- and 11-day-old conventional calves. Furthermore, the efa1 mutant strain induced no clinical signs in 4-day-old calves, whereas the wild-type strain caused bloody diarrhea lasting 3 to 4 days from 3 days postinoculation as reported previously (6, 21). Thus, Efa1 would appear to influence intestinal colonization and enteropathogenesis by STEC in calves. We were unable to clone the efa1 gene for the purpose of trans-complementation of the STEC O5 efa1 mutant. Cosmids containing efa1/lifA are underrepresented in cosmid libraries and are frequently unstable (32, 44), suggesting that overexpression of the gene may be toxic. A cosmid containing EPEC lifA is available (pIV-8-A) (32); however, we were unable to introduce this cosmid into the STEC O5 efa1 mutant by electroporation, possibly owing to incompatibility with one of the six endogenous plasmids (21). Rather, we tested an independent efa1 mutant in a different STEC serotype for its ability to colonize the bovine intestinal tract. The STEC O111 efa1::Kanr mutant (44) was also shed in significantly lower numbers than the corresponding wild-type strain. Thus, the possibility that mutations outside efa1 are responsible for the observed reduction in intestinal colonization is minimal. We were unable to introduce pIV-8-A into the STEC O111 efa1::kanr mutant since this strain is naturally resistant to the cosmid-encoded antibiotic resistance.
Consistent with the reduced fecal shedding of the STEC efa1 mutants, we recovered significantly fewer efa1 mutant bacteria from colonic mucosa 3 days after oral inoculation of calves compared to the wild type. Further, we observed very few adherent bacteria on the colonic epithelium of calves infected with the STEC O111 efa1 mutant by confocal microscopy. In contrast, the wild-type strain formed extensive microcolonies and AE lesions in the colon at 3 days postinoculation. Thus, Efa1 influences association of non-O157 STEC with bovine intestinal mucosa.
While E. coli O157:H7 lacks the full-length efa1 gene a truncated version of efa1 exists in the chromosome (24, 47). Recently, Tatsuno et al. (57) reported that a mini-Tn5Km2 transposon insertion upstream of the truncated efa1 gene reduces adherence of E. coli O157:H7 to Caco-2 cells (57). Further, E. coli O157:H7 strains encode a full-length homologue of efa1 (toxB/l7095) on the pO157 virulence plasmid (4, 30, 36, 44). The pO157 plasmid confers upon laboratory strains of E. coli the ability to inhibit IL-2 and IL-4 synthesis in mitogen-activated human PBMCs, and it has been assumed that this lymphostatin-like activity is determined by toxB (30). However, pO157 encodes several other putative virulence factors including enterohemolysin, an extracellular serine protease, catalase-peroxidase and a type II secretion system that may mediate the inhibitory effect.
The toxB gene, like efa1, has also been implicated in adherence. E. coli O157:H7 strains containing derivatives of pO157 that lack toxB exhibit reduced adherence to cultured epithelial cells (58). The authors reported that toxB indirectly influences adherence by modulating the production and secretion of LEE-encoded type III secreted proteins that are required for the formation of AE lesions (58). Mindful of the possibility of pleiotropic effects of efa1 mutations we analyzed the production and secretion of EspA and Tir by our EPEC and STEC O5 wild-type and mutant strains by Western blotting using specific antisera. We observed no reduction in production or secretion of EspA or Tir in the EPEC lifA mutant strain. However, a reduction in EspA and Tir production and secretion was observed in the STEC O5 efa1 mutant. This reduction in EspA and Tir secretion was insufficient to prevent the nucleation of filamentous actin under the sites of STEC attachment to cultured cells (this study, 44). Since actin condensation under adherent bacteria is dependent upon the translocation of LEE-encoded type III secreted effectors, our data would indicate that expression and secretion of Tir and EspA cannot be severely affected. Considerable natural variation in the level of secretion of the LEE-encoded Tir and EspD proteins has been observed among STEC O157:H7 strains isolated from cattle (40); however, it is not known if this correlates with the ability of the strains to efficiently colonize the bovine intestine.
It is unclear why mutation of EPEC lifA produced different phenotypes to mutation of the STEC O5 efa1 gene. EPEC lifA mutants do not show significantly reduced adherence to cultured cells (this study and reference 30); however, EPEC expresses type IV bundle forming pili which may mediate the initial interactions with cultured cells and mask the effect of other adhesins. The reasons why mutation of efa1 in STEC, but not lifA in EPEC, reduced expression and secretion of the LEE-encoded effectors EspA and Tir are obscure. Studies on the transcription and translation of LEE genes in EPEC and STEC efa1/lifA mutants are required to fully characterize this effect.
Efa1 is only the second STEC protein to be implicated in intestinal colonization in calves. Efa1 may influence colonization of the bovine intestine in several ways. It may act as an adhesin per se, as suggested by Nicholls et al. (44), it may promote STEC survival in the bovine gut by modulating mucosal immunity, or it may act indirectly by influencing the expression and secretion of LEE-encoded proteins or other membrane associated proteins that influence colonization. These activities are not necessarily mutually exclusive and are the subject of ongoing studies in our laboratory.
Acknowledgments
This work was supported by grants from the Biotechnology and Biological Sciences Research Council, United Kingdom (grant 201/D10261 to T.W., G.F., and A.P.) and the European Union (E.U. project QLK2-2000-00600).
We are grateful to Paul Monaghan and Pippa Hawes for assistance with the transmission electron microscopy.
Editor: A. D. O'Brien
REFERENCES
- 1.Abe, A., U. Heczko, R. G. Hegele, and B. B. Finlay. 1998. Two enteropathogenic Escherichia coli type III secreted proteins, EspA and EspB, are virulence factors. J. Exp. Med. 188:1907-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown, C. A., B. G. Harmon, T. Zhao, and M. P. Doyle. 1997. Experimental Escherichia coli O157:H7 carriage in calves. Appl. Environ. Microbiol. 63:27-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burland, V., Y. Shao, N. T. Perna, G. Plunkett, H. J. Sofia, and F. R. Blattner. 1998. The complete DNA sequence and analysis of the large virulence plasmid of Escherichia coli O157:H7. Nucleic Acids Res. 26:4196-4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Busch, C., F. Hofmann, J. Selzer, S. Munro, D. Jeckel, and K. Aktories. 1998. A common motif of eukaryotic glycosyltransferases is essential for the enzyme activity of large clostridial cytotoxins. J. Biol. Chem. 273:19566-19572. [DOI] [PubMed] [Google Scholar]
- 6.Chanter, N., G. A. Hall, A. P. Bland, A. J. Hayle, and K. R. Parsons. 1986. Dysentery in calves caused by an atypical strain of Escherichia coli (S102-9). Vet. Microbiol. 12:241-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chanter, N., J. H. Morgan, J. C. Bridger, G. A. Hall, and D. J. Reynolds. 1984. Dysentery in gnotobiotic calves caused by atypical Escherichia coli. Vet. Rec. 114:71.. [DOI] [PubMed] [Google Scholar]
- 8.Cray, Jr, W. C., and H. W. Moon. 1995. Experimental infection of calves and adult cattle with Escherichia coli O157:H7. Appl. Env. Microbiol. 61:1586-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean-Nystrom, E. A., B. T. Bosworth, H. W. Moon, and A. D. O'Brien. 1998. Escherichia coli O157:H7 requires intimin for enteropathogenicity in calves. Infect. Immun. 66:4560-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dean-Nystrom, E. A., B. T. Bosworth, W. C. Cray, Jr., and H. W. Moon. 1997. Pathogenicity of Escherichia coli O157:H7 in the intestines of neonatal calves. Infect. Immun. 65:1842-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeVinney, R., M. Stein, D. Reinscheid, A. Abe, S. Ruschkowski, and B. B. Finlay. 1999. Enterohemorrhagic Escherichia coli O157:H7 produces Tir, which is translocated to the host cell membrane but is not tyrosine phosphorylated. Infect. Immun. 67:2389-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Donnenberg, M. S. 2000. Pathogenic strategies of enteric bacteria. Nature 406:768-774. [DOI] [PubMed] [Google Scholar]
- 13.Donnenberg, M. S., and J. B. Kaper. 1991. Construction of an eaeA deletion mutant of enteropathogenic Escherichia coli by using a positive-selection suicide vector. Infect. Immun. 59:4310-4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ebel, F., T. Podzadel, M. Rohde, A. U. Kresse, S. Krämer, C. Deibel, C. A. Guzmán, and T. Chakraborty. 1998. Intial binding of Shiga toxin-producing Escherichia coli to host cells and subsequent induction of actin rearrangements depend on filamentous EspA-containing surface appendages. Mol. Microbiol. 30:147-161. [DOI] [PubMed] [Google Scholar]
- 15.Frankel, G., A. D. Phillips, L. Rosenshine, G. Dougan, J. B. Kaper, and S. Knutton. 1998. Enteropathogenic and enterohaemorrhagic Escherichia coli: more subversive elements. Mol. Microbiol. 30:911-921. [DOI] [PubMed] [Google Scholar]
- 16.Frankel, G., O. Lider, R. Hershkoviz, A. P. Mould, S. G. Kachalsky, D. C. A. Candy, L. Cahalon, M. J. Humphries, and G. Dougan. 1996. The cell-binding domain of intimin from enteropathogenic Escherichia coli binds to beta1 integrins. J. Biol. Chem. 271:20359-20364. [DOI] [PubMed] [Google Scholar]
- 17.Gansheroff, L. J., and A. D. O'Brien. 2000. Escherichia coli O157:H7 in beef cattle presented for slaughter in the U.S.: higher prevalence rates than previously estimated. Proc. Natl. Acad. Sci. USA 97:2959-2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goosney, D. L., J. Celli, B. Kenny, and B. B. Finlay. 1999. Enteropathogenic Escherichia coli inhibits phagocytosis. Infect. Immun. 67:490-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Griffin, P. M. 1995. Escherichia coli O157:H7 and other enterohaemorrhagic Escherichia coli, p. 739-761. In M. J. Blaser, P. D. Smith, J. I. Ravdin, H. B. Greenberg, and R. L. Guerrant (ed.), Infections of the gastrointestinal tract. Raven Press, New York, N.Y.
- 20.Hall, G. A., C. R. Dorn, N. Chanter, S. M. Scotland, H. R. Smith, and B. Rowe. 1990. Attaching and effacing lesions in vivo and adhesion to tissue culture cells of vero-cytotoxin-producing Escherichia coli belonging to serogroups O5 and O103. J. Gen. Microbiol. 136:779-786. [DOI] [PubMed] [Google Scholar]
- 21.Hall, G. A., D. J. Reynolds, N. Chanter, J. H. Morgan, K. R. Parsons, T. G. Debney, A. P. Bland, and J. C. Bridger. 1985. Dysentery caused by Escherichia coli (S102-9) in calves: natural and experimental disease. Vet. Pathol. 22:156-163. [DOI] [PubMed] [Google Scholar]
- 22.Hartland, E. L., M. Batchelor, R. M. Delahay, C. Hale, S. Matthews, G. Dougan, S. Knutton, I. Connerton, and G. Frankel. 1999. Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32:151-158. [DOI] [PubMed] [Google Scholar]
- 23.Hartland, E. L., S. J. Daniell, R. M. Delahay, B. C. Neves, T. Wallis, R. K. Shaw, C. Hale, S. Knutton, and G. Frankel. 2000. The type III protein translocation system of enteropathogenic Escherichia coli involves EspA-EspB protein interactions. Mol. Microbiol. 35:1483-1492. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi, T., K. Makino, M. Ohnishi, K. Kurokawa, K. Ishii, K. Yokoyama, C. G. Han, E. Ohtsubo, K. Nakayama, T. Murata, M. Tanaka, T. Tobe, T. Iida, H. Takami, T. Honda, C. Sasakawa, N. Ogasawara, T. Yasunaga, S. Kuhara, T. Shiba, M. Hattori, and H. Shinagawa. 2001. Complete genome sequence of enterohemorrhagic Escherichia coli O157:H7 and genomic comparison with a laboratory strain K-12. DNA Res. 8:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Heczko, U., C. M. Carthy, B. A. O'Brien, and B. B. Finlay. 2001. Decreased apoptosis in the ileum and ileal Peyer's patches: a feature after infection with rabbit enteropathogenic Escherichia coli O103. Infect. Immun. 69:4580-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hofmann, F., C. Busch, and K. Aktories. 1998. Chimeric clostridial cytotoxins: identification of the N-terminal region involved in protein substrate recognition. Infect. Immun. 66:1076-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and L. R. Pease. 1989. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 28.James, S. P., and J. M. Klapproth. 1996. Major pathways of mucosal immunity and inflammation: cell activation, cytokine production and the role of bacterial factors. Aliment. Pharmacol. Ther. 10(Suppl. 2):1-9. [DOI] [PubMed] [Google Scholar]
- 29.Just, I., F. Hofmann, and K. Aktories. 2000. Molecular mode of action of the large clostridial cytotoxins. Curr. Top. Microbiol. Immunol. 250:55-83. [DOI] [PubMed] [Google Scholar]
- 30.Klapproth, J. M., I. C. Scaletsky, B. P. McNamara, L. C. Lai, C. Malstrom, S. P. James, and M. S. Donnenberg. 2000. A large toxin from pathogenic Escherichia coli strains that inhibits lymphocyte activation. Infect. Immun. 68:2148-2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klapproth, J. M., M. S. Donnenberg, J. M. Abraham, and S. P. James. 1996. Products of enteropathogenic E. coli inhibit lymphokine production by gastrointestinal lymphocytes. Am. J. Physiol. 271:G841-G848. [DOI] [PubMed] [Google Scholar]
- 32.Klapproth, J. M., M. S. Donnenberg, J. M. Abraham, H. L. Mobley, and S. P. James. 1995. Products of enteropathogenic Escherichia coli inhibit lymphocyte activation and lymphokine production. Infect. Immun. 63:2248-2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Knutton, S., T. Baldwin, P. H. Williams, and A. S. McNeish. 1989. Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect. Immun. 57:1290-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levine, M. M., E. J. Berquist, D. R. Nalin, D. H. Waterman, R. B. Hornick, C. R. Young, S. Stoman, and B. Rowe. 1978. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet i:119-122. [DOI] [PubMed]
- 35.Mainil, J. 1999. Shiga/verocytotoxins and Shiga/verotoxigenic Escherichia coli in animals. Vet. Res. 30:235-257. [PubMed] [Google Scholar]
- 36.Makino, K., K. Ishii, T. Yasunaga, M. Hattori, K. Yokoyama, C. H. Yutsudo, Y. Kubota, Y. Yamaichi, T. Iida, K. Yamamoto, T. Honda, C. G. Han, E. Ohtsubo, M. Kasamatsu, T. Hayashi, S. Kuhara, and H. Shinagawa. 1998. Complete nucleotide sequences of 93-kb and 3.3-kb plasmids of an enterohemorrhagic Escherichia coli O157:H7 derived from Sakai outbreak. DNA Res. 5:1-9. [DOI] [PubMed] [Google Scholar]
- 37.Malstrom, C., and S. James. 1995. Inhibition of murine splenic and mucosal lymphocyte function by enteric bacterial products. Infect. Immun. 66:3120-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marchés, O., J.-P. Nougayrède, S. Boullier, J. Mainil, G. Charlier, I. Raymond, P. Pohl, M. Boury, J. De Rycke, A. Milon, and E. Oswald. 2000. Role of Tir and Intimin in the virulence of rabbit enteropathogenic Escherichia coli serotype O103:H2. Infect. Immun. 68:2171-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McKee, M. L., and A. D. O'Brien. 1996. Truncated enterohemorrhagic Escherichia coli (EHEC) O157:H7 intimin (EaeA) fusion proteins promote adherence of EHEC strains to HEp-2 cells. Infect. Immun. 64:2225-2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McNally, A., A. J. Roe, S. Simpson, F. M. Thomson-Carter, D. E. E. Hoey, C. Currie, T. Chakraborty, D. G. E. Smith, and D. L. Gally. 2001. Differences in levels of secreted locus of enterocyte effacement proteins between human disease-associated and bovine Escherichia coli O157. Infect. Immun. 69:5107-5114. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Menge, C., L. H. Wieler, T. Schlapp, and G. Baljer. 1999. Shiga toxin 1 from Escherichia coli blocks activation and proliferation of bovine lymphocyte subpopulations in vitro. Infect. Immun. 67:2209-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monaghan, P., P. R. Watson, H. Cook, L. Scott, T. S. Wallis, and D. Robertson. 2001. An improved method for preparing thick sections for immuno/histochemistry and confocal microscopy and its use to identify rare events. J. Microsc. 203:223-226. [DOI] [PubMed] [Google Scholar]
- 43.Nataro, J. P., and J. B. Kaper. 1998. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 11:142-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicholls, L., T. H. Grant, and R. M. Robins-Browne. 2000. Identification of a novel genetic locus that is required for in vitro adhesion of a clinical isolate of enterohaemorrhagic Escherichia coli to epithelial cells. Mol. Microbiol. 35:275-288. [DOI] [PubMed] [Google Scholar]
- 45.Paton, J. C., and A. W. Paton. 1998. Pathogenesis and diagnosis of Shiga-toxin producing Escherichia coli infections. Clin. Microbiol. Rev. 11:450-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, G. R., K. J. Bazeley, J. R. Jones, R. F. Gunning, M. J. Green, A. Cookson, and M. J. Woodward. 1999. Attaching and effacing lesions in the large intestine of an eight-month-old heifer associated with Escherichia coli O26 infection in a group of animals with dysentery. Vet. Record 145:370-373. [DOI] [PubMed] [Google Scholar]
- 47.Perna, N. T., G. Plunkett, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 48.Posfai, G., M. D. Koob, H. A. Kirkpatrick, and F. R. Blattner. 1997. Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J. Bacteriol. 179:4426-4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roe, A. J., and D. L. Gally. 2000. Enteropathogenic and enterohaemorrhagic Escherichia coli and diarrhoea. Curr. Opin. Infect. Dis. 13:511-517. [DOI] [PubMed] [Google Scholar]
- 50.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
- 51.Simon, R., U, Preifer, and A. Puhler. 1983. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in Gram-negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 52.Sinclair, J. F., and A. D. O'Brien. 2002. Cell surface-localized nucleolin is a eukaryotic receptor for the adhesin intimin-gamma of enterohemorrhagic Escherichia coli O157:H7. J. Biol. Chem. 277:2876-2885. [DOI] [PubMed] [Google Scholar]
- 53.Slutsker, L., A. A. Ries, K. D. Greene, J. G. Wells, L. Hutwagner, and P. M. Griffin. 1997. Escherichia coli O157:H7 diarrhea in the United States: clinical and epidemiological features. Ann. Intern. Med. 126:505-513. [DOI] [PubMed] [Google Scholar]
- 54.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. X. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, M. P., O. Marches, J. Campbell, V. Huter, G. Frankel, A. D. Phillips, E. Oswald, and T. S. Wallis. 2001. Intimin, Tir and Shiga toxin 1 do not influence enteropathogenic responses to Shiga toxin-producing Escherichia coli in bovine ligated intestinal loops. Infect. Immun. 70:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tarr, P. I., S. S. Bilge, J. C. Vary, Jr., S. Jelacic, R. L. Habeeb, T. R. Ward, M. R. Baylor, and T. E. Besser. 2000. Iha: a novel Escherichia coli O157:H7 adherence-conferring molecule encoded on a recently acquired chromosomal island of conserved structure. Infect. Immun. 68:1400-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tatsuno, I., H. Kimura, A. Okutani, K. Kanamaru, H. Abe, S. Nagai, K. Makino, H. Shinagawa, M. Yoshida, K. Sato, J. Nakamoto, T. Tobe, and C. Sasakawa. 2000. Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect. Immun. 68:5943-5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tatsuno, I., M. Horie, H. Abe, T. Miki, K. Makino, H. Shinagawa, H. Taguchi, S. Kamiya, T. Hayashi, and C. Sasakawa. 2001. toxB gene on pO157 of enterohaemorrhagic Escherichia coli O157:H7 is required for full epithelial cell adherence phenotype. Infect. Immun. 69:6660-6669. [DOI] [PMC free article] [PubMed] [Google Scholar]