Abstract
Ferric uptake regulator (Fur) and Fur-like proteins form an important family of transcriptional regulators in many bacterial species. In this work we have characterized a Fur-like protein, the peroxide regulator PerR, in an M1 serotype of Streptococcus pyogenes. To determine the role of PerR in S. pyogenes, we inactivated the gene by allelic replacement. PerR-deficient bacteria showed 48% reduction of 55Fe incorporation from the culture medium. Transcriptional analysis revealed that mtsA, encoding a metal-binding protein of an ABC transporter in S. pyogenes, was transcribed at lower levels than were wild-type cells. Although total iron accumulation was reduced, the growth of the mutant strain was not significantly hampered. The mutant showed hyperresistance to hydrogen peroxide, and this response was induced in wild-type cells by growth in aerobiosis, suggesting that PerR acts as an oxidative stress-responsive repressor. PerR may also participate in the response to superoxide stress, as the perR mutant was more sensitive to the superoxide anion and had a reduced transcription of sodA, which encodes the sole superoxide dismutase of S. pyogenes. Complementation of the mutation with a functional perR gene restored 55Fe incorporation, response to peroxide stress, and transcription of both mtsA and sodA to levels comparable to those of wild-type bacteria. Finally, the perR mutant was attenuated in virulence in a murine air sac model of infection (P < 0.05). These results demonstrate that PerR is involved in the regulation of iron homeostasis and oxidative stress responses and that it contributes to the virulence of S. pyogenes.
Oxidative stress is generated by exposure to reactive oxygen species (ROS), such as superoxide anion (O2−·), hydrogen peroxide (H2O2), and the highly toxic hydroxyl radical (OH·), which can damage nucleic acids, proteins, and cell membranes. As a defense, cells have evolved inducible responses to protect themselves against oxidative stress (18, 42, 51). In bacteria, a large body of evidence, originating mainly from studies with Escherichia coli and Salmonella, has shown that peroxide and superoxide stress responses are distinct (18). Superoxide dismutase (SOD) represents the first line of defense against superoxide stress by converting O2−· into H2O2 and O2, thereby protecting cells from the toxic effects of O2−· (21).
There is an intimate relationship between oxidative stress and iron metabolism. Iron is both an essential cofactor and a potentially hazardous metal participating in the production of ROS (27). Lack of iron regulation may impose oxidative stress upon cells (54), and many microorganisms have evolved systems that couple control of iron homeostasis to protection against ROS (7, 9, 16, 34, 35, 53, 56, 60). Bacterial iron metabolism is regulated by Fur (ferric uptake regulator) in gram-negative bacteria and by DtxR (diphtheria toxin repressor) in most gram-positive bacteria (for reviews see references 28 and 31). More recently, Fur and Fur-like proteins have also been characterized in gram-positive species (4, 22, 34, 35, 38, 39, 58). Both Bacillus subtilis and Staphylococcus aureus contain three distinct Fur homologues: Fur, which typically controls iron homeostasis (4, 35); the H2O2- and metal ion-responsive peroxide regulator PerR (4, 34); and Zur, which is involved in the regulation of zinc homeostasis (22, 39). In contrast, Streptococcus pyogenes seems to have a single PerR protein, which was recently reported to be involved in the resistance response to peroxides (38). Fur proteins generally act as transcriptional repressors by Fe2+-dependent binding to specific sequences (so-called iron boxes) in the promoters of iron-regulated genes (1, 14, 28). However, Fur can also regulate transcription of iron-activated genes (13) and can even behave as a positive regulator, either directly or indirectly (15, 20, 29, 44). Fur is now considered a pleiotropic regulator, as it can control not only iron acquisition systems but also the most diverse processes such as acid and oxidative stress responses, chemotaxis, swarming, bioluminescence, metabolic pathways, and production of virulence factors (reviewed in references 17, 28, and 48). With respect to oxidative stress, Fur regulation of SOD has been observed in several bacterial species (53). Interestingly, in E. coli, Fur represses transcription of manganese-dependent sodA (11, 52), but it is a positive regulator of iron-dependent sodB (15, 44).
S. pyogenes, or group A Streptococcus, is an important human pathogen causing various diseases, from mild suppurative throat and skin infections to life-threatening invasive diseases (12). S. pyogenes virulence determinants are often regulated in response to growth phase and environmental signals, such as temperature, osmolarity, pH, iron limitation, and O2 and CO2 tension (5, 24, 26, 40, 41, 50, 55, 57). S. pyogenes is a facultative anaerobe and lacks catalase. However, it has other defense mechanisms against oxidative stress, including an NADH-oxidase (25), a single Mn-dependent SOD (24), two peroxidases, and the PerR regulator (38).
Starting from a previous observation that PerR is involved in the inducible response to H2O2 in an S. pyogenes M6 serotype strain (38), in the present work we have characterized in more detail the phenotypes of a perR mutant in an M1 serotype. The results show that PerR affects iron metabolism and peroxide and superoxide stress responses and is necessary for full virulence.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. E. coli was cultured in Luria-Bertani medium (Difco, Detroit, Mich.) at 37°C under aerobic conditions. S. pyogenes was grown in Todd-Hewitt medium (Difco) supplemented with 0.2% yeast extract (THY) or in metal-depleted THY (Cx-THY) at 37°C with 5% CO2. Cx-THY was obtained by overnight (ON) treatment of THY with 5% (wt/vol) Chelex 100 resin (Sigma, St. Louis, Mo.), sterile filtration, and addition of 100 μM CaCl2 and 2 mM MgCl2. Aerobic growth was performed in rotating Erlenmeyer flasks at 37°C in THY medium, which was preequilibrated for at least 90 min prior to the inoculation of bacteria. For maximal cysteine proteinase expression, bacteria were cultivated in C medium (23). When appropriate, antibiotics were used at the following concentrations: kanamycin (Sigma) at 100 (for E. coli) or 175 (for S. pyogenes) μg/ml and spectinomycin (Sigma) at 100 (for E. coli) or 70 (for S. pyogenes) μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | recA1 endA1 hsdR17 | Invitrogenb |
| S. pyogenes | ||
| AP1 | Serotype M1, wild type | World Health Organization Prague collectionc |
| SR301 | perR::kan Kmr | This study |
| SR304 | perR::kan/pLU21 (perR+) Kmr Spr | This study |
| RJ1 | mtsA::kan Kmr | 37 |
| Plasmids | ||
| pFW13 | Allelic replacement vector, Kmr | 46 |
| pLZ12-Spec | E. coli-Streptococcus vector, Spr | 36 |
| pLU11 | perR::kan in pFW13, Kmr | This study |
| pLU21 | perR+ in pLZ12-Spec, Spr | This study |
Kmr and Spr indicate resistance to kanamycin and spectinomycin, respectively.
Invitrogen, Life Technologies, Inc., Carlsbad, Calif.
World Health Organization Collaborating Center for Reference and Research on Streptococci, Institute of Hygiene, Prague, Czech Republic.
Construction of strains.
The primers used for constructing the strains are shown in Table 2. Allelic replacement mutagenesis of perR was performed in two steps. First, an 878-bp fragment comprising the first 201 bp of perR together with a 677-bp region upstream of the gene, was amplified by PCR from AP1 chromosomal DNA with the primers PerR-forward (BglII) and PerR-reverse (SacI). This PCR product was digested with BglII and SacI and inserted into the BamHI and SacI sites of plasmid pFW13 (46), generating plasmid pLU10. For the second cloning, a 999-bp PCR product, including the last 105 bp of perR and the following 894 bp of the region downstream of the perR stop codon, was generated with the primers PerR-forward (ApaI) and PerR-reverse (BlnI). Following digestion with ApaI and BlnI, the PCR fragment was ligated into the corresponding restriction sites of pLU10. The new plasmid was denominated pLU11 (Table 1). Standard procedures for cloning, transformation, and analysis of E. coli clones were used (49). Plasmid pLU11 was used to transform competent cells of the S. pyogenes AP1 strain as previously described (30). Transformants were selected on THY plates containing kanamycin and analyzed by PCR and Southern blotting by using standard techniques (49). Genomic DNA was prepared as previously described (45), modified in the initial incubation step by the addition of 500 U of mutanolysin (Sigma)/ml and 15 mg of lysozyme (Sigma)/ml. A clone with the expected mutant perR allele was selected and denominated SR301. In order to verify that the mutation introduced in perR could be complemented in trans with a wild-type perR allele, a 1,295-bp fragment comprising the whole perR gene, including a putative promoter and a transcriptional terminator, was amplified with the primers PerR-forward (EcoRI) and PerR-reverse (SphI). The PCR product was digested with EcoRI and SphI and ligated to plasmid pLZ12-Spec (36) cut with the same enzymes, resulting in plasmid pLU21 (Table 1). Plasmid pLU21 was introduced into the mutant SR301 strain, and clones were selected on THY plates containing kanamycin and spectinomycin. The presence of the plasmid was verified by using the Concert Rapid Plasmid Miniprep system (GibcoBRL) and restriction mapping. One transcomplemented clone was chosen and denominated SR304.
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a | Target |
|---|---|---|
| PerR-forward (BglII) | GCAGATCTATTTCTCATTTTGACAAC | perR |
| PerR-reverse (SacI) | CCGGAGCTCATAGACTGTGGCAAGGC | perR |
| PerR-forward (ApaI) | ATTGGGCCCATTGCCAAAGAAGCCC | perR |
| PerR-reverse (BlnI) | ATCCTAGGTTGTTAAACTGTACTGTGC | perR |
| PerR-forward (EcoRI) | TCGAATTCTTTTCTTACTTGTTATTTCTC | perR |
| PerR-reverse (SphI) | ATGAGCATGCATGGTTCTTTCCTTTAATTCC | perR |
| MtsA-forward | TACGAACCATTACCAGAAGAT | mtsA |
| MtsA-reverse | CTTCTTCTTCGGTGTTAATTTCCCAG | mtsA |
| FhuD-forward | ATAGCGGGCTGAAAGATTGAGGTC | fhuD |
| FhuD-reverse | CAAGATTATTACCACCAAAACAGGG | fhuD |
| SodA-forward | TTTACCAGAACTTCCATACGCG | sodA |
| SodA-reverse | TCTTTTTGAGATTGGAAACCC | sodA |
| M1-forward | GGCTAACGGTGATGGTAATCC | emm1 |
| M1-reverse | CGCTGGTCTTCTAAGGCTTG | emm1 |
| 16S-forward | ATGTTAGTAATTTAAAAGGGG | 16S rRNA |
| 16S-reverse | TTTAAGAGATTAGCTTGCCGT | 16S rRNA |
Restriction sites for subsequent cloning of the PCR products are underlined. Primer sequences were chosen by using the complete genome of the M1 serotype strain SF370 (19).
Total 55Fe incorporation assays.
S. pyogenes strains AP1, SR301, and SR304 were grown ON in THY broth. Each ON culture was washed once in Cx-THY and inoculated 1:100 in 1 ml of fresh Cx-THY containing 0.25 μCi of 55FeCl3 (Amersham Pharmacia Biotech, Uppsala, Sweden). In parallel, 10-ml reference culture tubes were inoculated and their growth was carefully followed until the optical density at 620 nm (OD620) was ≅0.6, at which point the corresponding 1-ml samples were collected. To ensure that equivalent amounts of cells were harvested, viable counts from the reference tubes were performed. Then samples were spun down (10,000 × g, 3 min), and supernatants were collected. Bacteria were washed with 1 ml of fresh Cx-THY for a total of three washes. Finally, bacterial pellets were resuspended in 200 μl of fresh Cx-THY and mixed with 5 ml of Ready Safe scintillation cocktail (Beckman). Five milliliters of scintillation fluid was also added to the supernatants. Radioactivity was measured with a β-counter calibrated for 55Fe. The percentage of 55Fe associated with the bacterial pellet was calculated by dividing the number of counts per minute of the pellet by the number of counts per minute of the pellet plus the supernatant. The experiments were performed at least three times in duplicate or triplicate samples.
Oxidative stress assays.
ON cultures of S. pyogenes AP1, SR301, and SR304 strains were reinoculated 1:250 in fresh THY or Cx-THY and incubated at 37°C either with 5% CO2 or under aerobic conditions. When the OD620 of the cells reached ≅0.5, a 100-μl aliquot was removed (time zero) and kept on ice, and 5 mM H2O2 was added to the bacterial cultures. Samples were collected at 15, 30, 60, and 90 min after the addition of H2O2. Catalase (5 mg/ml; Sigma) was added, and the tubes were put on ice. Appropriate bacterial dilutions were plated onto THY agar plates. The cells were counted, and the percentage of survival was calculated by dividing the number of CFU at different time points with the initial number of CFU at time zero. Experiments were performed four to five times with duplicate samples, and the results were expressed as means ± standard deviations (SD). For growth in the presence of paraquat (methyl viologen; Sigma), ON cultures of AP1 and SR301 were inoculated 1:1,000 in fresh THY containing 2 or 10 mM paraquat. Samples with no paraquat were also used as controls. After 15 h of incubation at 37°C with 5% CO2, the OD620 was recorded. Experiments were performed five times with duplicate or triplicate samples.
Protein methods.
The presence of M1 protein and C5a peptidase at the surface of AP1 and SR301 bacteria was investigated by cleavage with cyanogen bromide (CNBr) and S. pyogenes cysteine proteinase (SCP), respectively. Digestion with CNBr was performed as described previously (10), and solubilized protein fragments were analyzed by sodium dodecyl sulfate (SDS)-10% polyacrylamide gel electrophoresis (PAGE) and Coomassie brilliant blue staining. Activation of SCP and proteolytic digestions were carried out as previously described (3). Surface proteins released by SCP were analyzed by SDS-10% PAGE, blotted onto polyvinylidene difluoride membranes (Immobilon-P; Millipore, Bedford, Mass.), and treated with anti-C5a peptidase rabbit serum (1:3,000). The release of SCP into the culture medium of wild-type and mutant strains grown ON in C medium was assayed by precipitation of culture supernatants with 5% trichloroacetic acid, subsequent protein separation by SDS-10% PAGE, and immunoblotting with anti-SCP rabbit polyclonal antibodies (1:50,000). Peroxidase-conjugated protein A (1:5,000; Sigma) was used as a secondary antibody in all immunoblots. After washing, proteins were detected by chemiluminescence as previously described (43). Membranes were dried and autoradiographed with X-Omat AR films (Kodak).
RNA preparation and Northern blotting.
To prepare total RNA, S. pyogenes strains AP1, SR301, and SR304 were cultured in 20 ml of THY and harvested in the mid-logarithmic growth phase (OD620 ≅ 0.5). Cells were centrifuged (5,000 × g, 10 min, 4°C); resuspended in 125 μl of 100 mM Tris-HCl, pH 8.0, containing 15 mg of lysozyme (Sigma)/ml, 100 U of mutanolysin (Sigma), and 20 mM ribonucleoside vanadyl complexes (Sigma); and incubated for 20 min at 37°C. Then, 2.5 ml of TriReagent (Sigma) were added to each sample and incubated for 5 min at room temperature before the addition of 0.5 ml of chloroform. Following extraction and precipitation, RNA was resuspended in 50 μl of 0.1% (vol/vol) diethylpyrocarbonate (Sigma)-treated H2O, spectrophotometrically quantified, and immediately frozen at −70°C. For Northern blot experiments, 10 μg of total RNA was separated on a 1% agarose gel in HEPES buffer (20 mM Na-HEPES, pH 7.0, 5 mM NaCOOH, 1 mM EDTA) containing 17% (vol/vol) formaldehyde. RNA was transferred onto Hybond-N filters (Amersham), cross-linked (Spectronics Corporation), and hybridized with different probes. To verify that the same amounts of RNA had been loaded, an identical gel was run in parallel and stained with ethidium bromide. In addition, all filters were also hybridized with a 16S rRNA-specific probe. The oligonucleotides used to obtain mtsA (487 bp)-, fhuD (470 bp)-, sodA (620 bp)-, and emm1 (195 bp)-specific probes are listed in Table 2. Chromosomal DNA of strain AP1 was used as a template. PCR products were purified on MicroSpin S-200 HR columns (Pharmacia) and labeled with 32P by using the Megaprime labeling kit (Amersham). Hybridizations were performed at 46 to 50°C ON in a rotating hybridization oven. Filters were washed extensively with 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.1% SDS. Membranes were then exposed on a BAS-III imaging plate, scanned with a Bio-Imaging Analyzer BAS-2000 (Fuji Photo Films, Tokyo, Japan), and autoradiographed with X-Omat AR films (Kodak). Northern blot experiments were repeated on two to three independent RNA preparations. Densitometric analysis of each transcript-specific signal was performed following normalization with the corresponding signals of 16S rRNA.
Animal experiments.
S. pyogenes strains AP1 and SR301 were grown in THY until the OD620 reached ≅0.4, washed once in sterile phosphate-buffered saline (pH 7.4), and diluted in phosphate-buffered saline to 107 CFU/ml. Viable counts were performed to verify the accuracy of the dilutions. Seven- to 9-week-old female BALB/c mice were used. The air sac skin infection model was used, and each mouse received subcutaneous injections of 900 μl of air together with 100 μl of the bacterial suspension. Two sets of experiments were performed. In the first set, four groups of mice (n = 3) were inoculated with 106 or 105 CFU of either the wild type or the mutant strain. The experiment was repeated with the same bacterial doses and groups of 10 animals each. Mice were checked at regular intervals three times per day for a total of 10 days. Moribund (ruffled fur, inactive, unresponsive to stimuli) animals were euthanized and recorded as dead 8 h later. Data from both experiments were pooled, and the results were expressed as percentages of mice surviving over time. The lethal dose killing 50% of the animals (LD50) was also calculated.
Bioinformatics and statistics.
Sequence analysis and comparisons were performed by using MacVector, version 6.5.3 (Oxford Molecular, Cambridge, United Kingdom). The complete genome sequence of S. pyogenes strain SF370 (19) was used to search for fur and perR boxes in S. pyogenes.
The effects of aerobic growth on H2O2 stress response were expressed as the induction index, meaning the relative increase of survival as a consequence of aerobic growth. This was calculated by forming the ratio for each time point between the percentage of survival of bacteria cultured in aerobiosis and that of bacteria grown in the presence of CO2, with a 95% confidence interval. The results from bacterial growth, 55Fe incorporation, and oxidative stress experiments were expressed as means ± SD. Doubling times in the exponential growth phase were calculated from data obtained from 6 to 10 independent experiments, and statistical significance was analyzed by using the unpaired Student t test. For animal data, statistical significance (P < 0.05) was calculated with both the Fisher exact test, by using InStat, version 3.0 (GraphPad Software, Inc., San Diego Calif.), and the Mann-Whitney U test.
RESULTS
Construction of strains.
Fur and Fur-like proteins are important regulators of various processes in several bacterial species (17). S. pyogenes contains a fur-like gene, which is denominated perR in a serotype M6 strain (38). The role of PerR in S. pyogenes was investigated by constructing a perR isogenic mutant in strain AP1 of the M1 serotype. A DNA fragment encompassing nucleotide residues 202 to 361 of perR, encoding part of the DNA- and metal-binding domains of PerR, was deleted and replaced with a kanamycin resistance cassette (46). Allelic exchange was used to replace the wild type with the perR mutant allele through a double-crossover recombination event. Appropriate gene disruption was confirmed by Southern blotting and PCR analysis (data not shown), and the mutant strain was denominated SR301 (perR::kan) (Table 1). For complementation of the perR mutation in SR301, the entire perR gene, including a putative promoter, was cloned in a plasmid able to replicate in S. pyogenes (36) and transformed into the SR301 strain. A complemented strain was designated SR304 [perR::kan/pLU21 (perR+)] (Table 1).
PerR affects iron metabolism in S. pyogenes.
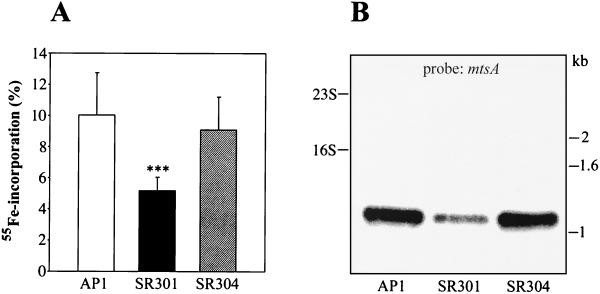
Fur, in the presence of Fe2+, generally acts as a transcriptional repressor of iron uptake systems (1, 14, 17). As a consequence, fur mutants often express iron import systems in a constitutive fashion (2, 28). Iron incorporation by AP1 and SR301 bacteria was investigated by measuring the amount of 55Fe accumulated in cells grown to exponential phase in Cx-THY. Interestingly, the amount of 55Fe detected in the mutant was reduced by 48% compared to both the wild type and the complemented SR304 strain (Fig. 1A). Little is known about metal ion uptake systems in S. pyogenes, and only two transporters have been annotated in the complete S. pyogenes genome: the MtsABC transporter with broad specificity for metal cations (37) and the putative ferrichrome uptake system FhuGBDA (19, 50). Therefore, in order to investigate whether expression of mtsABC and/or fhuGBDA was affected in PerR-deficient bacteria, Northern blot analysis on total RNA from strains AP1, SR301, and SR304 was performed. The probes used were mtsA, coding for the lipoprotein of MtsABC, and fhuD, encoding a putative ferrichrome transporter. We failed to detect any transcript in all samples when using the fhuD probe, whereas strong hybridization occurred with the mtsA probe. Densitometric analysis showed a 2.4-fold reduction of mtsA transcription in the SR301 mutant compared to that of the wild type (Fig. 1B). In accordance with the 55Fe incorporation results (Fig. 1A), transcription of mtsA was totally restored in the complemented strain (Fig. 1B). These data suggest that the reduced accumulation of iron by the perR mutant is at least partially due to a decreased transcription of mtsA.
FIG. 1.
Total iron incorporation by S. pyogenes strains AP1, SR301, and SR304. (A) AP1 (wild type), SR301 (perR), and SR304 (perR+) cells were cultured in Cx-THY in the presence of 55Fe until the mid-exponential growth phase. Cells were spun down, and supernatants were saved. After a total of three washes, the amount of radioactivity in the bacterial pellet was measured. The fraction of 55Fe associated with the pellet was calculated by dividing the number of counts per minute of the pellet by the number of counts per minute of the pellet plus the supernatant. Values are presented as means ± SD from at least three independent experiments with duplicate or triplicate samples. Statistical significance (P < 0.001) is indicated by three asterisks. (B) Transcriptional analysis of the mtsABC metal transporter operon. Total RNA was prepared from strains AP1, SR301, and SR304. Bacteria were grown until the mid-exponential phase, subjected to Northern blotting, and hybridized with an mtsA probe. The positions of 23S and 16S rRNA are shown to the left. Size markers (in kilobases) are indicated to the right.
Growth features of PerR-deficient S. pyogenes.
In order to analyze whether the decreased iron accumulation in the perR mutant affected bacterial growth in vitro, strains AP1 and SR301 were cultivated in rich medium (THY), metal-depleted THY (Cx-THY), and iron-repleted (100 μM ferric citrate) Cx-THY. No significant difference was observed when comparing generation times in the exponential phase of wild-type and mutant strains in THY (50 ± 11 and 53 ± 9 min, respectively) (data not shown). A small growth defect instead appeared in the perR mutant when grown in Cx-THY (Fig. 2). The SR301 strain showed an increased generation time (70 ± 14 min) compared to the AP1 strain (54 ± 17 min); however, this difference was not statistically significant with the unpaired Student t test (P = 0.073). The addition of ferric citrate to Cx-THY somewhat improved the mutant growth rate (63 ± 17 min). When AP1 and SR301 bacteria were cultivated aerobically in THY, the perR mutant grew at a slower rate (77 ± 25 min) than the wild type (60 ± 20 min), but again, the difference was not statistically significant (P = 0.12) (data not shown). Both the mutant and wild type grew very poorly in Cx-THY in aerobiosis. In conclusion, the growth of SR301 seems somewhat hampered in Cx-THY compared to the wild type, although the difference is not statistically significant. Overall, these results indicate that the lack of PerR does not dramatically affect the in vitro growth of S. pyogenes.
FIG. 2.
Growth of wild-type and perR mutant strains of S. pyogenes. AP1 (wild type) and SR301 (perR) strains were cultured in Cx-THY medium or Cx-THY medium supplemented with 100 μM ferric citrate. Experiments were performed 6 to 10 times, and results from a representative one are shown.
Further characterization of the response to peroxide stress in wild-type and perR mutant S. pyogenes.
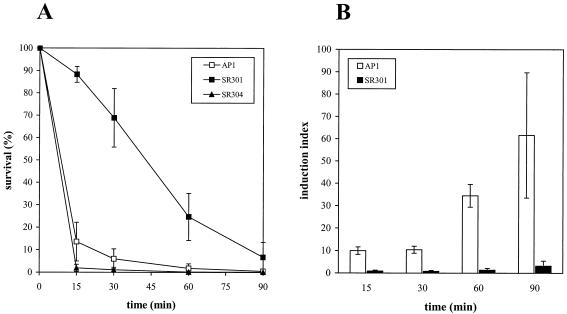
PerR has been described as a peroxide-responsive repressor in certain gram-positive bacteria (4, 34), including S. pyogenes (38). In order to further characterize S. pyogenes susceptibility to peroxide stress, strains AP1, SR301, and SR304 were grown until mid-log phase and then subjected to peroxide stress (5 mM H2O2). Fifteen minutes after challenge, 88.3% of the SR301 bacteria were still alive compared to 13.6% of the wild-type cells, whereas after 60 min, the percentages of surviving mutant and wild-type bacteria were 29.8 and 1.8, respectively. Sensitivity to H2O2 was completely restored in the transcomplemented SR304 strain (Fig. 3A). Since iron potentiates oxidative stress (53), the hyperresistance to H2O2 observed in the perR mutant could possibly be a result of reduced intracellular iron levels. Therefore, we performed the same peroxide stress assay on the S. pyogenes mtsA mutant RJ1 strain, which is hampered in iron uptake (37). However, RJ1 bacteria showed a hypersensitivity to H2O2 (R. Janulczyk, S. Ricci, and L. Björck, submitted for publication). In addition, to further verify that intracellular iron levels did not significantly influence the susceptibility to H2O2, the wild-type strain was grown in iron-limited conditions (Cx-THY) and challenged with H2O2. AP1 bacteria showed the same sensitivity to peroxide as when cultivated in THY (data not shown). Therefore, these results further support a crucial role for PerR in the regulation of the response to peroxide stress in S. pyogenes.
FIG. 3.
S. pyogenes sensitivity to peroxide stress. (A) Survival of strains AP1 (wild type), SR301 (perR), and SR304 (perR+) after the addition of 5 mM H2O2. Samples were collected 15, 30, 60, and 90 min postchallenge with H2O2, and viable counts were performed. The results are expressed as percentages of survival over time. Values indicate the means ± SD of four independent experiments. (B) O2-inducible resistance response to H2O2. Strains AP1 and SR301 were grown either in aerobiosis or with CO2 before adding H2O2. Samples were collected 15, 30, 60, and 90 min after challenge, and viable counts were performed from each culture. The results are expressed as the relative increases of survival induced by growth in the presence of O2 (induction index) with a 95% confidence interval at different time points. The experiments were performed five times.
Since the response to H2O2 can be induced by sublethal doses of H2O2 and other compounds (i.e., ethanol) (38), we decided to analyze whether aerobic growth could induce this adaptive response to hydrogen peroxide. Oxidative stress experiments were performed on strains AP1 and SR301 grown under aerobic conditions, and in parallel, control cultures were also cultivated in the presence of CO2. The numbers of CFU per milliliter of bacteria grown aerobically or in CO2 were determined 15, 30, 60, and 90 min after H2O2 challenge (Table 3). Percentages of survival were calculated, and the induction index was determined for each time point (Fig. 3B). In response to O2, wild-type bacteria were already able to mount an adaptive resistance to peroxide stress 15 min after the addition of H2O2. Moreover, after 60 and 90 min, this resistance was more than 30- and 60-fold larger in cells grown in aerobiosis than in uninduced cultures, respectively. Practically no induction was seen in the SR301 mutant, although a small (approximately threefold) additional level of resistance could be achieved 90 min after peroxide challenge (Fig. 3B). These results show that O2 is able to induce a resistance to H2O2 in wild-type bacteria. This response seems constitutive in the perR mutant, thus strengthening a role for PerR as an oxidative stress-responsive repressor in S. pyogenes.
TABLE 3.
Survival of strains AP1 and SR301 at different time points following peroxide challenge
| Strain (growth condition)b | CFU/ml at time (min)a:
|
||||
|---|---|---|---|---|---|
| 0 | 15 | 30 | 60 | 90 | |
| AP1 (CO2) | 2.5 × 108 ± 7.7 × 107 | 3.8 × 107 ± 3.6 × 107 | 1.5 × 107 ± 7.9 × 106 | 5.1 × 106 ± 4.2 × 106 | 1.2 × 106 ± 1 × 106 |
| SR301 (CO2) | 2.9 × 108 ± 1 × 108 | 2.7 × 108 ± 9.7 × 107 | 2.1 × 108 ± 9.5 × 107 | 7.3 × 107 ± 3.3 × 107 | 1.8 × 107 ± 1.5 × 107 |
| AP1 (O2) | 1.1 × 108 ± 8.6 × 107 | 1.6 × 108 ± 1.5 × 108 | 7.4 × 107 ± 6.4 × 107 | 7.3 × 107 ± 5.6 × 107 | 3.2 × 107 ± 2.2 × 107 |
| SR301 (O2) | 1.1 × 108 ± 6.9 × 107 | 9 × 107 ± 7.9 × 107 | 5.5 × 107 ± 3.9 × 107 | 4.8 × 107 ± 5 × 107 | 2.5 × 107 ± 3.1 × 107 |
CFU per milliliter of AP1 (wild type) and SR301 (perR) bacteria surviving H2O2 challenge. Bacterial dilutions were plated onto THY plates 0, 15, 30, 60, and 90 min after the addition of 5 mM H2O2. Results are expressed as means ± SD of four to five independent assays.
AP1 and SR301 strains were grown either under aerobic conditions (O2) or in the presence of 5% CO2 prior to challenge with H2O2.
Response to superoxide stress in wild-type and PerR-deficient bacteria.
Redox-cycling agents, such as paraquat, can cause oxidative stress by generating the superoxide radical, O2− ·, which is converted into H2O2 and O2 by SOD (21). In order to analyze S. pyogenes sensitivity to paraquat, strains AP1 and SR301 were cultured ON in the presence of 0, 2, and 10 mM paraquat. The perR mutant was more sensitive to the O2−·-generating agent than AP1 bacteria. At 2 mM, a small growth defect was observed in the mutant, whereas when using 10 mM, a concentration that moderately affected growth of the wild type, SR301 bacteria were highly hampered in growth (P < 0.001) (Fig. 4A). As SOD represents the major defense against the superoxide anion, we performed transcriptional analysis of sodA, which encodes the single SOD present in S. pyogenes (24). Total RNA was extracted from the wild type, perR mutant, and transcomplemented strains, subjected to Northern blotting, and probed with sodA. A reduced transcription of sodA (3.3-fold) was observed in the perR mutant, whereas transcomplementation of the perR mutation restored sodA transcription to the levels found with wild-type bacteria (Fig. 4B). The present data indicate that SR301 bacteria are more sensitive to O2−· than the wild type, and this is probably due to a decreased SOD expression in the mutant, suggesting a possible role for PerR in the regulation of SOD production in S. pyogenes.
FIG. 4.
Sensitivity to superoxide stress in S. pyogenes. (A) Growth of AP1 (wild type) and SR301 (perR) strains in the presence of 0, 2, and 10 mM paraquat. The OD620 was recorded after 15 h of incubation. The results are expressed as means ± SD of five independent experiments with duplicate or triplicate samples. Statistical significance (P < 0.001) is indicated by three asterisks. (B) Transcriptional analysis of sodA. Total RNA was prepared from strains AP1 (wild type), SR301 (perR), and SR304 (perR+) grown until the mid-exponential phase. RNA was subjected to Northern blotting and hybridized with a sodA probe. Positions of 23S, 16S, and 5S rRNA are shown on the left, and size markers (in kilobases) are presented on the right.
PerR is required for full virulence in S. pyogenes.
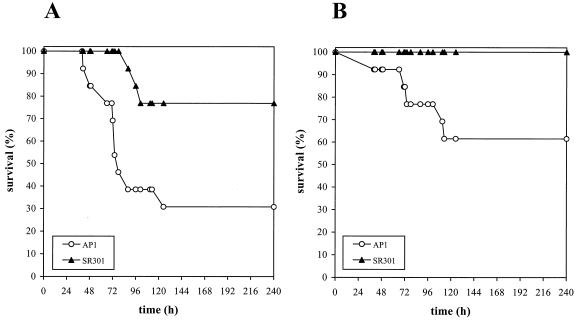
Both fur and perR mutants in S. aureus are attenuated in virulence (34, 35). Therefore, we tested the virulence of strains AP1 and SR301 in BALB/c mice. Skin air sacs were inoculated with either 106 or 105 CFU of bacteria, and mouse survival was monitored for 10 days. When using 106 CFU, 30.8% of mice infected with the wild type were alive at the end of the experiment, compared to 76.9% of the group inoculated with the perR mutant (P < 0.05). Moreover, mice injected with the perR mutant showed delayed symptoms of infection, and the first casualty was recorded after 88 h, in comparison to wild-type streptococci, which were lethal in less than 2 days (Fig. 5A). With the lower dose, all animals inoculated with the perR mutant survived, compared to 61.5% survival observed in the group infected with the AP1 strain (Fig. 5B). Analysis of these differences showed a statistical significance (P < 0.05) and suggested that the perR mutant had an approximately 10-fold reduction of virulence. The LD50 of mice infected with AP1 and SR301 bacteria were 2.4 × 105 and 3 × 106 CFU, respectively (data not shown).
FIG. 5.
Virulence of S. pyogenes wild-type and perR mutant strains. Skin air sacs of BALB/c mice (n = 13 per group) were injected with either 106 (A) or 105 (B) CFU of strain AP1 (wild type) or SR301 (perR). Animals were monitored for a total of 10 days. The results are expressed as percentages of survival over time. Statistical significance was determined with both the Mann-Whitney U test and the Fisher exact test (P < 0.05).
The expression of some known virulence determinants of S. pyogenes was also investigated by Northern and/or Western blot analysis. However, no significant differences in emm1 transcription or in the production of M1 protein, cysteine proteinase, and C5a peptidase were observed between the wild type and the perR mutant (data not shown).
Computer-based search for putative PerR boxes in S. pyogenes.
Using the complete genome of strain SF370 (19), we searched for putative iron boxes similar to the consensus sequences described for E. coli (fur), B. subtilis (fur and perR), and S. aureus (fur and perR) (8, 9, 14, 34, 35). No perfect matches were identified. With the threshold set at a minimum of 14 out of 19 nucleotides identical to the consensus sequence, imperfect matches were found; however, they lacked the minimum three repeats necessary for functional iron boxes (17) or were within predicted coding regions.
DISCUSSION
Fur is a well-known iron-responsive protein of many gram-negative bacteria (for a review, see reference 17), and recently Fur homologues have also been identified in gram-positive species (4, 22, 34, 35, 38, 39, 58). In the present study, an isogenic mutant of the gene encoding the peroxide-responsive Fur homologue PerR was constructed and characterized in an S. pyogenes strain of the M1 serotype. Fur is generally considered a transcriptional repressor (1, 14), and fur mutants typically exhibit a constitutive derepression of iron uptake systems, leading to intracellular iron overload and potential Fe-dependent lethality (54). In our case, the perR mutant strain did not behave as a typical fur mutant. Instead, it showed 48% reduction of 55Fe incorporation, which could be partially explained by the down-regulation of the mtsABC system, which is involved in uptake and transport of a variety of cations (37; Janulczyk et al., submitted). A fur mutant of Pseudomonas aeruginosa also showed a reduced siderophore-mediated iron uptake (32). Concerning fhuGBDA expression, the reason why we were not able to detect any fhuD transcript, even in the wild-type sample, is unclear. The fhuGBDA operon may be poorly expressed or transcribed at a specific growth phase. The metal transport deficiency observed in the perR mutant slightly affected growth in metal-depleted conditions, but it was not possible to calculate a convincing statistically significant difference between the mutant and the wild type.
In accordance with previous studies, which have demonstrated hyperresistance to H2O2 stress in perR mutants of B. subtilis (4), S. aureus (34), and in an M6 S. pyogenes strain (38), the present results show that PerR is involved in the response to H2O2 also in the M1 serotype of S. pyogenes. Iron, as a partner of the Fenton reaction, can potentiate oxygen toxicity by converting H2O2 into OH·. The possibility that H2O2 resistance was a consequence of low intracellular iron levels was excluded by the observations that an S. pyogenes mtsA mutant was hampered in iron uptake and hypersensitive to H2O2 (Janulczyk et al., submitted) and that the wild-type strain was equally sensitive to H2O2 both in iron-rich and iron-depleted culture medium. Therefore, resistance to H2O2 relies on PerR acting as a repressor of a gene or regulon involved in the defense against peroxide stress. Resistance to H2O2 can be induced by different stimuli, and while King et al. used H2O2 or ethanol as inducers (38), we obtained a 60-fold-higher survival by growing wild-type bacteria in the presence of oxygen prior to challenge.
The present work also suggests a role for PerR in the response to superoxide stress. The perR mutant was more sensitive to O2−· than the WT strain, as it grew very poorly in the presence of 10 mM paraquat. Consistent with the fact that SOD is the main defense against superoxide stress in many bacteria, including S. pyogenes (24), we observed an approximately threefold transcriptional reduction of sodA in the perR mutant compared to that in the wild-type strain. In E. coli, SOD is not strictly necessary for aerobic survival and half of the normal enzymatic activity is generally sufficient for aerobic growth; however, SOD becomes crucial under oxidative stress conditions (6). Also in the present case, the growth of the perR mutant was not significantly disturbed under aerobic conditions, but the mutant was sensitive to high levels of paraquat. Thus, SR301 bacteria are probably producing enough SOD to handle moderate levels of oxidative stress. In addition, the mutant was derepressed in the peroxide stress response and had low levels of intracellular iron, which might partially compensate for a lower production of SOD. A reduced total SOD activity together with a defective iron uptake was also described in a P. aeruginosa fur mutant (32). E. coli sod mutants are generally sensitive to both O2−· and H2O2 (6), whereas the perR mutant in S. pyogenes was resistant to H2O2 but sensitive to O2−·. This heterogeneity in ROS susceptibility is interesting but not unprecedented. King et al. showed that inactivation of two distinct peroxidase genes in S. pyogenes generated mutants which were more sensitive to paraquat and at the same time slightly more resistant than the wild type to H2O2 in a survival assay (38).
Fur proteins are generally operating as repressors; however, Fur-dependent positive regulation has also been reported (15, 20, 29, 44). In E. coli, sodA (Mn-SOD) is under the control of six different proteins (11), including a Fur-mediated negative regulation (52), while sodB (Fe-SOD) is subjected to positive control by Fur (15). When Fur acts as an activator, no iron boxes are identified in the promoters of target genes (53). Also in S. pyogenes, PerR may behave as a bifunctional regulator, acting as a repressor of the peroxide regulon while being a transcriptional activator of SOD. The fact that we could not find any Fur or PerR boxes upstream of sodA and mtsABC suggests that PerR might function as an enhancer of sodA and mtsABC transcription.
Analogous to Fur and PerR in S. aureus (34, 35), PerR is important for virulence in S. pyogenes. In a murine skin model of infection, we observed a statistically significant difference in survival time and LD50 after injection of wild-type or mutant bacteria. In particular, no mortality was recorded in the group infected with the perR mutant when using the lower bacterial dose. The hypothesis that known virulence factors were affected in the mutant was investigated. However, no significant variation in the production of M protein, cysteine proteinase, and C5a peptidase was revealed. Previous reports have shown that Streptococcus pneumoniae and Streptococcus agalactiae sodA mutants are attenuated in virulence (47, 59). Whether the attenuation described here is due to inactivation of perR, as reported in S. aureus (34, 35), or is a consequence of sodA down-regulation, is unclear. Moreover, since growth of the perR mutant showed a tendency to be affected in a metal-depleted medium, we cannot completely rule out that restricted iron availability in the host might hamper the mutant, resulting in virulence attenuation.
Iron metabolism and oxidative stress defenses are strictly interconnected in bacteria (53). In B. subtilis, S. aureus, and the gram-negative bacterium Campylobacter jejuni, there is a close relationship between PerR-mediated control of peroxide stress and intracellular metal ion homeostasis (9, 33-35, 56). In particular, B. subtilis PerR requires a metal cofactor (Fe, Mn) to bind DNA, and the presence of this metal ion is responsible for regulating the PerR response towards H2O2 (33). In the present case, when the levels of intracellular H2O2 are high, S. pyogenes PerR might (i) derepress the H2O2-responsive regulon, (ii) partially repress sodA in order to limit the production of H2O2, and (iii) lower the intracellular concentration of free Fe2+ that can participate in the Fenton reaction by down-regulation of mtsABC and possibly of other metal acquisition systems. In relation to virulence, the attenuation of the perR mutant may be a result of direct or indirect regulatory effects on virulence determinants, and/or of a disturbed balance between levels of intracellular iron and oxidative stress defenses. The effects observed in the mutant depend upon PerR, since all the phenotypes could be complemented in trans, and although the molecular mechanisms are not known, PerR appears to play a crucial role in the interplay between oxidative stress responses, metal homeostasis, and virulence of S. pyogenes.
Acknowledgments
The excellent technical assistance of Ingbritt Gustafsson is greatly appreciated. We thank A. Podbielski for plasmid pFW13 and P. Cleary for the C5a peptidase antiserum. We are grateful to A. Olsén for stimulating discussions and to R. Manganelli for a critical reading of the manuscript.
This work was supported by grants from the Swedish Research Council (projects 7480 and 14379), the Foundations of Crafoord, Kock, Lundberg, Österlund, and Wenner-Gren, and the Commission of the European Union (contract BIO4-CT98-5021 to S.R.).
Editor: E. I. Tuomanen
REFERENCES
- 1.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 2.Bagg, A., and J. B. Neilands. 1987. Molecular mechanisms of regulation of siderophore-mediated iron assimilation. Microbiol. Rev. 51:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berge, A., and L. Björck. 1995. Streptococcal cysteine proteinase releases biologically active fragments of streptococcal surface proteins. J. Biol. Chem. 270:9862-9867. [DOI] [PubMed] [Google Scholar]
- 4.Bsat, N., A. Herbig, L. Casillas-Martinez, P. Setlow, and J. D. Helmann. 1998. Bacillus subtilis contains multiple Fur homologues: identification of the iron uptake (Fur) and peroxide regulon (PerR) repressors. Mol. Microbiol. 29:189-198. [DOI] [PubMed] [Google Scholar]
- 5.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlioz, A., and D. Touati. 1986. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic growth? EMBO J. 5:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J. D., and J. D. Helmann. 1995. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative-stress gene. Mol. Microbiol. 18:295-300. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., L. P. James, and J. D. Helmann. 1993. Metalloregulation in Bacillus subtilis: isolation and characterization of two genes differentially repressed by metal ions. J. Bacteriol. 175:5428-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., L. Keramati, and J. D. Helmann. 1995. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc. Natl. Acad. Sci. USA 92:8190-8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collin, M., and A. Olsén. 2000. Generation of a mature streptococcal cysteine proteinase is dependent on cell wall-anchored M1 protein. Mol. Microbiol. 36:1306-1318. [DOI] [PubMed] [Google Scholar]
- 11.Compan, I., and D. Touati. 1993. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K12. J. Bacteriol. 175:1687-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cunningham, M. W. 2000. Pathogenesis of group A streptococcal infections. Clin. Microbiol. Rev. 13:470-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delany, I., G. Spohn, R. Rappuoli, and V. Scarlato. 2001. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol. Microbiol. 42:1297-1309. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aereobactin operon plasmid ColIV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubrac, S., and D. Touati. 2000. Fur positive regulation of iron superoxide dismutase in Escherichia coli: functional analysis of the sodB promoter. J. Bacteriol. 182:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dussurget, O., and I. Smith. 1998. Interdependence of mycobacterial iron regulation, oxidative-stress response and isoniazid resistance. Trends Microbiol. 6:354-358. [DOI] [PubMed] [Google Scholar]
- 17.Escolar, L., J. Perez-Martin, and V. de Lorenzo. 1999. Opening the iron box: transcriptional metalloregulation by the Fur protein. J. Bacteriol. 181:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farr, S. B., and T. Kogoma. 1991. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55:561-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fridovich, I. 1997. Superoxide anion radical (O2−·), superoxide dismutases and related matters. J. Biol. Chem. 272:18515-18517. [DOI] [PubMed] [Google Scholar]
- 22.Gaballa, A., and J. D. Helmann. 1998. Identification of a zinc-specific metalloregulatory protein, Zur, controlling zinc transport in Bacillus subtilis. J. Bacteriol. 180:5815-5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlach, D., H. Knöll, W. Köhler, J.-H. Ozegowski, and V. Hribalova. 1983. Isolation and characterization of erythrogenic toxins. V. Communication: identity of erythrogenic toxin type B and streptococcal proteinase precursor. Zentbl. Bakteriol. Mikrobiol. Hyg. Abt. 1 Orig. A. 255:221-233. [PubMed] [Google Scholar]
- 24.Gibson, C. M., and M. G. Caparon. 1996. Insertional inactivation of Streptococcus pyogenes sod suggests that prtF is regulated in response to a superoxide signal. J. Bacteriol. 178:4688-4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson, C. M., T. C. Mallett, A. Claiborne, and M. G. Caparon. 2000. Contribution of NADH oxidase to aerobic metabolism of Streptococcus pyogenes. J. Bacteriol. 182:448-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffiths, B. B., and O. McClain. 1988. The role of iron in the growth and hemolysin (streptolysin S) production in Streptococcus pyogenes. J. Basic Microbiol. 7:427-436. [DOI] [PubMed] [Google Scholar]
- 27.Griffiths, E. 1999. Iron in biological systems, p. 1-26. In J. J. Bullen and E. Griffiths (ed.), Iron and infection. Wiley, Chichester, United Kingdom.
- 28.Griffiths, E., and H. Chart. 1999. Iron as a regulatory signal, p. 213-253. In J. J. Bullen and E. Griffiths (ed.), Iron and infection. Wiley, Chichester, United Kingdom.
- 29.Gruer, M. J., and J. R. Guest. 1994. Two genetically distinct and differentially regulated aconitases (AcnA and AcnB) in Escherichia coli. Microbiology 140:2531-2541. [DOI] [PubMed] [Google Scholar]
- 30.Hanski, E., G. Fogg, A. Tovi, N. Okada, I. Burstein, and M. Caparon. 1995. Molecular analysis of Streptococcus pyogenes adhesion. Methods Enzymol. 253:269-305. [DOI] [PubMed] [Google Scholar]
- 31.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 32.Hassett, D. J., P. A. Sokol, M. L. Howell, J.-F. Ma, H. T. Schweizer, U. Ochsner, and M. L. Vasil. 1996. Ferric uptake regulator (Fur) mutants of Pseudomonas aeruginosa demonstrate defective siderophore-mediated iron uptake, altered aerobic growth, and decreased superoxide dismutase and catalase activities. J. Bacteriol. 178:3996-4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Herbig, A. F., and J. D. Helmann. 2001. Roles of metal ions and hydrogen peroxide in modulating the interaction of Bacillus subtilis PerR peroxide regulon repressor with operator DNA. Mol. Microbiol. 41:849-859. [DOI] [PubMed] [Google Scholar]
- 34.Horsburgh, M. J., M. O. Clements, H. Crossley, E. Ingham, and S. J. Foster. 2001. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect. Immun. 69:3744-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Husmann, L. K., J. R. Scott, G. Lindahl, and L. Stenberg. 1995. Expression of the Arp protein, a member of the M protein family, is not sufficient to inhibit phagocytosis of Streptococcus pyogenes. Infect. Immun. 63:345-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Janulczyk, R., J. Pallon, and L. Björck. 1999. Identification and characterization of a Streptococcus pyogenes ABC transporter with multiple specificity for metal cations. Mol. Microbiol. 34:596-606. [DOI] [PubMed] [Google Scholar]
- 38.King, K. Y., J. A. Horenstein, and M. J. Caparon. 2000. Aerotolerance and peroxide resistance in peroxide and PerR mutants of Streptococcus pyogenes. J. Bacteriol. 182:5290-5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindsay, J. A., and S. J. Foster. 2001. Zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology 147:1259-1266. [DOI] [PubMed] [Google Scholar]
- 40.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation on emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miller, R. A., and B. E. Britigan. 1997. Role of oxidants in microbial pathophysiology. Clin. Microbiol. Rev. 10:1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nesbitt, S. A., and M. A. Horton. 1992. A non radioactive biochemical characterization of membrane proteins using enhanced chemiluminescence. Anal. Biochem. 206:267-272. [DOI] [PubMed] [Google Scholar]
- 44.Niederhoffer, E. C., C. M. Naranjo, K. L. Bradley, and J. A. Fee. 1990. Control of Escherichia coli superoxide dismutase (sodA and sodB) genes by the ferric uptake regulation (fur) locus. J. Bacteriol. 172:1930-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 46.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lütticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 47.Poyart, C., E. Pellegrini, O. Gaillot, C. Boumaila, M. Baptista, and P. Trieu-Cuot. 2001. Contribution of Mn-cofactored superoxide dismutase (SodA) to the virulence of Streptococcus agalactiae. Infect. Immun. 69:5098-5106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ratledge, C., and L. G. Dover. 2000. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 54:881-941. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 50.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. L. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storz, G., and J. A. Imlay. 1999. Oxidative stress. Curr. Opin. Microbiol. 2:188-194. [DOI] [PubMed] [Google Scholar]
- 52.Tardat, B., and D. Touati. 1993. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol. Microbiol. 9:53-63. [DOI] [PubMed] [Google Scholar]
- 53.Touati, D. 2000. Iron and oxidative stress in bacteria. Arch. Biochem. Biophys. 373:1-6. [DOI] [PubMed] [Google Scholar]
- 54.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Heyningen, T., G. Fogg, D. Yates, E. Hanski, and M. G. Caparon. 1993. Adherence and fibronectin binding are environmentally regulated in the group A streptococci. Mol. Microbiol. 9:1213-1222. [DOI] [PubMed] [Google Scholar]
- 56.van Vliet, A. H. M., M.-L. A. Baillon, C. W. Penn, and J. M. Ketley. 1999. Campylobacter jejuni contains two Fur homologs: characterization of iron-responsive regulation of peroxide stress defense genes by the PerR repressor. J. Bacteriol. 181:6371-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wessels, M. R. 1999. Regulation of virulence factor expression in group A Streptococcus. Trends Microbiol. 7:428-430. [DOI] [PubMed] [Google Scholar]
- 58.Xiong, A., V. K. Singh, G. Cabrera, and R. K. Jayaswal. 2000. Molecular characterization of the ferric-uptake regulator, Fur, from Staphylococcus aureus. Microbiology 146:659-668. [DOI] [PubMed] [Google Scholar]
- 59.Yesilkaya, H., A. Kadioglu, N. Gingles, J. E. Alexander, T. J. Mitchell, and P. W. Andrew. 2000. Role of manganese-containing superoxide dismutase in oxidative stress and virulence of Streptococcus pneumoniae. Infect. Immun. 68:2819-2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng, M., B. Doan, T. D. Schneider, and G. Storz. 1999. OxyR and SoxRS regulation of fur. J. Bacteriol. 181:4639-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]